Abstract
We recently assessed the metabolism of Synechocystis sp PCC6803 through a constraints-based reconstruction and analysis approach and identified its main metabolic properties. These include reduced metabolic robustness, in contrast to a high photosynthetic robustness driving the optimal autotrophic metabolism. Here, we address how these metabolic features affect biotechnological capabilities of this bacterium. The search for growth-coupled overproducer strains revealed that the carbon flux re-routing, but not the electron flux, is significantly more challenging under autotrophic conditions than under mixo- or heterotrophic conditions. We also found that the blocking of the light-driven metabolism was required for carbon flux re-routing under mixotrophic conditions. Overall, our analysis, which represents the first systematic evaluation of the biotechnological capabilities of a photosynthetic organism, paradoxically suggests that the light-driven metabolism itself and its unique metabolic features are the main bottlenecks in harnessing the biotechnological potential of Synechocystis.
Keywords: Synechocystis sp. PCC6803, genome-scale modeling, COBRA methods, biosustainability, metabolic engineering, photosynthetic robustness
Introduction
The development of renewable energy sources has received significant interest in recent years owing to the depletion of fossil fuels, ever-increasing demand for energy, and concerns over climate change. A promising source of renewable energy is the recycling of CO2 into usable fuels and fine chemicals by photosynthetic organisms using solar energy. There are, however, increasing concerns over the methods currently in use for producing biodiesel from crops and biomass. The problems include high production costs and a reduction in the amount of land available for growing edible crops. These issues highlight the need for a new generation of biofuel technology.1,2 Cyanobacteria possess several properties, which make them promising candidates for sustainable bioenergy generation. They are the only prokaryotes capable of carrying out oxygenic CO2-fixation photosynthesis with higher efficiency than vascular plants.3-5 The cultivation of cyanobacteria is simple, inexpensive and it does not compete directly with agricultural crops for land or water. In addition, they are a source of natural high-value products, such as carotenoids, lipids, and vitamins.6,7 They are also amenable to genetic manipulation.8,9 These features have motivated recent engineering efforts of cyanobacteria for producing valuable chemicals and biofuel-like compounds from the main biosynthetic building blocks, establishing a proof of concept of direct biofuel production from oxygenic photosynthesis.10-18 However, with a few exceptions,19,20 the productivity has been very low compared with heterotrophic organisms.21
We recently reconstructed a genome-scale metabolic model of the cyanobacteria Synechocystis sp PCC6803 (iJN678).22 The model was used to study in detail the photosynthetic process under different light and inorganic carbon conditions as well as under genetic perturbations. The systems analysis identified two main states of the photosynthetic apparatus: A CO2 limited state (CLS) and a light-limited state (LLS). In addition, it was shown that optimal photosynthetic performance requires high photosynthetic robustness, including multiple lipids, photosynthetic pigments and alternate electron flow pathways (AEF), and that this photosynthetic robustness comes at a cost of reduced metabolic robustness. In order to explore the impact of these unique metabolic features and to obtain a better understanding of the opportunities and bottlenecks offered by cyanobacteria in biotechnology, we present here the first analysis of the metabolic engineering capabilities of Synechocystis using iJN678. Employing an approach analogous to those previously used for in silico-driven metabolic engineering in heterotrophic organisms,23 we analyzed how the electron and carbon flux can be funneled to the overproduction of both native (fumarate, ethanol, sucrose, and H2) and non-native compounds (L-lactate and 1-butanol).
The above metabolites were chosen as representatives of key points in the metabolism and/or because they have already been overproduced in Synechocystis (Fig. 1). Growth-coupled production designs were attempted since they represent a stable phenotype, allowing for an easy selection of the overproducing strains.23 Flux balance analysis (FBA)24,25 was used to predict flux values in both the wild type and the mutants. The search for mutants was performed using a randomized version of the strategy described in Nogales et al.26 Using gene-protein-reaction associations, which specify via Boolean rules the gene product(s) catalyzing a reaction, a mutant was created by randomly knocking out a fixed number of genes and therefore, disabling flux through the affected reaction(s). An FBA was performed to determine the maximum growth rate of the mutant, followed by another FBA to determine the maximum product rate. Finally, a third FBA was performed to determine the minimum product rate, while enforcing the maximal growth rate; thus, revealing growth-coupled designs. By repeating this process many times, a list of different knockout designs was obtained. The number of knockouts was varied between 3 and 25, and the number of repeats was between 5 × 106 and 5 × 107. The knockout search was performed under both autotrophic and mixotrophic conditions, employing a light-limited state (LLS) where the photon uptake rate was fixed to 30 mmol.gDW−1.h−1 and a carbon-limited state (CLS) where the photon uptake rate was fixed to 100 mmol.gDW−1.h−1. The HCO3- uptake rate was set to 3.7 mmol.gDW−1.h−1 in all cases and for mixotrophic conditions, the glucose uptake rate was set to 1 mmol.gDW−1.h−1. Heterotrophic conditions were analyzed by setting the photon uptake rate to zero. The iJN678 model includes 678 genes, which results in a very large search space and to make the search more manageable, a pre-processing step for reducing the number of target genes was performed. By identifying essential genes, genes corresponding to blocked reactions and genes, which participated only in conjunctive gene-protein-reaction rules, the number of target genes was reduced to 217, 222 and 239 for autotrophic, heterotrophic, and mixotrophic conditions, respectively. The theoretical maximum production rate for each metabolite was estimated by maximizing the production of the target metabolite while fixing the growth rate to 5% of the maximal growth achieved under each growth condition.
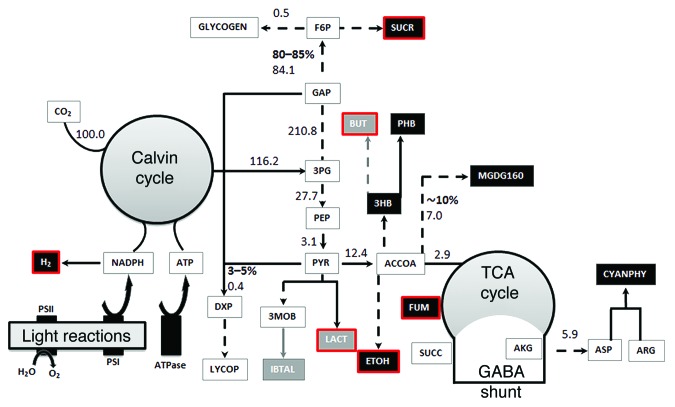
Figure 1. A depiction of the central metabolism of Synechocystis. Native and non-native experimentally overproduced metabolites in Synechocystis are represented by black and gray squares, respectively and the metabolites analyzed in this study are indicated by red lines. The carbon partitioning (in %) to sugar, lipids and terpenoid biosynthesis together with the predicted carbon flux distribution (normalized to the CO2 uptake rate) -under autotrophic conditions is also shown. The non-native metabolites are 1-butanol (BUT), lactate (LAC), isobutyraldehyde (IBTAL). The abbreviations for the native metabolites are given in Nogales et al.22
Results
Under autotrophic conditions, we found that the photosynthetic states determined the theoretical maximum production of the metabolites under consideration. In the LLS, the yield per carbon of molecule of metabolites with neutral or positive oxidation states, e.g., fumarate, sucrose, and lactate, was significantly higher (Fig. 2; Table 1). Under the CLS, the yields were predicted to be almost identical (≈0.95 moles of carbon of metabolite/mole of CO2), however, the theoretical maximal production rate of both 1-butanol and ethanol increased significantly suggesting that the production of the reduced compounds is limited by the light availability under the LLS. The maximum production of H2 was much higher in the CLS, which is consistent with the excess of light and the photolytic origin of the electrons used to reduce the protons.
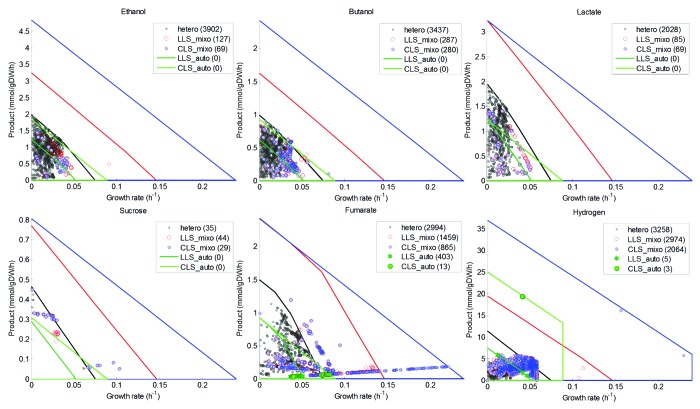
Figure 2. Production envelopes for wild-type and knockout Synechocystis strains. The production envelopes for each metabolite is shown as a function of the biomass production rate of the wild-type Synechocystis network under heterotrophic (black lines), autotrophic LLS (dark green lines), autotrophic CLS (light green lines), mixotrophic LLS (red lines) and mixotrophic CLS (blue lines), as well as the growth-coupled deletion mutants identified (dots). The number of growth-coupled knockouts found in each condition is shown in brackets.
Table 1. Properties of the growth-coupled overproducer designs under autotrophic conditions.
| |
Autotrophic conditions |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Light limiting state (0.0522) | Carbon limiting state (0.0884) | |||||||||
| Metabolite |
Maximun production rate |
Number of knockouts |
Growth rate |
Production rate (mmol.gDW−1h−1) |
BPCY |
Maximun production rate |
Number |
Growth rate |
Production rate |
BPCY |
| (mmol.gDW−1h−1) |
(h−1) |
(mmol.gDW−1h−1) |
Knockouts |
(h−1) |
(mmol.gDW−1h−1) |
|||||
| Fumarate (4 C) |
0.897 |
1 |
0.05 |
0.043 |
0.0022 |
0.878 |
1 |
0.082 |
0.069 |
0.0057 |
| Ethanol (2 C) |
1.192 |
1 |
0.0519 |
0 - 0.0101 |
- |
1.757 |
1 |
0.0875 |
0 - 0.0170 |
- |
| 1-Butanol (4 C) |
0.596 |
None found |
- |
- |
- |
0.878 |
None found |
- |
- |
- |
| Sucrose (12 C) |
0.276 |
None found |
- |
- |
- |
0.292 |
None found |
- |
- |
- |
| Lactate (3 C) |
1.185 |
None found |
- |
- |
- |
1.171 |
None found |
- |
- |
- |
| H2 | 7.154 | 12 | 0.041 | 1.744 | 0.0723 | 24.416 | 8 | 0.041 | 19.345 | 0.8001 |
LLS, Light limiting state; CLS, Carbon limiting state; BPCY, Biomass-product coupled yield. Numbers inside parenthesis represent the wild-type growth rate.
The search for knockout mutants for overproducing fumarate, ethanol, 1-butanol, sucrose, or L-lactate under autotrophic conditions was very challenging. For fumarate, a maximal production rate of 0.043 mmol.gDW−1.h−1 in the LLS and of 0.069 mmol.gDW−1.h−1 in the CLS were achieved with a deletion of the gene slr0018, which encodes for fumarase (FUM) (Table 1). These predicted production rates are significantly lower than those reported in computational studies with heterotrophic bacteria,23 but similar to the in vivo yields found for other overproduced metabolites in cyanobacteria.21 While cyanobacteria have successfully been engineered to produce ethanol, 1-butanol, sucrose, and L-lactate under autotrophic conditions, by expressing heterologous enzymes,11,13,14 our search did not reveal any growth-coupled mutants overproducing these metabolites. However, mutant strains able to produce small amounts of metabolites at their maximum growth rates were identified in some cases. For instance, the deletion of either slr2132, which encodes for phosphate acetyltransferase (PTAr), or sll1299 encoding for acetate kinase (ACKr), resulted in a theoretical maximum production of ethanol of 0.010 mmol.gDW−1.h−1 under the LLS and 0.017 mmol.gDW−1.h−1 under the CLS (Table 1). Since, pyruvate decarboxylase and alcohol dehydrogenase II genes from the obligatory ethanol producing Zymomonas mobilis have previously been introduced in cyanobacteria for overproducing ethanol,14,27 our results suggest that the production rate in these recombinant strains could be improved by blocking PTAr and/or ACKr. Taken together, the results obtained under autotrophic conditions and the low yields found experimentally strongly suggest that a re-routing of the carbon flux is more difficult to achieve in photosynthetic organisms than in heterotrophs, such as E. coli.23
To give additional support to this hypothesis, and to investigate whether the autotrophic metabolism itself and/or the overall metabolic network of Synechocystis are responsible for this phenomenon, we searched for overproducing mutants under heterotrophic conditions with glucose as the sole carbon and energy source. Several growth-coupled knockouts were identified for all the metabolites analyzed. In addition, very high yields were predicted, ranging from 75% of the maximum production rate for fumarate and sucrose to more than 95% for ethanol, 1-butanol, and lactate (Table 2; Fig. 2). In fact, the maximum yields for ethanol, lactate, and fumarate were 1.9, 1.85 and 1.14 mmol/mmol of glucose, respectively, in the same range as those predicted in silico for E. coli and by using a similar number of knockouts.23 These findings indicate that it is the light-driven metabolism, rather than the metabolic network itself, that is responsible for the lack of success in obtaining growth coupled mutants under autotrophic conditions.
Table 2. Properties of the growth-coupled overproducer designs under heterotrophic conditions.
| Heterotrophic conditions (0.0743) | |||||
|---|---|---|---|---|---|
| Metabolite |
Maximun production rate |
Number of knockouts |
Growth rate |
Production rate (mmol.gDW−1h−1) |
BPCY |
| (mmol.gDW−1h−1) |
(h−1) |
||||
| Fumarate (4C) |
1.462 |
4 |
0.045 |
0.719 |
0.0324 |
| Ethanol (2 C) |
1.901 |
2 |
0.035 |
1.218 |
0.0426 |
| 1-Butanol (4 C) |
0.951 |
4 |
0.036 |
0.559 |
0.0199 |
| Sucrose (12 C) |
0.443 |
5 |
0.062 |
0.053 |
0.0033 |
| Lactate (3 C) |
1.867 |
3 |
0.023 |
1.444 |
0.0366 |
| H2 | 10.926 | 7 | 0.019 | 3.195 | 0.062 |
LLS, Light limiting state; CLS, Carbon limiting state; BPCY, Biomass-product coupled yield. Numbers inside parenthesis represent the wild-type growth rate.
Synechocystis is able to grow mixotrophically with the auto- and heterotrophic metabolism occurring concurrently. We simulated this condition in order to analyze the effects of the simultaneous presence of glucose and light on the production yields. The mixotrophic metabolism behaved similarly to the heterotrophic metabolism and provided much more flexibility for re-routing the carbon flux, compared with the autotrophic metabolism. High yielding growth-coupled knockouts were found for all the metabolites, in both the LLS and the CLS (Table 3; Fig. 2). However, these yields, ranging from 20 - 50% of the maximum production under the LLS and from 10 - 40% under the CLS, were markedly lower than those found under heterotrophic conditions. The mixotrophic mutants were found to share several interesting features: First, many equivalent overproducing mutants were predicted in the two photosynthetic states but the excess of light in CLS led to a significant decrease in the number of overproducing mutants. Under the CLS, the AEF pathways are essential for growth due to their role in redox balancing and they cannot be blocked simultaneously.22 This could indicate that the essentiality of the AEF pathways under the CLS limits the biotechnological potential of Synechocystis in this state. Second, the blocking of key photosynthetic reactions, including photosystems I (PSI) and II (PSII), the cytochrome b6f (CBFC) or ferredoxin NADP+ oxidoreductase (FNOR), as well as several AEFs (leading to reduced photosynthetic robustness) was required to couple the production of the target metabolite to growth. Third, in most of the cases, the overproducing mutants were non-viable under autotrophic conditions and glucose was used as the sole carbon source. Consistently and with few exceptions, the theoretical maximum production and the growth rates of the mixotrophic mutants were within the range predicted for the heterotrophic condition (Fig. 2). Fumarate was a notable exception and several mutants were found to be in the range corresponding to the LLS and the CLS mixotrophic states.
Table 3. Properties of the growth-coupled overproducer designs under mixotrophic conditions.
| |
Mixotrophic conditions |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Light limiting state (0.145) | Carbon limiting state (0.238) | |||||||||
| Metabolite |
Maximun production rate |
Number of knockouts |
Growth rate |
Production rate (mmol.gDW−1h−1) |
BPCY |
Maximun production rate |
Number Knockouts |
Growth rate |
Production rate |
BPCY |
| (mmol.gDW−1h−1) |
(h−1) |
(mmol.gDW−1h−1) |
(h−1) |
(mmol.gDW−1h−1) |
||||||
| Fumarate (4 C) |
2.351 |
6 |
0.061 |
0.88 |
0.0533 |
2.303 |
6 |
0.061 |
0.88 |
0.0533 |
| Ethanol (2 C) |
3.092 |
10 |
0.091 |
0.487 |
0.0441 |
4.607 |
7 |
0.037 |
0.682 |
0.0256 |
| 1-Butanol (4 C) |
1.546 |
5 |
0.038 |
0.591 |
0.0222 |
2.303 |
5 |
0.038 |
0.591 |
0.0222 |
| Sucrose (12 C) |
0.732 |
5 |
0.03 |
0.295 |
0.0088 |
0.767 |
5 |
0.03 |
0.295 |
0.0088 |
| Lactate (3 C) |
3.085 |
6 |
0.029 |
1.008 |
0.0291 |
3.071 |
6 |
0.029 |
1.008 |
0.0291 |
| H2 | 18.554 | 11 | 0.112 | 2.897 | 0.3248 | 35.452 | 8 | 0.156 | 16.12 | 2.507 |
LLS, Light limiting state; CLS, Carbon limiting state; BPCY, Biomass-product coupled yield. Numbers inside parenthesis represent the wild-type growth rate.
In summary, overproducing mutants under the mixotrophic conditions were obtained by avoiding the light-driven metabolism and by reducing the photosynthetic robustness. This could indicate that the photosynthetic processes do indeed limit the possibilities in re-routing of the carbon flux and consequently the overproduction of the target metabolites.
Photohydrogen production has previously been reported in Synechocystis and other cyanobacteria.10,11 In order to explore how the electron flux can be re-routed under different growth conditions, we extended our analysis to search for H2 overproducing mutants. Several mutants with high production rates were found in all the growth conditions. The yields of H2 were highest under auto- and mixotrophic conditions and the excess of light under the CLS increased the production rates almost 4-fold (Tables 1–3). This was expected since the photolysis of H2O is the main source of electrons in presence of glucose and it is the sole electron source under autotrophic conditions. An interesting finding was that the H2 overproducing mutants were achieved by blocking the AEF pathways almost exclusively, thus reducing the photosynthetic robustness. Consequently the H2 production remained as the main electron sink in the network (Tables 1 and 3).
This study is a first step toward evaluating how the particular metabolic features of Synechocystis that we revealed in our previous work, may affect the biotechnological potential of this organism. It must be noted that since the random search strategy is unable to test all possible knockouts (unless the number of knockouts is very small), the existence of growth-coupled autotrophic mutants for some of the compounds under investigation cannot be ruled out. The multiple overproducing mutants found under heterotrophic and mixotrophic conditions indicate, however, that the lack of growth-coupled autotrophic mutants is rather associated with the properties of light-driven metabolism than the search strategy itself. Another caveat is the limited number of compounds examined in this study and the conclusions may not apply to other metabolites or compounds of interest. In addition, it must be taken into account that we have mainly explored the growth-coupled biotechnological capabilities of Synechocystis and that alternative production strategies were beyond the scope of this study. Nevertheless, the analysis of the Synechocystis network under different growth condition presented here and the comparison of the results with those obtained previously with heterotrophic bacteria, offers a general view of the biotechnological capabilities of this cyanobacterium.
Several conclusions can be inferred from this study. First, we have found that the re-routing of the carbon flux in Synechocystis under autotrophic conditions is significantly more challenging than under hetero- or mixotrophic conditions. The constrained carbon flux distribution, in which up to 80 - 85% of the total fixed CO2 is funneled to sugar biosynthesis18 together with the metabolic peculiarities of Synechocystis under autotrophic conditions,22 could be the primary contributing factors. Mixotrophic conditions could be used in order to bypass this limitation, while allowing net CO2 fixation. However, we found that the blocking of the light-driven metabolism and that reduction of the photosynthetic robustness was a prerequisite for coupling the production of the target metabolites to growth. In addition, the mixotrophic metabolism in these mutant strains resembled the heterotrophic metabolism, with (net) CO2 fixation absent in the most of the cases. Finally, we have noted that, in contrast to the carbon flux, the electron flux can be manipulated more easily.10,11,28
Growth-coupled production is an attractive strategy in metabolic engineering. It is achieved by reducing the metabolic robustness of the host organism by deleting competing pathways, while the biosynthetic pathway of the target metabolite remains as the sole carbon and/or electron sink in the network. This way, the overproduction of the target compound is required for the organism to grow. The presented results strongly suggest that, while the high photosynthetic robustness required for optimal autotrophic metabolism allows flexible re-routing of the electron flux, it might also act as a non-desirable electron and carbon sink. Combined with low metabolic robustness inherent to cyanobacteria networks, this may hamper the possibilities for re-routing the carbon flux, thus, limiting the biotechnological capabilities of Synechocystis.
Disclosure of Potential Conflicts of Interest
The authors declare that they have no conflict of interest.
Acknowledgments
We thank Jóhanna Björk Pálsdóttir for assistance with the computational analysis. This work was supported by the US Department of Energy, Offices of Advanced Scientific Computing Research and Biological and Environmental Research as part of the Scientific Discovery Through Advanced Computing program Grant DE-SC0002009. J.N. was funded by the Spanish MEC through National Plan of I-D+i 2008–2011.
Footnotes
Previously published online: www.landesbioscience.com/journals/bioe/article/22792
References
- 1.Delucchi MA. Impacts of biofuels on climate change, water use, and land use. Ann N Y Acad Sci. 2010;1195:28–45. doi: 10.1111/j.1749-6632.2010.05457.x. [DOI] [PubMed] [Google Scholar]
- 2.Melillo JM, Reilly JM, Kicklighter DW, Gurgel AC, Cronin TW, Paltsev S, et al. Indirect emissions from biofuels: how important? Science. 2009;326:1397–9. doi: 10.1126/science.1180251. [DOI] [PubMed] [Google Scholar]
- 3.Quintana N, Van der Kooy F, Van de Rhee MD, Voshol GP, Verpoorte R. Renewable energy from Cyanobacteria: energy production optimization by metabolic pathway engineering. Appl Microbiol Biotechnol. 2011;91:471–90. doi: 10.1007/s00253-011-3394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waterbury JB, Watson SW, Guillard RRL, Brand LE. Widespread occurrence of a unicellular, marine, planktonic, cyanobacterium. Nature. 1979;277:293–4. doi: 10.1038/277293a0. [DOI] [Google Scholar]
- 5.Dismukes GC, Carrieri D, Bennette N, Ananyev GM, Posewitz MC. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr Opin Biotechnol. 2008;19:235–40. doi: 10.1016/j.copbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Abed RMM, Dobretsov S, Sudesh K. Applications of cyanobacteria in biotechnology. J Appl Microbiol. 2009;106:1–12. doi: 10.1111/j.1365-2672.2008.03918.x. [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Fallon S, Sheng J, Curtiss R., 3rd CO2-limitation-inducible Green Recovery of fatty acids from cyanobacterial biomass. Proc Natl Acad Sci U S A. 2011;108:6905–8. doi: 10.1073/pnas.1103016108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koksharova OA, Wolk CP. Genetic tools for cyanobacteria. Appl Microbiol Biotechnol. 2002;58:123–37. doi: 10.1007/s00253-001-0864-9. [DOI] [PubMed] [Google Scholar]
- 9.Heidorn T, Camsund D, Huang H-H, Lindberg P, Oliveira P, Stensjö K, et al. Chapter Twenty-Four - Synthetic Biology in Cyanobacteria: Engineering and Analyzing Novel Functions. In: Chris V, ed. Methods in Enzymology: Academic Press, 2011:539-79. [DOI] [PubMed] [Google Scholar]
- 10.Baebprasert W, Jantaro S, Khetkorn W, Lindblad P, Incharoensakdi A. Increased H2 production in the cyanobacterium Synechocystis sp. strain PCC 6803 by redirecting the electron supply via genetic engineering of the nitrate assimilation pathway. Metab Eng. 2011;13:610–6. doi: 10.1016/j.ymben.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Ducat DC, Sachdeva G, Silver PA. Rewiring hydrogenase-dependent redox circuits in cyanobacteria. Proc Natl Acad Sci U S A. 2011;108:3941–6. doi: 10.1073/pnas.1016026108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yacoby I, Pochekailov S, Toporik H, Ghirardi ML, King PW, Zhang S. Photosynthetic electron partitioning between [FeFe]-hydrogenase and ferredoxin:NADP+-oxidoreductase (FNR) enzymes in vitro. Proc Natl Acad Sci U S A. 2011;108:9396–401. doi: 10.1073/pnas.1103659108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niederholtmeyer H, Wolfstädter BT, Savage DF, Silver PA, Way JC. Engineering cyanobacteria to synthesize and export hydrophilic products. Appl Environ Microbiol. 2010;76:3462–6. doi: 10.1128/AEM.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dexter J, Fu P. Metabolic engineering of cyanobacteria for ethanol production. Energy & Environmental Science. 2009;2:857–64. doi: 10.1039/b811937f. [DOI] [Google Scholar]
- 15.Lan EI, Liao JC. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab Eng. 2011;13:353–63. doi: 10.1016/j.ymben.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Liu X, Sheng J, Curtiss R., 3rd Fatty acid production in genetically modified cyanobacteria. Proc Natl Acad Sci U S A. 2011;108:6899–904. doi: 10.1073/pnas.1103014108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan X, Yao L, Gao Q, Wang W, Qi F, Lu X. Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab Eng. 2011;13:169–76. doi: 10.1016/j.ymben.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 18.Lindberg P, Park S, Melis A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab Eng. 2010;12:70–9. doi: 10.1016/j.ymben.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Sheehan J. Engineering direct conversion of CO(2) to biofuel. Nat Biotechnol. 2009;27:1128–9. doi: 10.1038/nbt1209-1128. [DOI] [PubMed] [Google Scholar]
- 20.Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat Biotechnol. 2009;27:1177–80. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- 21.Ducat DC, Avelar-Rivas JA, Way JC, Silver PA. Rerouting carbon flux to enhance photosynthetic productivity. Appl Environ Microbiol. 2012;78:2660–8. doi: 10.1128/AEM.07901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nogales J, Gudmundsson S, Knight EM, Palsson BO, Thiele I. Detailing the optimality of photosynthesis in cyanobacteria through systems biology analysis. Proc Natl Acad Sci U S A. 2012;109:2678–83. doi: 10.1073/pnas.1117907109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feist AM, Zielinski DC, Orth JD, Schellenberger J, Herrgard MJ, Palsson BØ. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab Eng. 2010;12:173–86. doi: 10.1016/j.ymben.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fell DA, Small JR. Fat synthesis in adipose tissue. An examination of stoichiometric constraints. Biochem J. 1986;238:781–6. doi: 10.1042/bj2380781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savinell JM, Palsson BO. Network analysis of intermediary metabolism using linear optimization. II. Interpretation of hybridoma cell metabolism. J Theor Biol. 1992;154:455–73. doi: 10.1016/S0022-5193(05)80162-6. [DOI] [PubMed] [Google Scholar]
- 26.Nogales J, Gudmundsson S, Thiele I. An in silico re-design of the metabolism in Thermotoga maritima for increased biohydrogen production. Int J Hydrogen Energy. 2012;37:12205–18. doi: 10.1016/j.ijhydene.2012.06.032. [DOI] [Google Scholar]
- 27.Deng M-D, Coleman JR. Ethanol synthesis by genetic engineering in cyanobacteria. Appl Environ Microbiol. 1999;65:523–8. doi: 10.1128/aem.65.2.523-528.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cournac L, Guedeney G, Peltier G, Vignais PM. Sustained photoevolution of molecular hydrogen in a mutant of Synechocystis sp. strain PCC 6803 deficient in the type I NADPH-dehydrogenase complex. J Bacteriol. 2004;186:1737–46. doi: 10.1128/JB.186.6.1737-1746.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]