Abstract
Background and objectives
Following small bowel resection (SBR), the luminal environment is altered, which contributes to clinical manifestations of short bowel syndrome (SBS) including malabsorption, mucosal inflammation and bacterial overgrowth. However, the impact of SBR on the colon has not been well-defined. The aims of this study were to characterize the colonic microbiota following SBR and to assess the impact of SBR on mucosal inflammation in the colon.
Results
Analysis of the colonic microbiota demonstrated that there was a significant level of dysbiosis both two and six weeks post-SBR, particularly in the phylum Firmicutes, coupled with a decrease in overall bacterial diversity in the colon. This decrease in diversity was associated with an increase in colonic inflammation six weeks post-surgery.
Methods
Female (4-week old) piglets (5−6/group) received a 75% SBR, a transection (sham) or no surgery. Compositional analysis of the colonic microbiota was performed by high-throughput sequencing, two- and six-weeks post-surgery. The gene expression of the pro-inflammatory cytokines interleukin (IL)-1β, IL-6, IL-8, IL-18 and tumor necrosis factor (TNF)-α in the colonic mucosa was assessed by qRT-PCR and the number of macrophages and percentage inducible nitric oxide synthase (iNOS) staining in the colonic epithelium were quantified by immunohistochemistry.
Conclusions
SBR significantly decreased the diversity of the colonic microbiota and this was associated with an increase in colonic mucosal inflammation. This study supports the hypothesis that SBR has a significant impact on the colon and that this may play an important role in defining clinical outcome.
Keywords: short bowel syndrome, mucosal immunology, colonic microbiota, high-throughput sequencing, bacterial diversity, small bowel resection
Introduction
Short bowel syndrome (SBS) is the clinical state of malabsorption and malnutrition that occurs following small bowel resection (SBR).1 Surgical resection of the small bowel may be required for the treatment of a range of conditions including congenital bowel abnormalities and necrotising enterocolitis in children and Crohn disease, trauma and malignancy in adults.2 Although the underlying reason for SBR between adults and children may differ, the clinical manifestations and consequences of SBS are similar. The symptoms of SBS reflect the loss of absorptive surface area and functional disturbance of the remaining intestine and include diarrhea, vomiting, malabsorption, dehydration and electrolyte imbalance, malnutrition and bacterial overgrowth.
Most cases of SBS occur in the neonatal age group and the mortality associated with the condition ranges from 15−25%.3,4 The gut microbiota is known to be a major factor in determining the clinical outcome in children.5 The effects of an alteration in the microbiota on the small intestine are well-studied in this cohort and include increased risk of bacterial overgrowth, bacteraemia, villous atrophy and small intestinal mucosal inflammation, which can result in a loss of intestinal epithelial barrier function and can lead to sepsis.6-8 In both adult patients with SBS and a piglet SBS model, the colon is known to play a critical role in determining clinical outcome, and significant morphological adaptation of the colonic mucosa has been observed following resection.9-11 However, the actual impact of changes in the microbiota on the colon itself is poorly understood. Anecdotal evidence suggests that changes in the composition of the colonic microbiota contribute to the generation of symptoms including malnutrition due to decreased bile acid deconjugation, insufficient breakdown of nutrients and diarrhea and play a key role in development of serious complications such as septicaemia. However, very little is known about specific changes to the colonic microbiota following SBR or the impact these changes may have on colonic mucosal inflammation, the adaptive response and severity of symptoms in SBS.
Despite an obvious clinical need, there are a number of barriers to the study of gut function and immune regulation in SBS. Patients with SBS are a complex, heterogeneous group and access to tissue is limited. Thus, many studies are based on animal models. Rodent SBS models, although useful for molecular and cellular analysis, may not accurately reflect the physiology and pathophysiology of the human intestine. The pig is accepted as the best model for the study of human intestinal biology and diet12-14 and, with a similar intestinal microbiota to that of humans,15-17 it is an ideal model for the study of complex gastrointestinal diseases.
The aims of this study were: (1) to utilize high-throughput sequencing to characterize changes in the colonic diversity and microbial composition following SBR, (2) to examine the influence of surgical resection on colonic mucosal inflammation and (3) to examine whether there is a correlation between microbial differences and inflammation in a pre-clinical piglet model of SBS in children.
Results
Piglets that received a small bowel resection had a poorer clinical outcome
Food intake was monitored throughout the study and all diets were isocaloric and isonitrogenous. Piglets that received a small bowel resection (SBR) had suboptimal weight gain compared with the non-operation control (NOC) and sham-operated piglets (Fig. 1A). This was significant from the two-week time-point (p = 0.004), up until and including the six-week time-point (p < 0.0001). The sham group also had a significantly lower mean weight than the NOC group at the five-week and six-week time-points (p = 0.041 and p < 0.0001).18 Examination of feed intake showed that, although piglets in the SBR group consumed equal energy per kg per day, their total energy consumption was reduced when compared with the NOC and sham groups due to their decreased weight. Piglets in the SBR group failed to resolve diarrhea at either the two-week (p = 0.002 and p = 0.005) or six-week (p < 0.0001) time-points (Fig. 1B).
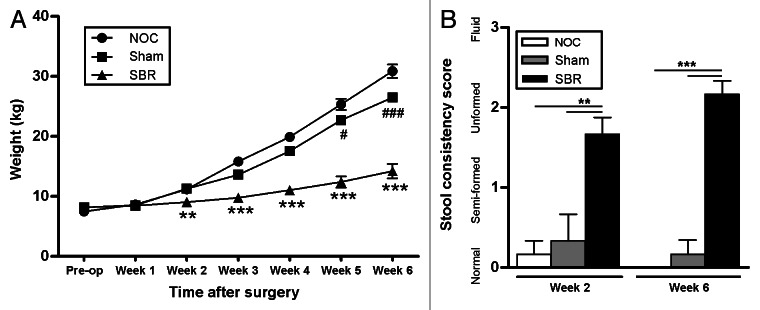
Figure 1. Clinical effects of small bowel resection. (A) The weight of piglets that received a small bowel resection (SBR) was significantly lower two weeks post-surgery than those that received a sham surgery (Sham) or no surgery (NOC; **p < 0.01). This suboptimal weight gain was sustained for the duration of the experiment (***p < 0.001 at week 3, p < 0.0001 at weeks 4, 5 and 6). The sham piglets also had a significantly lower weight gain at weeks 5 (#p < 0.05) and 6 (###p < 0.0001) compared with the NOC group. Values are expressed as mean ± SEM; n = 12 NOC, n = 10 sham, n = 12 SBR at pre-op, week 1 and week 2; n = 6 NOC, n = 5 sham, n = 6 SBR at weeks 3, 4, 5 and 6. (B) Piglets in the SBR group had a significantly higher stool consistency score than the sham and NOC groups two weeks (**p < 0.01) and six weeks (***p < 0.001) post-surgery. Values are expressed as mean ± SEM; n = 5−6/group/time-point.
Total colonic bacterial number is unaltered following small bowel resection
Absolute quantification, using qPCR, was used to determine the impact of surgery on total numbers of bacteria. The results indicate that there were no significant differences in the total 16S rRNA gene copies (representative of total bacteria numbers) between any of the groups at either week two (p = 0.7845) or week six (p = 0.8784; data not shown).
Surgical resection of the small intestine decreases bacterial diversity in the colon
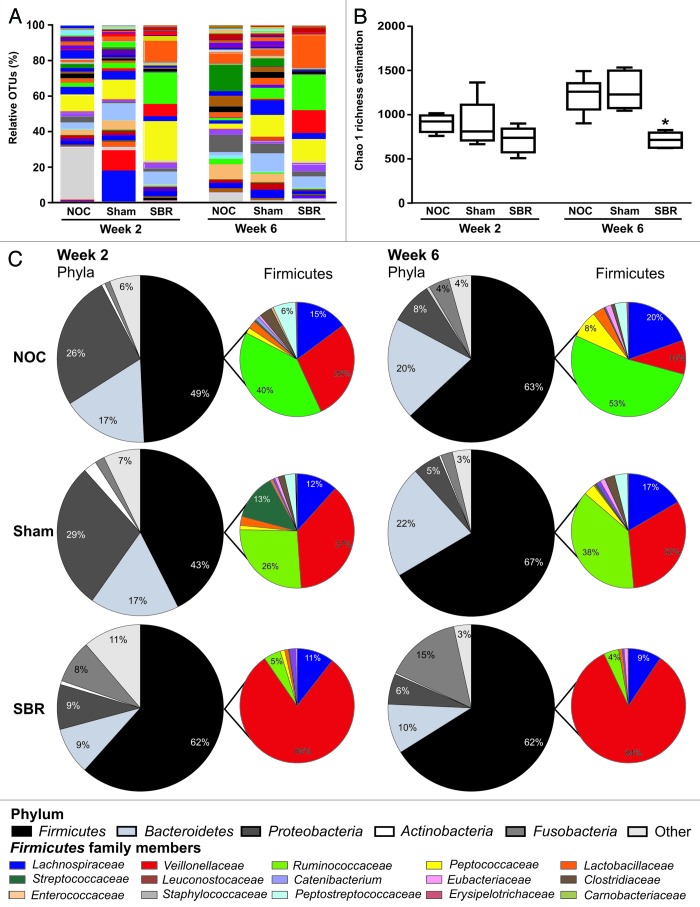
The colonic content from NOC, sham and SBR piglets was sequenced two- and six-weeks post-surgery. A total of 318,784 V4 16S sequence reads were generated, averaging at 9,376 per sample. Species richness, coverage and diversity estimations were calculated for each data set (Table 1). Rarefaction curves for each sample were parallel or approaching parallel, with the x axis indicating total bacteria diversity was well represented (Fig. S1A). There was a trend toward a decrease in the level of diversity in the SBR group compared with the NOC and sham groups at the two-week time-point as assessed by the Chao 1 calculation (Fig. 2A and B and Table 1) and the Simpson and Shannon indices (Table 1). By six weeks, however, there was a significant decrease in diversity in the SBR group compared with the NOC and sham groups as demonstrated by the Chao 1 calculation (p = 0.004; Fig. 2A and B), the Simpson index (p = 0.008) and the Shannon index (p = 0.003; Table 1). Principle component analysis revealed that the SBR groups at week two and week six clustered together and were distinct from the NOC and sham groups, which clustered together at both times (Fig. S1B).
Table 1. Surgical resection decreases the diversity of colonic bacteria. Estimation of diversity within the non-operation control (NOC), sham and small bowel resection (SBR) groups at two- and six- weeks post surgery as assessed by Chao 1 richness estimation, Shannon’s index for diversity, number of observed species and Simpson’s diversity index.
| Data set | NOC Week 2 |
Sham Week 2 |
SBR Week 2 |
NOC Week 6 |
Sham Week 6 |
SBR Week 6 |
|---|---|---|---|---|---|---|
| Chao 1 richness estimation |
905 ± 41 |
892 ± 123 |
718 ± 59 |
1224 ± 82 |
1276 ± 97 |
716 ± 34 |
| Shannon’s index for diversity |
6.9 ± 0.2 |
6.5 ± 0.6 |
5.7 ± 0.3 |
7.1 ± 0.1 |
7.4 ± 0.2 |
5.5 ± 0.2 |
| Number of observed species |
552 ± 16 |
571 ± 80 |
475 ± 44 |
726 ± 46 |
777 ± 63 |
428 ± 15 |
| Simpson’s diversity index | 0.96 ± 0.01 | 0.95 ± 0.02 | 0.93 ± 0.01 | 0.98 ± 0.01 | 0.98 ± 0.00 | 0.93 ± 0.01 |
Figure 2. Surgical resection of the small intestine decreases the diversity of colonic bacteria and alters the relative proportion of families in the Firmicutes phylum. (A) There is a decrease in the number of distinct operational taxonomic units (OTUs) representing bacterial genera two and six weeks post-surgery in the small bowel resection (SBR) group compared with the non-operation control (NOC) and sham group. This is translated into a decrease in overall bacterial diversity as calculated by the Chao 1 richness estimation (B). (C) Pie charts representing the major bacterial phyla and the relative proportion of families in the Firmicutes family at the two- and six-week time-point in the NOC, sham and SBR groups.
The composition of the pig microbiota is altered two weeks after small bowel resection
Two weeks post-SBR, the majority of changes were detected at the family and genus levels. There were significant differences at the family level between the SBR group and the NOC and sham groups (Fig. 2C and Table 2). These included a significant increase in Veillonellaceae (p = 0.0018 and p = 0.0115) and a significant decrease in Ruminococcaceae (p = 0.0012 and p = 0.0244) in the SBR group compared with the NOC and sham groups (Fig. 2C and Table 2). Peptostreptococcaceae was undetectable in any of the piglets in the SBR group at week two (Table 2). At the genus level, there was a significant increase in Acidaminococcus in the SBR group compared with the NOC and sham groups (p = 0.0015 and p = 0.0033; Table 2).
Table 2. Small bowel resection alters the composition of the colonic microbiota over time. The composition of the colonic microbiota is altered two and six weeks post-surgery in the small bowel resection (SBR) group. Values are relative proportions of operational taxonomic units (OTUs) ± SEM, n = 5−6/group/time-point. Statistical significance was determined using a general linear model ANOVA; *p < 0.05; **p < 0.01; ***p < 0.001 compared with non-operation control (NOC) group; †p < 0.05, ††p < 0.01, †††p < 0.001 compared with sham group.
| Weeks post-surgery | Week 2 | Week 6 | ||||
|---|---|---|---|---|---|---|
|
Group |
NOC |
Sham |
SBR |
NOC |
Sham |
SBR |
|
Phylum: |
|
|
|
|
|
|
| Firmicutes |
52.6 ± 5.0 |
52.1 ± 10.6 |
68.5 ± 6.1 |
61.7 ± 6.5 |
67.7 ± 3.6 |
68.2 ± 4.2 |
| Proteobacteria |
27.8 ± 5.1 |
23.1 ± 14.6 |
11.9 ± 4.6 |
10.6 ± 3.3 |
6.0 ± 0.6 |
6.2 ± 1.8 |
| Bacteroidetes |
17.9 ± 2.5 |
19.1 ± 5.8 |
10.1 ± 1.7 |
22.3 ± 2.6 |
23.2 ± 1.9 |
10.0 ± 1.6**†† |
| Fusobacteria |
0.9 ± 0.4 |
3.8 ± 3.1 |
8.5 ± 2.7 |
4.7 ± 2.3 |
2.8 ± 1.4 |
15.4 ± 2.4**†† |
| Actinobacteria |
0.7 ± 0.2 |
2.0 ± 1.5 |
1.0 ± 0.4 |
0.7 ± 0.2 |
0.3 ± 0.1 |
0.3 ± 0.1 |
|
Family: |
|
|
|
|
|
|
| Enterobacteriaceae |
31.8 ± 6.9 |
5.8 ± 2.5** |
1.0 ± 0.5*** |
6.7 ± 2.7 |
1.9 ± 0.7 |
4.0 ± 2.1 |
| Ruminococcaceae |
28.1 ± 3.1 |
27.0 ± 4.2 |
5.2 ± 2.5**† |
41.5 ± 5.2 |
36.1 ± 4.1 |
3.9 ± 1.6***††† |
| Veillonellaceae |
19.5 ± 4.9 |
36.7 ± 8.4 |
70.5 ± 7.1**† |
9.9 ± 2.9 |
28.0 ± 6.5 |
64.2 ± 6.6***††† |
| Peptostreptococcaceae |
4.6 ± 1.5 |
2.1 ± 1.5 |
0 |
2.8 ± 0.6 |
3.7 ± 1.4 |
0*† |
| Prevotellaceae |
3.9 ± 0.6 |
14.4 ± 2.8* |
7.1 ± 0.5 |
2.0 ± 0.9 |
9.7 ± 1.8** |
5.8 ± 0.7** |
| Bacteroidaceae |
3.5 ± 0.8 |
1.1 ± 0.6 |
2.4 ± 1.1 |
9.0 ± 2.1 |
6.0 ± 0.9 |
2.9 ± 1.2 |
| Erysipelotrichaceae |
2.5 ± 0.9 |
1.4 ± 0.7 |
0.4 ± 0.2 |
10.0 ± 6.0 |
3.8 ± 1.3 |
0 |
| Clostridiaceae |
2.1 ± 0.5 |
1.0 ± 0.5 |
0 |
0.7 ± 0.1 |
2.7 ± 1.3 |
0.1 ± 0.0 |
| Lactobacillaceae |
1.5 ± 0.6 |
2.1 ± 1.1 |
1.2 ± 0.3 |
2.9 ± 0.8 |
0.2 ± 0.1* |
0.8 ± 0.4 |
| Fusobacteriaceae |
1.4 ± 0.6 |
7.5 ± 3.9 |
11.9 ± 4.1 |
8.3 ± 4.3 |
4.8 ± 2.5 |
18.2 ± 3.0**†† |
| Peptococcaceae |
1.1 ± 0.5 |
0.9 ± 0.3 |
0.4 ± 0.4 |
6.3 ± 1.9 |
3.2 ± 0.7 |
0** |
|
Genus: |
|
|
|
|
|
|
| Enterobacteriaceae family members |
48.8 ± 8.7 |
9.4 ± 3.7*** |
1.4 ± 0.7*** |
11.6 ± 2.8 |
3.6 ± 1.1 |
5.7 ± 2.9 |
| Bacteroides |
5.8 ± 1.3 |
2.1 ± 1.1 |
3.5 ± 1.5 |
21.2 ± 3.2 |
11.5 ± 1.6 |
4.3 ± 1.7 |
| Parabacteroides |
5.1 ± 1.2 |
10.9 ± 3.3 |
1.6 ± 1.1 |
19.2 ± 4.5 |
10.9 ± 3.4 |
0.6 ± 0.2* |
| Prevotella |
6.7 ± 1.6 |
23.2 ± 4.5* |
10.4 ± 1.2 |
3.1 ± 1.5 |
18.9 ± 3.4** |
9.6 ± 0.9* |
| Clostridium |
3.4 ± 0.7 |
1.8 ± 0.9 |
0 |
1.8 ± 0.6 |
5.1 ± 2.6 |
0.1 ± 0.1† |
| Fusobacterium |
2.7 ± 1.4 |
12.4 ± 6.6 |
17.7 ± 6.2 |
11.9 ± 5.1 |
9.0 ± 4.8 |
29.9 ± 4.3**†† |
| Megasphaera |
17.1 ± 6.0 |
27.3 ± 4.4 |
31.4 ± 6.1 |
6.2 ± 2.5 |
23.5 ± 7.7 |
18.3 ± 3.0* |
| Acidaminococcus |
4.5 ± 1.2 |
8.0 ± 2.8 |
31.7 ± 6.7**†† |
3.6 ± 2.3 |
11.1 ± 6.5 |
30.2 ± 5.1***†† |
| Peptococcus |
1.9 ± 0.9 |
1.5 ± 0.6 |
0.7 ± 0.6 |
14.9 ± 5.7 |
6.1 ± 1.7 |
0.1 ± 0.1 |
| Lactobacillus | 4.0 ± 1.8 | 3.4 ± 1.8 | 1.7 ± 0.4 | 6.4 ± 1.9 | 0.4 ± 0.2* | 1.3 ± 0.6 |
Small bowel resection leads to dysbiosis of the gut microbiota six weeks post-surgery
Six weeks post-surgery, there were significant decreases in the proportions of Bacteroidetes (p = 0.0018 and p = 0.0013) and significantly increased proportions of Fusobacteria (p = 0.0072 and p = 0.0032) in the SBR group compared with the NOC and sham groups at this time-point (Fig. 2C; Table 2). The majority of differences were observed at the level of the family, particularly in members of the Firmicutes phylum (Fig. 2C and Table 2). At six weeks, there was a significant decrease in members of the Peptostreptococcaceae (p = 0.0477 and p = 0.0154) and Ruminococcaceae (p = 0.00001 and p = 0.0002) families in the six-week SBR group compared with the six-week NOC and sham groups. Peptococcaceae were undetectable in the majority of SBR piglets at week six. Conversely, there was a significant increase in the proportions of Veillonellaceae (p = 0.00001 and p = 0.0007) and Fusobacteriaceae (p = 0.0072 and p = 0.0032) in the SBR group compared with the NOC and sham groups at the six-week time-point. Increases at the family level were reflected at the genus level, with increases in the proportion of Megasphaera (p = 0.0156) in the SBR group compared with the NOC group, and Acidaminococcus (p = 0.0006 and p = 0.0063) and Fusobacterium (p = 0.0073 and p = 0.0032) compared with both the NOC and sham groups. Clostridium proportions were significantly decreased in the SBR group compared with the sham group (p = 0.0433).
There is an increase in colonic inflammation following small bowel resection, which is associated with decreased diversity
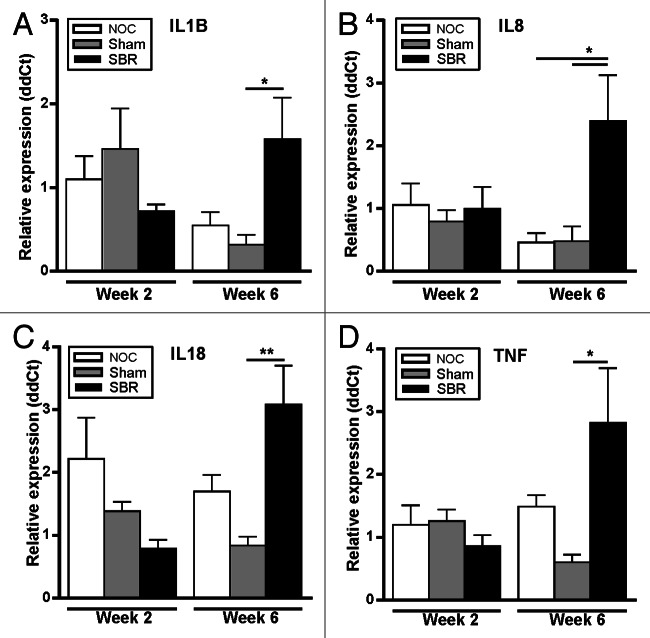
The gene expression of a panel of pro-inflammatory cytokines was assessed in the colon. Two weeks after surgery, there was no difference in inflammatory gene expression among groups (Fig. 3A−D). There was, however, a significant increase in the expression of IL-1β (p = 0.045), IL-18 (p = 0.006) and TNF-α (p = 0.036) in the SBR group compared with the sham group six-weeks post-surgery (Fig. 3A, C and D), and IL-8 was significantly increased in the SBR group compared with both the NOC and sham groups (p = 0.025 and p = 0.034; Fig. 3B).
Figure 3. Surgical resection alters the expression of pro-inflammatory cytokines in the colon. In the colon, the gene expression of IL-1β (A), IL-18 (C) and TNF-α (D) was significantly higher in the SBR group compared with the sham group at six weeks (*p < 0.05, **p < 0.01). IL8 (B) was significantly higher in the colon in the small bowel resection (SBR) group compared with the non-operation control (NOC) and sham group at six weeks (*p < 0.05). Values are expressed as mean ± SEM; n = 5−6/group/time-point.
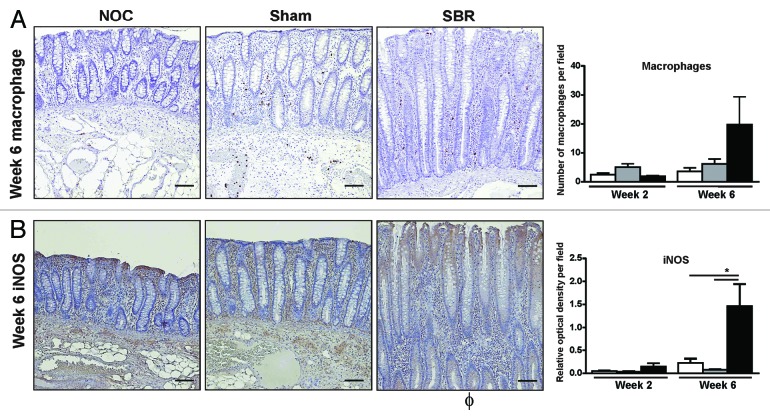
In the case of IL-1β and IL-8, the increase in these pro-inflammatory cytokines correlated with the observed decrease in colonic bacterial diversity (r = -0.829 and r = -0.771, respectively; Table 3). The increase in colonic pro-inflammatory cytokines was also associated with a concomitant increase in the total number of macrophages present in the colonic epithelium of the SBR group compared with the NOC and sham groups at six weeks (Fig. 4A). At this time-point, there was also a significant increase in the amount of the inflammatory marker iNOS in the colonic epithelium of the SBR group compared with both the NOC and the sham (p = 0.03; Fig. 4B).
Table 3. A decrease in colonic bacterial diversity is associated with an increase in inflammatory cytokines. The expression of colonic IL-1β and IL-8 negatively correlate with colonic bacterial diversity as assessed by Spearman’s rank correlation coefficient.
| Bacterial α diversity | IL-1β | IL-8 | IL18 | TNF-α |
|---|---|---|---|---|
| Spearman r |
-0.8286 |
-0.7714 |
-0.2571 |
-0.4857 |
| p value | 0.0583 | 0.1028 | 0.6583 | 0.3556 |
Figure 4. There is an increase in inflammatory cells and mediators in the colonic epithelium following small bowel resection. There was an increase in the number of macrophages (A) and the percentage of cytoplasmic inducible nitric oxide synthase (iNOS) staining (B) in the colonic epithelium six weeks post-surgery. Immunohistochemistry images illustrate representative staining six weeks post-surgery. Scale bar represents 100 µm. Values are expressed as mean ± SEM; n = 5−6/group.
Discussion
This study provides the first comprehensive description of the changes in the colonic microbiota following small bowel resection. Not only does this study take advantage of high-throughput sequencing to identify and characterize the dysbiosis that occurs following SBR using an established preclinical piglet model of SBS in children, it also links these changes with local mucosal inflammatory responses.
Following SBR, the luminal environment becomes altered due to rapid shunting of luminal content from the upper intestine into the lower intestine, due to shortened bowel length and increased intestinal transit.19 This exposes the colon and its resident bacteria to both digesta and intestinal and pancreatico-biliary secretions that would normally be broken down or absorbed in the upper intestinal tract, which could potentially impact on the composition of bacteria in the lumen.20 Evidence on the impact of SBR on bacterial composition is largely anecdotal; however any changes in bacterial composition are likely to have important implications for patients with SBS, in terms of management of their condition.
This study has established that there is a significant decrease in overall bacterial diversity in the colon in a piglet model of SBS that is particularly evident six weeks post-resection. This is consistent with other inflammatory conditions including Crohn disease,21 ulcerative colitis22 and antibiotic-associated diarrhea23 and has previously been described in adult patients with SBS.24 A decrease in microbial diversity also corresponds to decreased metabolic diversity, which has potential implications for SBS patients including insufficient breakdown of dietary components such as complex polysaccharides leading to malabsorption, reduced energy availability and decreased production of short-chain fatty acids.25
This decrease in diversity is due to certain species becoming dominant in the colon, to the detriment of others. At two weeks post-surgery, members of the Firmicutes phylum underwent the most significant alterations, with an increase in the proportion of the Veillonellaceae family and a decrease in the proportion of the Ruminococcaceae family in the SBR group relative to the NOC and sham group. Major differences in bacterial composition were observed six weeks post-surgery. The phylum Fusobacteria was significantly increased in the SBR group relative to the NOC and sham groups. Gram negative bacteria in this phylum express pro-inflammatory molecules such as lipopolysaccharide, which is known to trigger inflammation in the gut.26-28 At the family level, the majority of changes were again within the Firmicutes phylum, with a decrease in the proportion of Clostridiaceae, Peptostreptococcaceae and Peptococcaceae in the SBR group compared with the sham group at six weeks. These Gram positive anaerobic bacteria may provide CpG DNA with immunomodulatory activities and their decrease or absence is postulated to disrupt the crosstalk between the host mucosal immune system and the microbiota.29 The increase in the Veillonellaceae family and the decrease in Ruminococcaceae family observed in the SBR group at two weeks was sustained at the six week time-point. Several studies have linked alterations in each of these families with systemic and local inflammation.30,31 As the delicate balance between host epithelial cells and resident bacteria is already disturbed in patients with SBS due to the unfavorably altered luminal environment, these specific changes in key bacterial families may further exacerbate the clinical manifestations of SBS.
Interestingly, there were also alterations in some bacterial families in the sham surgery group. The proportion of Enterobacteriaceae was significantly decreased and the proportion of Prevotellaceae was significantly increased in the sham group compared with the NOC group at the two week time-point. This increase in Prevotellaceae was sustained at the six week time-point and there was also a decrease in the proportion of Lactobacillaceae at this time. It is known that surgical trauma often results in postoperative ileus and in the activation of resident macrophages and the upregulation of functional activity of pro-inflammatory cytokines.32 Thus, the sham surgery, although relatively minor compared with the massive small bowel resection surgery, would also have an effect on the gut microbiota.
A limited number of studies have examined the bacterial community in SBS patients using culture- and microscopy-based techniques. These studies suggest that there is a shift in the microbiota and that the microbiota of SBS patients is composed primarily of Gram positive organisms such as lactobacilli.33 More recent studies that used quantitative PCR to identify changes in the microbiota in adult patients with SBS, confirmed that there is enrichment in lactobacilli in these patients.24,34 Using high-throughput DNA sequencing, we observed no significant difference in the proportion of Lactobacillus spp at either time-point. Given that a purported 70−80% of gut bacteria are unculturable, our results may reflect the more comprehensive identification of previously unidentified bacterial content35 and may also indicate that there are important differences between adult and pediatric SBS patients in terms of their microbiota.
Despite significant differences in pro-inflammatory bacteria in the SBR group two weeks post-surgery, there was no increase in pro-inflammatory colonic cytokine expression at this time-point. However, six weeks post-SBR, there were significant increases in the pro-inflammatory cytokines IL-1β, IL-8, IL-18 and TNF-α in the SBR group. This was coupled with an increase in the number of macrophages in the colon, as well as an increase in colonic iNOS in the SBR group. This increase in mucosal inflammation coincided with a marked decrease in diversity and dysbiosis at the later time-point. Consistent with this observation, there was a correlation between a decrease in bacterial diversity and an increase in colonic IL-8 and IL-1β. Several studies have shown a link between diversity and inflammation, although there is debate as to which is the causative agent.21,29,36 In our study, the decrease in diversity was evident two weeks post-surgery, but significant markers of inflammation did not occur until six weeks post-SBR. Thus, it is our hypothesis that the colonic microbiota is negatively impacted by the altered luminal environment following the surgical resection, resulting in a decrease in diversity, which initiates a pro-inflammatory response in the colon.
This study has identified and characterized dysbiosis in the colon that occurs following SBR and challenges current accepted theories regarding specific alterations in the microbiota of patients with SBS. We have shown a reduction in diversity following SBR due to certain bacterial species establishing dominance and described inflammation in the colon that persists for six weeks after SBR. We suggest that this inflammation is a consequence, rather than a trigger of the dysbiosis and that the overall composition of the microbiota may be more relevant than the presence or absence of a single species. This study has highlighted that the colon is significantly impacted by proximal SBR and the role that the colon may play in determining the clinical outcome following a resection has been underappreciated.
Materials and Methods
Animals
This study was approved by the Animal Ethics Committee of the Murdoch Childrens Research Institute. Weaned female 3-week-old piglets (Landrace/Large White cross; Aussie Pride Pork) were transported to The University of Melbourne Centre for Animal Biotechnology and acclimatised prior to surgery. Piglets were fed a polymeric infant formula diet (Karicare De-Lact, Nutricia) supplemented to meet the daily requirements for piglets as described previously.11,37,38 The diets were isocaloric and isonitrogenous among the groups and were administered on a per kilogram basis. Water was given twice daily. Piglets were housed separately throughout the study to allow accurate daily monitoring of food and water intake and stool output.
Clinical assessment and growth
Piglet weight was measured weekly before feeding. Faecal samples were collected weekly and stool consistency was scored by the Royal Children's Hospital Laboratory Services, Melbourne, Australia using the following scale: 0 = formed, 1 = semi-formed, 2 = unformed and 3 = fluid.
Experimental design
At four weeks of age, piglets underwent either a 75% proximal small bowel resection (SBR, n = 12) or a transection and re-anastomosis (sham, n = 10) operation. One group of piglets did not receive any surgery (non-operation control; NOC, n = 12). The 75% SBR included the removal of the small bowel from 90 cm distal to the ligament of Treitz to 225 cm proximal to the ileocecal valve. During the sham procedure the intestine was transected and re-anastomosed at a site 225 cm proximal to the ileocecal valve. Piglets received intramuscular amoxicillin (70 mg/kg; CSL Limited) 24 h pre-surgery. On the day of surgery, piglets were anesthetized and given amoxicillin. Piglets received amoxicillin and oral rehydration salts (Sanofi-Aventis Australia) for three days post-surgery in line with current clinical practice. Water and the polymeric infant formula diet were re-introduced from the third day post-operation. All piglets in the NOC group followed the same feeding regime, with piglets in the week 2 NOC group receiving antibiotics in line with the surgical groups.
Sample collection
Animals in the SBR and sham groups were sacrificed either two- or six-weeks post-surgery and at age-matched times in the NOC group. Colonic tissue was collected 3 cm and 10 cm distal to the cecum in the two-week and six-week groups, respectively, at locations optimised for age, as based on a previous study.37 A 3 cm section from each site was divided in half longitudinally and fixed in 10% neutral buffered formalin (Australian Biostain Pty Ltd) or snap frozen in liquid nitrogen. Colonic content was collected from the excised colonic tissue sample.
DNA extraction and amplicon sequencing
DNA was extracted from colonic content using the standard QIAamp DNA Stool Mini Kit protocol (Qiagen, 51504), with the addition of an initial bead beating step. The 16S rRNA amplicons were generated using a previously outlined approach.39 Amplicons were generated using one forward primer and a combination of four reverse primers as described previously.40 Each primer contained a distinct multiple identifier (MID) allowing pooling of the amplicons and subsequent separation of the results for analysis. Duplicate PCR products were pooled and cleaned using Agencourt AMPure kit (Beckman Coulter, A63880). Quantification was completed using Quant-iT Picogreen quantification kit (Invitrogen, P7589) and the Nanodrop 3300 (Thermo Scientific). The V4 region of the 16S rRNA was sequenced at the Teagasc 454-Sequencing facility on a Genome Sequencer FLX platform (Roche Diagnostics Ltd.).
Bioinformatic analysis
Raw sequencing reads were quality trimmed using the RDP Pyrosequencing Pipeline applying the following criteria: (1) exact matches to primer sequences and barcode tags, (2) no ambiguous bases (Ns) and (3) read-lengths no shorter than 150 base pairs. Trimmed FASTA sequences were then BLASTED41 against the SILVA (v100) database for 16S reads.42 Phylum, family and genus counts were extracted from MEGAN43 using a bit score cut-off of 86.42 Clustering into operational taxonomical units (OTUs), alignments, chimera-checking and alpha diversities were implemented using the Qiime suite of tools.44 A phylogenetic tree was generated using the FastTree package45 and principal coordinate analysis (PCoA), measuring dissimilarities at phylogenetic distances based on unweighted Unifrac analysis, was performed with Qiime suite of tools(Ref). PCoA plots were visualized with KiNG software package (http://kinemage.biochem.duke.edu/software/king.php).
Quantitative PCR
Absolute quantification was completed using the Roche LightCycler 480 platform (Roche Diagnostics). Samples consisted of 2 µl PCR grade water, 1 µl forward primer F1 (5′ AYTGGGYDTAAAGNG; 0.15 µM), 1 µl reverse primer R1 (5′TACCRGGGTHTCTAATCC; 0.15 µM), 1 µl template DNA and 5 µl SYBR green (Roche Diagnostics, 04887352001). Bacteria were quantified using 16S rRNA counts based on a standard curve, using a previously outlined calculation.46 All reactions were run in triplicate.
Real-time reverse-transcription PCR (qRT-PCR)
The muscle layer was stripped from 100 mg of colonic tissue leaving the colonic mucosa, which comprised the epithelium and the lamina propria. Total RNA was extracted from the mucosa using TRIzol (Invitrogen, 15596–026). cDNA was synthesized with the Transcriptor First Strand cDNA Synthesis Kit (Roche Applied Science, 04897030001). PCR primers were designed against pig gene sequences using Roche Universal ProbeLibrary Assay Design Centre (Roche Applied Science, 04683633001). Primer sequences and probe combinations are listed in supplementary Table 1. PCR reactions were performed in triplicate on the LightCycler 480. The 2-ΔΔCt method47 was used to calculate relative changes in gene expression using RPL32 as a housekeeping gene and relative to a pre-operation group (n = 6).
Microscopic assessment of inflammation
Immunohistochemistry was performed on formalin-fixed, paraffin-embedded colon tissues sectioned at 5 µm. To identify macrophages, antigen retrieval was performed with proteinase K followed by addition of primary antibody (MAC387; 1:1000; Abcam, ab22506) for two hours at room temperature. Staining of inducible nitric oxide synthase (iNOS) was performed by performing antigen retrieval in citrate buffer (pH 6.0) and adding primary antibody (iNOS; 1:100; Abcam, ab15323) overnight at 4°C. Secondary staining and 3,3-Diaminobenzidine (DAB) detection was performed using Histostain-Plus 3rd Gen IHC Detection Kit (Invitrogen, 85–9073). Negative controls were included in which the primary antibody was substituted with antibody diluent. Slides were viewed under an apochromat 10x objective lens using a Nikon Eclipse 80i microscope (Nikon Instruments Inc.), equipped with a DS-Ri1 CCD camera (Nikon) and controlled by NIS-Elements acquisition software version 4.00 (Nikon). For each set of analyses, ten fields of view of the epithelium were collected. Macrophages were quantified by counting the number of positively-stained cells in the colonic epithelium using ImageJ.48 The amount of iNOS epithelial cytoplasmic staining was quantified by color deconvolution using ImageJ.49
Statistical analysis
Data are presented as mean values with their standard error (SEM). Statistical analysis was performed using one-way ANOVA at each time-point, followed by Tukey’s post-hoc test (GraphPad Prism Software 5.0). Sequencing analysis was completed using Minitab Release 15.1.1.0 (Minitab Inc. 2007). To identify if significant differences occurred at phylum, family and genus levels between the three groups, non-parametric Kruskal-Wallis or Mann-Whitney tests were performed followed by Tukey’s post-hoc test. This test was also used for comparing the changes in total bacterial counts in the three groups. The non-parametric Spearman rank correlation was used to determine relationships between diversity and inflammation. Statistical significance was accepted at p < 0.05.
Supplementary Material
Acknowledgments
This work was supported by the Victorian Government's Operational Infrastructure Support Program. F.F. is in receipt of an Irish Research Council for Science, Engineering and Technology EMBARK scholarship and is a Teagasc Walsh fellow. Research in the Ross, Fitzgerald, Stanton and Cotter laboratories is funded by Science Foundation Ireland (in the form of a research center grant to the Alimentary Pharmabiotic Centre and a PI award to P.C.). The authors wish to thank Magdy Sourial, Shane Osterfield and Dr Andrew French for expert technical assistance with the animals and Dr Jean-Pierre Scheerlinck and The University of Melbourne Centre for Animal Biotechnology for the use of their facilities. The authors would also like to thank Dr Eva Rosberg-Cody for high-throughput DNA sequencing services.
Glossary
Abbreviations:
- ANOVA
analysis of variance
- cDNA
complementary deoxyribonucleic acid
- DAB
3,3-diaminobenzidine
- IL
interleukin
- iNOS
inducible nitric oxide synthase
- LPS
lipopolysaccharide
- NOC
non-operation control
- qPCR
quantitative polymerase chain reaction
- qRTPCR
real-time reverse-transcription polymerase chain reaction
- SBR
small bowel resection
- SBS
short bowel syndrome
- TNF
tumor necrosis factor
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/24372
References
- 1.Sukhotnik I, Siplovich L, Shiloni E, Mor-Vaknin N, Harmon CM, Coran AG. Intestinal adaptation in short-bowel syndrome in infants and children: a collective review. Pediatr Surg Int. 2002;18:258–63. doi: 10.1007/s003830100695. [DOI] [PubMed] [Google Scholar]
- 2.Quirós-Tejeira RE, Ament ME, Reyen L, Herzog F, Merjanian M, Olivares-Serrano N, et al. Long-term parenteral nutritional support and intestinal adaptation in children with short bowel syndrome: a 25-year experience. J Pediatr. 2004;145:157–63. doi: 10.1016/j.jpeds.2004.02.030. [DOI] [PubMed] [Google Scholar]
- 3.Höllwarth ME. Short bowel syndrome: pathophysiological and clinical aspects. Pathophysiology. 1999;6:1–19. doi: 10.1016/S0928-4680(98)00035-2. [DOI] [Google Scholar]
- 4.Schalamon J, Mayr JM, Höllwarth ME. Mortality and economics in short bowel syndrome. Best Pract Res Clin Gastroenterol. 2003;17:931–42. doi: 10.1016/S1521-6918(03)00079-9. [DOI] [PubMed] [Google Scholar]
- 5.Goulet O, Joly F. [Intestinal microbiota in short bowel syndrome] Gastroenterol Clin Biol. 2010;34(Suppl 1):S37–43. doi: 10.1016/S0399-8320(10)70019-1. [DOI] [PubMed] [Google Scholar]
- 6.Cole CR, Hansen NI, Higgins RD, Ziegler TR, Stoll BJ, Eunice Kennedy Shriver NICHD Neonatal Research Network Very low birth weight preterm infants with surgical short bowel syndrome: incidence, morbidity and mortality, and growth outcomes at 18 to 22 months. Pediatrics. 2008;122:e573–82. doi: 10.1542/peds.2007-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole CR, Frem JC, Schmotzer B, Gewirtz AT, Meddings JB, Gold BD, et al. The rate of bloodstream infection is high in infants with short bowel syndrome: relationship with small bowel bacterial overgrowth, enteral feeding, and inflammatory and immune responses. J Pediatr. 2010;156:941–7, 947, e1. doi: 10.1016/j.jpeds.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaufman SS, Loseke CA, Lupo JV, Young RJ, Murray ND, Pinch LW, et al. Influence of bacterial overgrowth and intestinal inflammation on duration of parenteral nutrition in children with short bowel syndrome. J Pediatr. 1997;131:356–61. doi: 10.1016/S0022-3476(97)80058-3. [DOI] [PubMed] [Google Scholar]
- 9.Goulet O, Colomb-Jung V, Joly F. Role of the colon in short bowel syndrome and intestinal transplantation. J Pediatr Gastroenterol Nutr. 2009;48(Suppl 2):S66–71. doi: 10.1097/MPG.0b013e3181a118ef. [DOI] [PubMed] [Google Scholar]
- 10.Joly F, Mayeur C, Messing B, Lavergne-Slove A, Cazals-Hatem D, Noordine M-L, et al. Morphological adaptation with preserved proliferation/transporter content in the colon of patients with short bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2009;297:G116–23. doi: 10.1152/ajpgi.90657.2008. [DOI] [PubMed] [Google Scholar]
- 11.Healey KL, Bines JE, Thomas SL, Wilson G, Taylor RG, Sourial M, et al. Morphological and functional changes in the colon after massive small bowel resection. J Pediatr Surg. 2010;45:1581–90. doi: 10.1016/j.jpedsurg.2010.02.040. [DOI] [PubMed] [Google Scholar]
- 12.Kararli TT. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm Drug Dispos. 1995;16:351–80. doi: 10.1002/bdd.2510160502. [DOI] [PubMed] [Google Scholar]
- 13.Miller ER, Ullrey DE. The pig as a model for human nutrition. Annu Rev Nutr. 1987;7:361–82. doi: 10.1146/annurev.nu.07.070187.002045. [DOI] [PubMed] [Google Scholar]
- 14.Moughan PJ, Birtles MJ, Cranwell PD, Smith WC, Pedraza M. The piglet as a model animal for studying aspects of digestion and absorption in milk-fed human infants. World Rev Nutr Diet. 1992;67:40–113. doi: 10.1159/000419461. [DOI] [PubMed] [Google Scholar]
- 15.Buzoianu SG, Walsh MC, Rea MC, O’Sullivan O, Crispie F, Cotter PD, et al. The effect of feeding Bt MON810 maize to pigs for 110 days on intestinal microbiota. PLoS One. 2012;7:e33668. doi: 10.1371/journal.pone.0033668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–11. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 18.Nagy ÈS, Paris MCJ, Taylor RG, Fuller PJ, Sourial M, Justice F, et al. Colostrum protein concentrate enhances intestinal adaptation after massive small bowel resection in juvenile pigs. J Pediatr Gastroenterol Nutr. 2004;39:487–92. doi: 10.1097/00005176-200411000-00008. [DOI] [PubMed] [Google Scholar]
- 19.Compher C, Rubesin S, Kinosian B, Madaras J, Metz D. Noninvasive measurement of transit time in short bowel syndrome. JPEN J Parenter Enteral Nutr. 2007;31:240–5. doi: 10.1177/0148607107031003240. [DOI] [PubMed] [Google Scholar]
- 20.Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–51. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Fölsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–93. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaughlin SD, Walker AW, Churcher C, Clark SK, Tekkis PP, Johnson MW, et al. The bacteriology of pouchitis: a molecular phylogenetic analysis using 16S rRNA gene cloning and sequencing. Ann Surg. 2010;252:90–8. doi: 10.1097/SLA.0b013e3181e3dc8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–8. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 24.Joly F, Mayeur C, Bruneau A, Noordine M-L, Meylheuc T, Langella P, et al. Drastic changes in fecal and mucosa-associated microbiota in adult patients with short bowel syndrome. Biochimie. 2010;92:753–61. doi: 10.1016/j.biochi.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 25.Gerritsen J, Smidt H, Rijkers GT, de Vos WM. Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 2011;6:209–40. doi: 10.1007/s12263-011-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, Niebergall J, et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol. 2006;177:8785–95. doi: 10.4049/jimmunol.177.12.8785. [DOI] [PubMed] [Google Scholar]
- 27.Ma CY, Shi GY, Shi CS, Kao YC, Lin SW, Wu HL. Monocytic thrombomodulin triggers LPS- and gram-negative bacteria-induced inflammatory response. J Immunol. 2012;188:6328–37. doi: 10.4049/jimmunol.1102266. [DOI] [PubMed] [Google Scholar]
- 28.Lapthorne S, Macsharry J, Scully P, Nally K, Shanahan F. Differential intestinal M-cell gene expression response to gut commensals. Immunology. 2012;136:312–24. doi: 10.1111/j.1365-2567.2012.03581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562–72. doi: 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 31.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux J-J, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proceedings of the National Academy of Sciences 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalff JC, Türler A, Schwarz NT, Schraut WH, Lee KK, Tweardy DJ, et al. Intra-abdominal activation of a local inflammatory response within the human muscularis externa during laparotomy. Ann Surg. 2003;237:301–15. doi: 10.1097/01.SLA.0000055742.79045.7E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bongaerts GP, Severijnen RS, Tangerman A, Verrips A, Tolboom JJ. Bile acid deconjugation by Lactobacilli and its effects in patients with a short small bowel. J Gastroenterol. 2000;35:801–4. doi: 10.1007/s005350070016. [DOI] [PubMed] [Google Scholar]
- 34.Mayeur C, Gratadoux JJ, Bridonneau C, Chegdani F, Larroque B, Kapel N, et al. Faecal d/l lactate ratio is a metabolic signature of microbiota imbalance in patients with short bowel syndrome. PLoS One. 2013;8:e54335. doi: 10.1371/journal.pone.0054335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson KH, Blitchington RB, Greene RC. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J Clin Microbiol. 1990;28:1942–6. doi: 10.1128/jcm.28.9.1942-1946.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Craven M, Egan CE, Dowd SE, McDonough SP, Dogan B, Denkers EY, et al. Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn’s disease. PLoS One. 2012;7:e41594. doi: 10.1371/journal.pone.0041594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira-Fantini PM, Thomas SL, Wilson G, Taylor RG, Sourial M, Bines JE. Short- and long-term effects of small bowel resection: a unique histological study in a piglet model of short bowel syndrome. Histochem Cell Biol. 2011;135:195–202. doi: 10.1007/s00418-011-0778-2. [DOI] [PubMed] [Google Scholar]
- 38.Stephens AN, Pereira-Fantini PM, Wilson G, Taylor RG, Rainczuk A, Meehan KL, et al. Proteomic analysis of the intestinal adaptation response reveals altered expression of fatty acid binding proteins following massive small bowel resection. J Proteome Res. 2010;9:1437–49. doi: 10.1021/pr900976f. [DOI] [PubMed] [Google Scholar]
- 39.Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–42. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- 40.Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59:1635–42. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–96. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–86. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106:2365–70. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Rasband WS. Image J. U. S. National Institutes of Health, Bethesda, Maryland, USA, 1997-2012.
- 49.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.