Abstract
We recently investigated how post-natal microbial gut colonization is important for the development of the immune system, especially in the systemic compartments. This addendum presents additional data which in accordance with our previous findings show that early life microbial colonization is critical for a fine-tuned immune homeostasis to develop also in the intestinal environment. A generalized reduction in the expression of immune signaling related genes in the small intestine may explain previously shown increased systemic adaptive immune reactivity, if the regulatory cross-talk between intra- and extra-intestinal immune cells is immature following a neonatal germ-free period. These findings are furthermore discussed in the context of recently published results on how lack of microbial exposure in the neonatal life modifies disease expression in rodents used as models mimicking human inflammatory diseases. In particular, with a focus on how these interesting findings could be used to optimize the use of rodent models.
Keywords: microbial variation, germ-free, mucosal immunity, animal models, early life
Microbial Variation and Host Phenotype
Bacteriological examinations of the gut microbiota have revealed a substantial inter-individual and inter-vendor variation among commercially available laboratory rodent.1 This has been suggested, to some extent, to explain variability in responses to experimental treatments, when using rodents as experimental models.2,3 Increasing amounts of evidence have demonstrated an important role for the gut microbiota in modifying the immune system and as an environmental factor in the development of several immune-mediated diseases.4-7 In accordance, previous studies have further linked differences in gut microbiota profiles to the variation observed in disease parameters in immune-mediated rodent models.8,9 Microbial standardization is, thus, required to control inter-individual variation, or at least it is necessary for researchers to be able to select the most suitable experimental mice based on their microbiological status.10
In infection studies, one approach to circumvent the colonization resistance to pathogens conveyed by the gut microbiota was to utilize antibiotics. After recolonization, the resistance was transferable to germ-free (GF) mice, and thus, characterizing specific strains colonization resistance abilities makes such gut microbiotas useful in order to better control the variation in infection studies.11,12 By modifying the gut microbiota composition in rodent models, it might also become possible to manipulate host immunity in order to promote or prevent certain disease traits and produce laboratory rodents with controlled and well characterized individual phenotypes. However, only little is known about the impacts that different bacterial epitopes or compositions of microbes may have on the immune system in different periods of life. Epidemiological data suggest that microbial exposure in the neonatal life is crucial to the development of an immune system that can prevent atopic and other inflammatory diseases, and this can be influenced by early life events such as mode of delivery, use of formula to supplement breast milk and antibiotic therapy. A deviant gut microbiota with less bifidobacteria and more clostridia has for example been reported in both children born by cesarean section (CS) and those with allergies.13,14 In addition, higher levels of interleukin (IL)-13 in stimulated cord blood cells,15 lower counts of neutrophils, monocytes and natural killer (NK) cells16,17 and reduced production of IL-4r, IL-1β, IL-6 and tumor necrosis factor-α have all been associated with CS compared with vaginal delivery.18 Neonatal antibiotic treatment also confers persistent differences in immune components that may add to a higher risk of immune disorders later in life. Exacerbated allergic inflammation was for example associated with lower levels of regulatory T cells, elevated immunoglobulin E and enhanced basophil-mediated T helper type 2 cell responses as a result of antibiotics.19,20 In addition, specific strains of lactobacilli were demonstrated to suppress Staphylococcus aureus stimulated IL-4, IL-10 and interferon (IFN)-γ production in blood mononuclear cells which also in vivo showed a clear correlation in the infant gut in relation to cytokine secretion at two years of age.21 This might explain previously reported results of a lower risk of allergies in children colonized with lactobacilli whereas the presence of S. aureus seems to have the opposite effect.
To investigate the impact of a postnatal GF period on gut-related and systemic immunity later in life, we delayed the colonization of GF mice until one or three weeks of age and subsequently kept the mice in our less protected rodent facility until eight weeks of age.22 Furthermore, we looked into the possibility of permanently modifying the gut microbiota and host immunity by inoculating GF mice in early life with a microbiota from Taconic mice which was different from the microbiota in our facility. A postnatal GF period clearly affected several systemic immune cell populations, which is highly indicative of the postnatal period to be a critical time point for the acquisition of a gut microbiota, and it offers a tool to direct immune system development in this period of time. Accordingly, inoculations of the microbiota suspension at three weeks of age permanently determined the gut microbiota composition and subsequently skewed the immune system toward a pro-inflammatory state. However, higher relative amounts of NK-, NKT- and IFN-γ-producing T cells following an early GF period were only observed in the spleen and not in the mesenteric lymph node (MLN). Similarly, in a mouse model of oral sensitization to cow milk allergens, a recent study showed that late colonized ex-GF mice had a comparable cytokine production to conventional mice in MLN cells reactivated with cow’s milk β-lactoglobulin and casein whereas the reactivated splenocytes were significantly different.23
Window of Opportunity
Method improvements in the search of molecular mechanisms explaining the hygiene hypothesis using GF animal models are advancing our understanding of how a deprived early-life microbial exposure confers risk of persistent unbalanced immune responses beyond intestinal borders. A recently published paper by Olszak et al.24 demonstrated that only microbial exposure of GF mice in early life could abrogate an increased accumulation of invariant natural killer T (iNKT) cells in the lungs and related pathology of an allergic asthma model in contrast to late colonized GF mice. Systemic NKT cell accumulation was similarly detected in our ex-GF mice colonized only after one and three weeks of age, indicating that this effect might be regulated already in the very first few days of life. Interestingly, they also reported an increased iNKT accumulation and associated increase in chemokine (C-X-C motif) ligand 16 (CXCL16) gene expression in the colonic lamina propria in late colonized ex-GF mice compared with specific pathogen free (SPF) mice which implicates the existence of a close cross-talk between intra- and extra-intestinal immunity.
Another recent study investigated the impact of an early GF period on the immune system and a microarray analysis revealed a prominent decrease in especially intestinal microbial signaling toll-like receptors (TLRs) in GF and late colonized (five weeks) ex-GF mice compared with SPF mice.25 Furthermore, the list of overrepresented signal pathways was different between late colonized ex-GF and SPF mice but the list for the small intestine was the same as that of the large intestine, as previously seen in GF mice but not SPF mice.
To address the impact of gut microbes on some of the above mentioned intestinal immune-related genes in an even smaller time-window and to compare mucosal immune responses with the increased proportion of several adaptive immune cells in the spleen detected in our study, we present here some additional data generated on intestinal samples previously collected from our study in the co-housing experiment. In our extended analysis, gene expression of several immune-regulating receptors and cytokines were decreased in GF mice and mice colonized at either one or three weeks of age compared with SPF mice (Fig. 1) despite that the gut microbiota was similar in all groups at this stage as illustrated in the original paper (Fig. 2 in Hansen et al.).22 Both the pro-inflammatory cytokine IFN-γ and the regulatory cytokines IL-10 and transforming growth factor β (TGF-β) were significantly less expressed in the ex-GF and GF groups than in the SPF mice. Accordingly, mucosal T cell homeostasis is regulated by the epithelial cell-derived cytokine thymic stromal lymphopoietin (TSLP) by dendritic cell activation and signaling,26 and this cytokine was also significantly less expressed in the GF mice compared with the SPF mice; however a strong but non-significant trend (p = 0.09) existed also for the ex-GF groups. In resemblance to the study by Yamamoto et al.,25 the TLR gene expression was only high subsequent to microbial exposure during the neonatal period. Supportive of this, a low expression of TLRs was similarly seen also after early life antibiotic treatment.27 Regenerating islet-derived protein 3 γ (Reg3γ) is an antimicrobial peptide produced by intestinal paneth cells upon TLR stimulation,28 and not surprising, Reg3γ was expressed in a similar pattern as the TLRs. In contrast to Olszak et al.,24 the expression of CXCL16 was actually less in the intestine of the GF and late colonized ex-GF mice than in the SPF mice, but these data were generated on ileal and not colonic samples. In conclusion, microbial exposure already in the first week of life seems to be essential for several immune signaling related molecules and responses in the mucosal environment. This is a very interesting observation as some publications, including ours, also showed increased systemic adaptive immune reactivity in several immune cell compartments following a neonatal GF period.23,24,29 Most likely, the cross-talk between the mucosal and systemic immune cells during normal homeostatic conditions regulates and suppresses unwanted excessive immune responses outside the gut. However, lack of microbial signaling in the critical “window of opportunity” and a persistently immature intestinal immune system might disrupt this fine-tuned balanced.
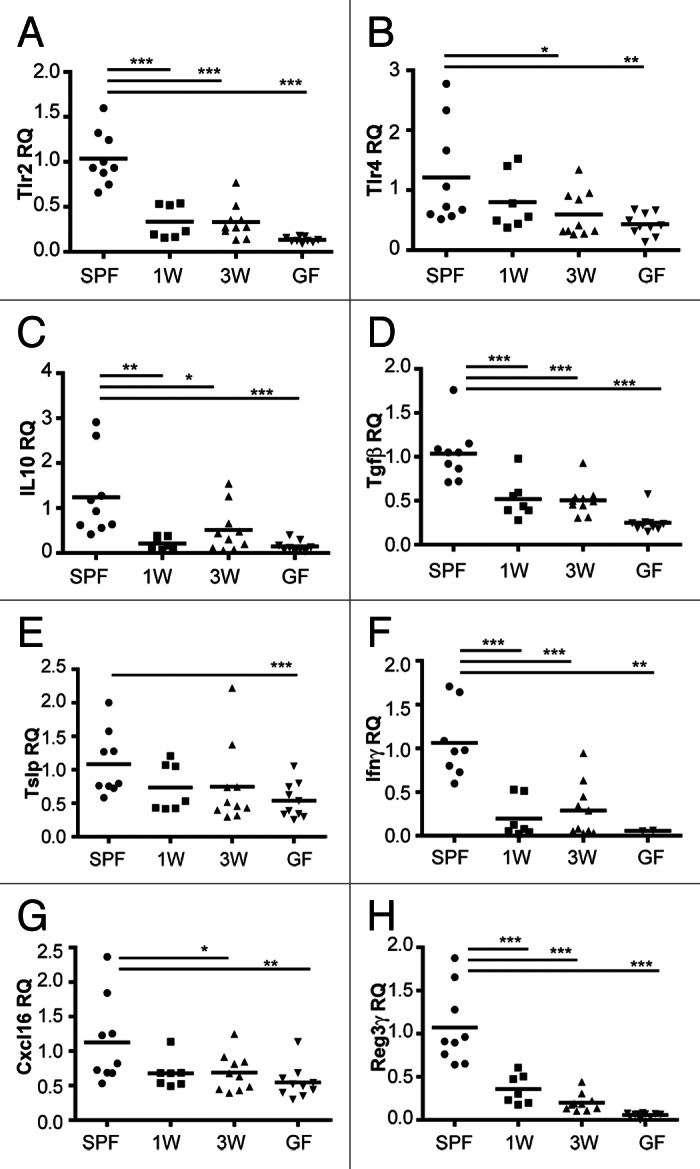
Figure 1. Microbial exposure in the first week of life is central for expression of several immune signaling- and response-related genes in the mucosal environment. Relative quantification (RQ) of gene expression of immune-regulating receptors (TLR2, TLR4) and cytokines (IL-10, TGF-β, TSLP, IFN-γ), the chemokine CXCL16 and the defensin Reg3γ were measured by quantitative real time PCR (qPCR) in ileal tissues of specific pathogen-free (SPF), germ-free (GF) and ex-GF mice colonized by co-housing with SPF mice at one (1W) or three weeks (3W) of age. TaqMan assays were used for qPCR, and fold change was calculated as 2-ΔΔCT where ΔΔCT = ΔCT(sample) - ΔCT(calibrator), where the average ΔCT of samples from SPF mice was used as calibrator. The mean is depicted. ***p < 0.001, **p < 0.01, *p < 0.05, compared with the SPF group.
Future Application
Understanding the gut microbiota and its direct or indirect interactions with the immune system will provide new ways of manipulating host biology; not only in humans with the perspective of preventing inflammatory diseases, but it also provides a rationale for manipulating intestinal bacterial communities in prospects of controlling phenotypic variation in immune-mediated animal models. In support of this, studies in mouse models of type 1 diabetes clearly show that diabetes incidence and onset time can be attenuated or accelerated by modifying bacterial and dietary antigen exposure. Already in the first week of life, oral feeding of a diabetogenic diet decreased the pro-/anti-inflammatory cytokine ratio in the gut which later in life significantly inhibited diabetes in bio-breeding rats.30 Furthermore, the optimal prevention of diabetes by oral administration of a bacterial extract was observed before six weeks of age in non-obese diabetic (NOD) mice.31 These interventions are most prominent in the pre-diabetic stage, suggesting that early life events during microbial colonization and development of the immune system are relevant in modifying autoimmune diabetes risk. In addition to this, we have previously demonstrated that vancomycin treatment of NOD mice attenuated diabetes development when they are treated only before weaning,32 and in a recent study neonatal vancomycin treatment was also shown to enhance susceptibility to murine allergic asthma.20 Manipulating the gut microbiota composition in early life can thus not only decrease but also increase disease severity; i.e., mice receiving subtherapeutic antibiotic therapy in early life accumulate fat.33 Hormone levels and expression of genes related to lipid metabolism were also increased in these mice, likely as a result of the observed shift in the composition of the gut microbiota.
In a vendors’ perspective, laboratory rodents are shipped off to the researchers already from three weeks of age with a health certificate solely focused on eradication of specific infectious agents.34 However, if manipulation of the early gut microbiota with for examples antibiotics, diets or by inoculating GF rodents with specific bacterial strains, can have lifelong impacts on immunity and phenotype of the model as discussed above, it offers an interesting opportunity to include characterizations of the gut microbiota together with the hygiene standardization performed at the vendor in order to produce different sub-colonies of rodent models fitted to its experimental purpose and with the aim to reduce their inter-individual variation. In light of these results, our vision for future production of laboratory rodents is that pups are born under gnotobiotic conditions and kept for a period of time during which a desired and durable immune response of the mice is primed by inoculating a well characterized gut microbiota before they are shipped off to the experimental units in which they are less susceptible to microbial impact on the immune system. Rodent models with a controlled and described disease expression may then be used systematically in multi-factorial studies in which the animals most fit for the given type of research are included in the experiment. By characterizing the gut microbiota of laboratory rodents in early life, researchers are able to incorporate this knowledge into their data evaluation.
Today, we do not know the proper microbiota composition best suited for a certain immune response. Differences in the initial microbial exposure guide the developing immune system in different directions,35 and more research is needed into the impact that different microbial components have on immune profiling. Thus, understanding how early life gut microbiota influence disease expression later in life is necessary to fully realize our vision. The underlying mechanisms are most likely numerous and complex.
Acknowledgments
This work was performed as a part of the research program of the UNIK: Food, Fitness and Pharma for Health and Disease. The UNIK project is supported by the Danish Ministry of Science, Technology and Innovation. It was further supported by the Center for Applied Laboratory Animal Research, CALAR.
Glossary
Abbreviations:
- NK
natural killer
- iNKT
invariant natural killer T
- IFN-γ
interferon γ
- NOD
non-obese diabetic
- CXCL16
chemokine (C-X-C motif) ligand 16
- MLN
mesenteric lymph node
- TLR
toll-like receptor
- GF
GF
- SPF
specific pathogen free
- IL
interleukin
- TGF-β
transforming growth factor β
- TSLP
thymic stromal lymphopoietin
- Reg3γ
regenerating islet-derived protein 3 γ
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/gutmicrobes/article/23999
References
- 1.Hirayama K, Endo K, Kawamura S, Mitsuoka T. Comparison of the intestinal bacteria in specific pathogen free mice from different breeders. Jikken Dobutsu. 1990;39:263–7. doi: 10.1538/expanim1978.39.2_263. [DOI] [PubMed] [Google Scholar]
- 2.Holmes E, Nicholson J. Variation in gut microbiota strongly influences individual rodent phenotypes. Toxicol Sci. 2005;87:1–2. doi: 10.1093/toxsci/kfi259. [DOI] [PubMed] [Google Scholar]
- 3.Tlaskalová-Hogenová H, Stepánková R, Hudcovic T, Tucková L, Cukrowska B, Lodinová-Zádníková R, et al. Commensal bacteria (normal microflora), mucosal immunity and chronic inflammatory and autoimmune diseases. Immunol Lett. 2004;93:97–108. doi: 10.1016/j.imlet.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101:15718–23. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–31. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernández-Sueiro JL, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–64. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–53. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–41. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- 10.Bleich A, Hansen AK. Time to include the gut microbiota in the hygienic standardisation of laboratory rodents. Comp Immunol Microbiol Infect Dis. 2012;35:81–92. doi: 10.1016/j.cimid.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Miller CP, Bohnhoff M. Changes in the mouse’s enteric microflora associated with enhanced susceptibility to salmonella infection following streptomycin treatment. J Infect Dis. 1963;113:59–66. doi: 10.1093/infdis/113.1.59. [DOI] [PubMed] [Google Scholar]
- 12.Wensinck F, Ruseler-van Embden JG. The intestinal flora of colonization-resistant mice. J Hyg (Lond) 1971;69:413–21. doi: 10.1017/S0022172400021665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129–34. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 14.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–21. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 15.Ly NP, Ruiz-Pérez B, Onderdonk AB, Tzianabos AO, Litonjua AA, Liang C, et al. Mode of delivery and cord blood cytokines: a birth cohort study. Clin Mol Allergy. 2006;4:13. doi: 10.1186/1476-7961-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grönlund MM, Nuutila J, Pelto L, Lilius EM, Isolauri E, Salminen S, et al. Mode of delivery directs the phagocyte functions of infants for the first 6 months of life. Clin Exp Immunol. 1999;116:521–6. doi: 10.1046/j.1365-2249.1999.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thilaganathan B, Meher-Homji N, Nicolaides KH. Labor: an immunologically beneficial process for the neonate. Am J Obstet Gynecol. 1994;171:1271–2. doi: 10.1016/0002-9378(94)90145-7. [DOI] [PubMed] [Google Scholar]
- 18.Malamitsi-Puchner A, Protonotariou E, Boutsikou T, Makrakis E, Sarandakou A, Creatsas G. The influence of the mode of delivery on circulating cytokine concentrations in the perinatal period. Early Hum Dev. 2005;81:387–92. doi: 10.1016/j.earlhumdev.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 19.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–46. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–7. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansson MA, Saghafian-Hedengren S, Haileselassie Y, Roos S, Troye-Blomberg M, Nilsson C, et al. Early-life gut bacteria associate with IL-4-, IL-10- and IFN-γ production at two years of age. PLoS One. 2012;7:e49315. doi: 10.1371/journal.pone.0049315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen CH, Nielsen DS, Kverka M, Zakostelska Z, Klimesova K, Hudcovic T, et al. Patterns of early gut colonization shape future immune responses of the host. PLoS One. 2012;7:e34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin S, Fischer R, Przybylski-Nicaise L, Bernard H, Corthier G, Rabot S, et al. Delayed bacterial colonization of the gut alters the host immune response to oral sensitization against cow’s milk proteins. Mol Nutr Food Res. 2012;56:1838–47. doi: 10.1002/mnfr.201200412. [DOI] [PubMed] [Google Scholar]
- 24.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto M, Yamaguchi R, Munakata K, Takashima K, Nishiyama M, Hioki K, et al. A microarray analysis of gnotobiotic mice indicating that microbial exposure during the neonatal period plays an essential role in immune system development. BMC Genomics. 2012;13:335. doi: 10.1186/1471-2164-13-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ziegler SF, Artis D. Sensing the outside world: TSLP regulates barrier immunity. Nat Immunol. 2010;11:289–93. doi: 10.1038/ni.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimmitt RA, Staley EM, Chuang G, Tanner SM, Soltau TD, Lorenz RG. Role of postnatal acquisition of the intestinal microbiome in the early development of immune function. J Pediatr Gastroenterol Nutr. 2010;51:262–73. doi: 10.1097/MPG.0b013e3181e1a114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–30. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fink LN, Metzdorff SB, Zeuthen LH, Nellemann C, Kristensen MB, Licht TR, et al. Establishment of tolerance to commensal bacteria requires a complex microbiota and is accompanied by decreased intestinal chemokine expression. Am J Physiol Gastrointest Liver Physiol. 2012;302:G55–65. doi: 10.1152/ajpgi.00428.2010. [DOI] [PubMed] [Google Scholar]
- 30.Scott FW, Rowsell P, Wang GS, Burghardt K, Kolb H, Flohé S. Oral exposure to diabetes-promoting food or immunomodulators in neonates alters gut cytokines and diabetes. Diabetes. 2002;51:73–8. doi: 10.2337/diabetes.51.1.73. [DOI] [PubMed] [Google Scholar]
- 31.Alyanakian MA, Grela F, Aumeunier A, Chiavaroli C, Gouarin C, Bardel E, et al. Transforming growth factor-beta and natural killer T-cells are involved in the protective effect of a bacterial extract on type 1 diabetes. Diabetes. 2006;55:179–85. doi: 10.2337/diabetes.55.01.06.db05-0189. [DOI] [PubMed] [Google Scholar]
- 32.Hansen CH, Krych L, Nielsen DS, Vogensen FK, Hansen LH, Sørensen SJ, et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55:2285–94. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 33.Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–6. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nicklas W, Deeny A, Diercks P, Gobbi A, Illgen-Wilcke B, Seidelin M. FELASA guidelines for the accreditation of health monitoring programs and testing laboratories involved in health monitoring. Lab Anim (NY) 2010;39:43–8. doi: 10.1038/laban0210-43. [DOI] [PubMed] [Google Scholar]
- 35.Karlsson H, Hessle C, Rudin A. Innate immune responses of human neonatal cells to bacteria from the normal gastrointestinal flora. Infect Immun. 2002;70:6688–96. doi: 10.1128/IAI.70.12.6688-6696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


