Abstract
Autophagy is an evolutionarily conserved cellular recycling mechanism that occurs at a basal level in all cells. It can be further induced by various stimuli including starvation, hypoxia, and treatment with cytokines such as IFNG/IFNγ and TGFB/TGFβ. Type I IFNs are proteins that induce an antiviral state in cells. They also have antiproliferative, proapoptotic and immunomodulatory activities. We investigated whether type I IFN can also induce autophagy in multiple human cell lines. We found that treatment with IFNA2c/IFNα2c and IFNB/IFNβ induces autophagy by 24 h in Daudi B cells, as indicated by an increase of autophagy markers MAP1LC3-II, ATG12–ATG5 complexes, and a decrease of SQSTM1 expression. An increase of MAP1LC3-II was also detected 48 h post-IFNA2c treatment in HeLa S3, MDA-MB-231, T98G and A549 cell lines. The presence of autophagosomes in selected cell lines exposed to type I IFN was confirmed by electron microscopy analysis. Increased expression of autophagy markers correlated with inhibition of MTORC1 in Daudi cells, as well as inhibition of cancer cell proliferation and changes in cell cycle progression. Concomitant blockade of either MTOR or PI3K-AKT signaling in Daudi and T98G cells treated with IFNA2c increased the level of MAP1LC3-II, indicating that the PI3K-AKT-MTORC1 signaling pathway may modulate IFN-induced autophagy in these cells. Taken together, our findings demonstrated a novel function of type I IFN as an inducer of autophagy in multiple cell lines.
Keywords: autophagy, human cancer cells, type I interferon, MTORC1, signal transduction, AKT, PI3K
Introduction
Type I interferons (IFNs) are a pleiotropic family of cytokines with known antiviral, antiproliferative and immunomodulatory functions.1,2 These activities have been demonstrated on different cell lines.3,4 Human Burkitt lymphoma Daudi cells are highly sensitive to antiproliferative and antiviral function of IFN3-6 and exhibit a prolonged biological response to IFN mediated by the continuous transcription of ISGs.7
Although most IFN activities are due to activation of JAK-STAT signaling pathways, there is increasing evidence that MAP kinase signaling pathways play an important role in activation of IFN-regulated genes.8,9 Additionally, several studies demonstrate that type I and type II IFNs can modulate the PI3K-AKT-MTOR signaling pathway.10,11
Autophagy is an evolutionarily conserved cellular process that mediates degradation of intracellular material. There are three types of autophagy: chaperone-mediated autophagy, microautophagy and macroautophagy, the latter in the context of this paper referred to as autophagy.12 During autophagy, cytoplasmic constituents including protein aggregates and organelles, are first sequestered in a double-membraned phagophore. Once the double membrane fuses and the autophagosome matures, it is transported along tubulin tracks to a perinuclear location, fusing with a lysosome. This generates an autolysosomal structure capable of degrading its contents.12-14 A low basal level of autophagy occurs in all cells, playing a role in maintenance of homeostasis, defense against intracellular pathogens, and MHC class II antigen presentation.13,14 Autophagy increases under various stressful conditions including starvation, hypoxia, genomic and ER stress.12 Thus, autophagy is predominantly a cell survival mechanism.15 Autophagy can be monitored by detection of relevant autophagy markers such as MAP1LC3 processing. MAP1LC3 is a cytosolic protein which is cleaved by ATG4 and then conjugated to phosphatidylethanolamine, creating an identifiable form (MAP1LC3-II) which closely correlates with formation of autophagosomes.16 Autophagy is also marked by increased expression of ATG5 and ATG12, which are covalently linked and essential for autophagosome formation,12,17 and SQSTM1 which colocalizes with MAP1LC3 and plays a role in degradation of protein aggregates by the autophagosome.18 One critical regulator of autophagy is MTORC1, which links nutrient abundance to cell growth and proliferation.19,20 The activity of MTORC1 can be regulated by different signaling pathways, including the PI3K-AKT axis. This pathway can be modulated by type I and type II IFNs.10,11 Type II IFN induces autophagy in macrophages, as well as in human cervical and mammary carcinoma cells.21-23 To date, there have been no published studies evaluating type I IFN’s ability to induce autophagy.
Here, we report for the first time that human type I IFNs induce autophagy in Daudi lymphoma cells as detected by changes in the expression of critical autophagy markers (MAP1LC3-II, ATG12–ATG5, SQSTM1), after 24 h treatment. We also detected increased levels of MAP1LC3-II 48 h post IFNA2c treatment in HeLa S3, MDA-MB-231, T98G and A549 cell lines. Autophagy markers were detected by western blot, and morphological changes associated with autophagy were confirmed using TEM. Moreover, we show that IFN-induced autophagy correlated with inhibition of MTORC1 activity in Daudi cells. Indeed, interfering with the PI3K-AKT-MTORC1 signaling axis using pharmacological inhibitors and siRNA further enhanced IFN-induced autophagy. Importantly, IFN-induced autophagy also correlated with cell cycle perturbation and inhibition of cellular proliferation in Daudi, HeLaS3, MDA-MB-231, T98G and A549. Thus, we provided evidence for a previously unrecognized function of type I IFN in autophagy induction.
Given the importance of autophagy in cancer as well as its role in both the pathogenesis and response to infectious and neurodegenerative diseases,24 our findings about the ability of type I IFN to regulate autophagy are of substantial clinical importance.
Results
Autophagy markers are detected in multiple human cell types after treatment with type I IFNs
To determine whether type I IFNs induce autophagy, Daudi cells were treated with 3.6 ng/mL (approximately 700 IU/mL) IFNA2c or IFNB for 6, 24 and 48 h. We observed increased levels of MAP1LC3-II in IFN-treated cells compared with mock-treated cells at 24 and 48 h (Fig. 1, ratios of MAP1LC3 were calculated as division of the ratio of induced MAP1LC3-I to induced MAP1LC3-II by the ratio of basal MAP1LC3-I to basal MAP1LC3-II. Numbers are shown below MAP1LC3 lanes). Furthermore, western blot analyses revealed increased formation of ATG12–ATG5 complexes at 24 and 48h (Fig. 1), and a decrease of SQSTM1 at 48 h (Fig. 1, lanes 9, 10) in IFN-treated cells, which is consistent with the induction of autophagy. Changes in the levels of autophagy markers were even more pronounced at 48 h, correlating with inhibition of proliferation (Table 1), and the accumulation of Daudi cells in the G1 phase of the cell cycle (Table 1). Cleavage of MAP1LC3-I to MAP1LC3-II, and an increase in formation of ATG12–ATG5 complexes, as well as the decrease of SQSTM1 were blocked in IFN-treated Daudi cells that were simultaneously treated with anti-IFNAR2 monoclonal antibody (A10), demonstrating that induction of autophagy is due to treatment with IFN (Fig. 1). In addition to time course experiments, treatment of Daudi cells for 48 h with different doses of IFNA2c increased the levels of autophagy markers in a concentration-dependent manner (Fig. S1).

Figure 1. Autophagy markers in IFN-treated Daudi cells. Lane: (1) Molecular weight marker; (2) negative control (NC), untreated cells, 6 h; (3) IFNA2c (3.6 ng/mL), 6 h; (4) IFNB (3.6 ng/mL), 6 h; (5) NC 24 h; (6) IFNA2c (3.6 ng/mL), 24 h; (7) IFNB (3.6 ng/mL), 24 h; (8) NC 48 h; (9) IFNA2c (3.6 ng/mL) 48 h; (10) IFNB (3.6 ng/mL), 48 h; (11) IFNA2c (3.6 ng/mL) 48 h + anti-IFNAR2 – mAb A10; (12) IFNB (3.6 ng/mL) 48 h + anti-IFNAR2 – mAb A10. Data are representative of three individual experiments. Ratios of MAP1LC3 were calculated as the division of the ratio of induced MAP1LC3-I to induced MAP1LC3-II by the ratio of basal MAP1LC3-I to basal MAP1LC3-II, and the numbers are shown below the MAP1LC3 lanes.
Table 1. Effects of IFNA2c on cell cycle, induction of autophagy and inhibition of proliferation.
| Cell line | Cell cycle analysis (% of total cells) | Autophagy activity factor (AAF) (mean ± SD) | Growth inhibition (%) (mean ± SD) |
||||
|---|---|---|---|---|---|---|---|
| |
|
Apoptotic |
G0/G1 |
S |
G2/M |
|
|
| Group 1 |
Daudi |
0.1 ± 0.1 |
42.6 ± 2.5 |
20.2 ± 1.0 |
24.8 ± 2.8 |
27.8 ± 8.8 |
66 ± 15** |
| Daudi + IFNA2c |
0.6 ± 1.0 |
53.9 ± 3.8 |
15.3 ± 3.0 |
14.0 ± 6.4 |
|||
| T98G |
5.5 ± 1.9 |
50.2 ± 7.7 |
22.4 ± 3.0 |
14.3 ± 1.4 |
27.7 ± 9.5 |
30 ± 8** |
|
| T98G + IFNA2c |
5.7 ± 5.3 |
61.9 ± 14.8 |
14.3 ± 6.0 |
6.9 ± 1.6 |
|||
| MDA-MB-231 |
1.3 ± 0.3 |
46.0 ± 5.5 |
19.8 ± 2.3 |
22.9 ± 2.6 |
9.5 ± 2.1 |
23 ± 5*** |
|
| MDA-MB-231 + IFNA2c |
1.1 ± 0.6 |
62.8 ± 12.7 |
17.5 ± 3.0 |
18.2 ± 1.8 |
|||
| Group 2 |
HeLa S3 |
1.6 ± 1.4 |
53.3 ± 6.2 |
20.2 ± 1.7 |
16.9 ± 1.6 |
27.6 ± 3.6 |
54 ± 11*** |
| HeLa S3 + IFNA2c |
0.9 ± 1.1 |
37.7 ± 7.6 |
33.7 ± 6.6 |
20.4 ± 4.9 |
|||
| BJAB |
0.3 ± 0.4 |
51.2 ± 10.8 |
24.6 ± 6.8 |
17.8 ± 10.3 |
5.3 ± 1.4 |
42 ± 5** |
|
| BJAB + IFNA2c |
0. Nine ± 1.4 |
30.8 ± 8.2 |
42.5 ± 12.6 |
13.8 ± 2.0 |
|||
| A549 |
0.9 ± 0.4 |
56.7 ± 3.8 |
20.4 ± 1.0 |
16.6 ± 9.4 |
10.5 ± 0.7 |
24 ± 7* |
|
| A549 + IFNA2c |
0.4 ± 0.2 |
38.3 ± 5.3 |
26.6 ± 6.5 |
17.8 ± 2.6 |
|||
| Group 3 | U937 |
2.2 ± 2.0 |
53.6 ± 3.5 |
23.9 ± 1.6 |
17.9 ± 2.1 |
8.7 ± 5.3 | 8 ± 6 ns |
| U937 + IFNA2c | 2.1 ± 1.4 | 49.1 ± 9.2 | 28.1 ± 1.6 | 17.7 ± 4.2 | |||
Different cell lines were incubated in the presence or absence of IFNA2c (3.6 ng/mL) for 48 h. To study changes in cell cycle, cells were stained with propidium iodide (PI), and signals were measured using a Cellometer image-based cytometer. To measure autophagic signals in living cells, Cellometer image-based cytometry in combination with Cyto-ID Green dye was used. To determine inhibition of proliferation, cell numbers were counted using Cellometer image-based cytometer in combination with Trypan Blue staining. Phase populations were reported as Apoptotic, G0/G1-, S- or G2/M-phase. Data shown are averages of three individual experiments, ± SD of experimental replicates. For inhibition of proliferation, we determined two-tailed p values by using a paired t-test that compared each treatment group relative to untreated control. Statistical significance was reported as follows: *p < 0.05 (significant); **p < 0.01 (very significant); ***p < 0.001 (extremely significant); ns: p > 0.05 (not significant).
To confirm that the observed effect is due to treatment with IFN and not the presence of secondary intermediates, Daudi cells were cultured in media from mock-treated Daudi cells or cells treated with IFNA2c for 24 h in the presence or absence of anti-IFNAR2 monoclonal antibody A10. An increase of MAP1LC3-II was only observed in cells cultured in media that contained IFNA2c (Fig. 2A), and this increase was blocked by anti-IFNAR2-mAb A10. The ability of IFNA2c to induce autophagy was also tested in STAT2-defective Daudi cells, a mutant cell line that is highly resistant to antiviral and antiproliferative activities of type I IFN.25 When STAT2-defective Daudi cells were treated with IFNA2c, no increase of MAP1LC3-II and a decrease in SQSTM1 was observed (Fig. 2B). Sensitivity of this cell line to type I IFN was previously demonstrated by detection of p-STAT1,25 and our results support that observation (Fig. 2B). Thus, our findings demonstrate that autophagy is a direct result of IFN signaling (Fig. 2B), and STAT2 may play a role in IFN-induced autophagy.
Figure 2. IFNA2c induced autophagy in Daudi cells, but not in STAT2-deficient Daudi. (A) Direct effect of IFNA2c on induction of autophagy. Daudi cells were cultured for 24 h in supernatants from either mock-treated Daudi cells or cells treated with 0.36 ng/mL of IFNA2c for 24 h in the presence or absence of anti-IFNAR2-mAb A10. Lanes: (1) molecular weight marker; (2) negative control, cells plus supernatant from mock-treated cells; (3) negative control, cells plus supernatant from mock-treated cells and anti-IFNAR2 – mAb A10; (4) cells plus supernatant from IFNA2c-treated cells; (5) cells plus supernatant from IFNA2c-treated cells and anti-IFNAR2 – mAb. Data are representative of two individual experiments. Ratios of MAP1LC3 were calculated as the division of the ratio of induced MAP1LC3-I to induced MAP1LC3-II by the ratio of basal MAP1LC3-I to basal MAP1LC3-II, and the numbers are shown below MAP1LC3 lanes. (B) Detection of MAP1LC3-I and MAP1LC3-II, SQSTM1, p-STAT1, p-STAT2, p-RPS6 after 48 h treatment of STAT2-defective mutant Daudi cells with IFNA2c. STAT2-defective mutant Daudi cells were incubated with or without IFNA2c for 48 h. Lanes: (1) molecular weight marker; (2) negative control, untreated cells; (3) IFNA2c (3.6 ng/mL). Data are representative of two individual experiments. Ratios of MAP1LC3 were calculated as the division of the ratio of induced MAP1LC3-I to induced MAP1LC3-II by the ratio of basal MAP1LC3-I to basal MAP1LC3-II, and the numbers are shown below the MAP1LC3 lanes.
We next asked whether type I IFN-induced autophagy was exclusive to Daudi cells, or if it can be induced in other cell lines. To answer this question, we screened several different cancer cell lines—BJAB, U937, MDA-MB-231, T98G, HeLa S3 and A549—for their ability to induce autophagy upon type I IFN treatment. We observed an increase of MAP1LC3-II in MDA-MB-231, T98G, HeLa S3 and A549 cells 48 h post IFNA2c treatment (Fig. 3). However, the expression of SQSTM1, ATG12–ATG5 complexes and ATG5 remained unchanged, with the exception of A549, where we observed a decrease in the level of ATG5. Interestingly, we observed differences in the level of SQSTM1 among different cell types. These changes were independent of IFN treatment (Fig. 3). A high level of SQSTM1 had been described in several tumor types and can be associated with dysfunction of Ras.26 Further studies will be required to explain the higher levels of SQSTM1 in various cancer cell types.
Figure 3. Detection of MAP1LC3-I and MAP1LC3-II, SQSTM1, ATG5 and p-RPS6 after 48 h treatment of different cell lines with IFNA2c. Different cell lines were incubated with or without IFNA2c for 48 h. Lanes: (1) molecular weight marker; (2) untreated BJAB; (3) BJAB plus IFNA2c (3.6 ng/mL); (4) untreated U937; (5) U937 plus IFNA2c (3.6 ng/mL); (6) untreated MDA-MB-231; (7) MDA-MB-231 plus IFNA2c (3.6 ng/mL); (8) untreated T98G; (9) T98G plus IFNA2c (3.6 ng/mL); (10) untreated HeLa S3; (11) HeLa S3 plus IFNA2c (3.6 ng/mL); (12) untreated A549; (13) A549 plus IFNA2c (3.6 ng/mL). Data are representative of two individual experiments. Ratios of MAP1LC3 were calculated as the division of the ratio of induced MAP1LC3-I to induced MAP1LC3-II by the ratio of basal MAP1LC3-I to basal MAP1LC3-II, and the numbers are shown below the MAP1LC3 lanes.
To confirm our western blot results, we labeled cells using a Cyto-ID Green dye to preferentially label autophagic vacuoles, and quantified this signal using a Cellometer image-based cytometer. Results showed an increase in autophagy activity factor (AAF) in all tested cell lines in response to IFNA2c, however the highest AAF values were calculated for Daudi, T98G and HeLa S3 (Table 1). Sensitivity of cell lines to type I IFN was demonstrated based on detection of p-STAT1 and p-STAT2 by western blot (WB). Although the levels of detected signal vary between cell lines, all tested cell lines showed sensitivity to IFNA2c. In U937 cells, STAT2 was below the level of detection (Fig. S2).
Demonstration of type I IFN induced autophagy by TEM
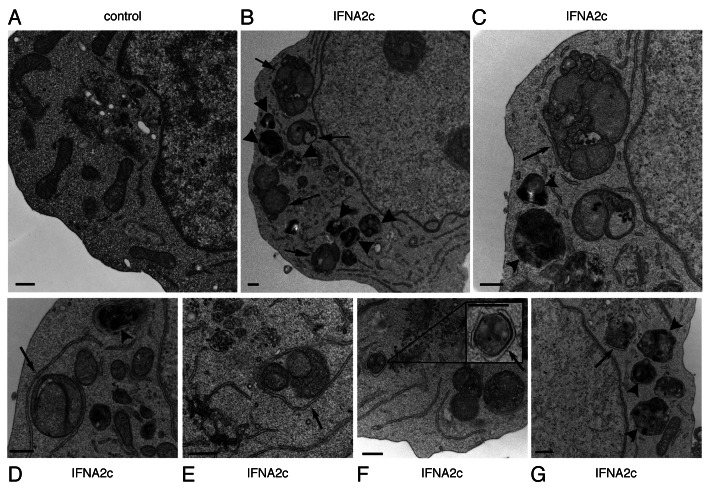
To confirm that our observations of MAP1LC3 processing and ATG12–ATG5 induction were truly indicative of autophagy, Daudi cells treated with 3.6 ng/mL of IFNA2c or IFNB were analyzed by TEM for evidence of autophagy (Figs. 4 and 5). Following treatment, IFNA2c-treated Daudi cells exhibited morphological changes consistent with autophagy as previously reported.27-29 Representing early stages of autophagy, double-membrane structures were frequently observed in association with several degrading organelles, including mitochondria as visualized in Figures 4 B–G (arrows) compared with typical untreated controls (Fig. 4A). Mature autophagosomes containing electron dense regions were also observed (Fig. 4C–E, arrowheads) suggesting that these autophagosomes were actively degrading their contents.
Figure 4. Transmission electron micrographs of IFNA2c-induced autophagy in Daudi cells. Cells were incubated with 3.6 ng/mL IFNA2c. (A) represents typical untreated control cells and (B–G) represent cell populations 48 h after treatment. Arrows and inset indicate regions with tight association between double-membrane structures and mitochondria. Arrowheads indicate mature autophagosomes containing highly degraded contents. Scale bars: 500 nm.
Figure 5. Transmission electron micrographs of IFNB-treated Daudi cells presenting morphological evidence of autophagy. All treated cells were incubated with 3.6 ng/mL IFNB. (A) represents typical untreated control cells and panels (B–G) represent cell populations 48 h after treatment. Arrows and inset indicate regions with tight association between double-membrane structures and mitochondria. Scale bars: 500 nm.
Likewise, IFNB-treated Daudi cells exhibited similar structures (Fig. 5B–F) with tight association between SER and mitochondria (arrows) and evidence of mature autophagosomes compared with typical untreated control specimens (Fig. 5A). Interestingly, as shown in the insets in Figure 4F and Figure 5B, electron dense zones were often apparent in regions of tight contact between the SER and mitochondria, perhaps due to membrane fusion.
Similar autophagic structures were observed in Daudi cells treated with bafilomycin A1 after 48 h of incubation with IFNA2c under starvation conditions (data not shown). Finally we confirmed the presence of similar autophagy features more frequently in MDA-MB-231, T98G and HeLa S3 cells treated with 3.6 ng/mL of IFNA2c for 48 h (Fig. 6). Together, these results confirm that type I IFN induces autophagy in a variety of cell lines.
Figure 6. Transmission electron micrographs of IFNA2c-treated adherent cells. Transmission electron micrographs of IFNA2c-treated adherent cells (MDA-MB-231, T98G and HeLa S3) presenting morphological evidence of autophagy demonstrated by tight association of double-membrane structure with degrading organelles (arrows), (A–C) respectively, as compared with the corresponding untreated controls, (D–F), where typical ER is shown (asterisks) adjacent to mitochondria. All treated cells were incubated with 36 ng/mL IFNA2c for 48 h. Scale bars: 500 nm.
Type I IFN induces autophagic flux
To address whether IFN-induced autophagy represents true autophagic flux (the complete process of autophagy that includes the delivery of cytosolic materials to lysosomes30), or merely an accumulation of nonfunctional autophagic machinery, we treated Daudi cells with bafilomycin A1 (V-ATPase inhibitor), which prevents maturation of autophagic vacuoles31,32 under nutrient-rich or starvation conditions. Treatment with bafilomycin A1 inhibited degradation of MAP1LC3-II associated with autolysosome activity, resulting in accumulation of MAP1LC3-II under both nutrient-rich and starvation conditions compared with untreated control (Fig. 7). Importantly, the changes in MAP1LC3-II levels were greater in IFN-treated samples compared with treatment with bafilomycin A1 alone, suggesting that IFNA2c induces autophagic flux. Many cancer cell lines are defective in autophagic flux.33 We therefore included starvation conditions in these experiments to show that Daudi cells are autophagy competent.

Figure 7. Detection of autophagy flux in Daudi cells. Daudi cells were incubated with 3.6 ng/mL IFNA2c for 48 h followed by a 4 h incubation with 0.161 µM bafilomycin A1 under nutrient-rich (RPMI 1640 media supplemented with 10% FBS) or starvation (Hank’s balanced salt solution) conditions. Lanes: (1) molecular weight marker; (2) untreated cells; (3) DMSO; (4) bafilomycin A1; (5) IFNA2c (3.6 ng/ml); (6) IFNA2c (3.6 ng/ml) plus DMSO; (7) IFNA2c (3.6 ng/ml) plus bafilomycin A1. Data are representative of three individual experiments. Ratios of MAP1LC3 were calculated as the division of the ratio of induced MAP1LC3-I to induced MAP1LC3-II by the ratio of basal MAP1LC3-I to basal MAP1LC3-II, and the numbers are shown below the MAP1LC3 lanes.
Effects of IFNA2c on cell cycle, and inhibition of cells proliferation
We next explored if IFN-induced autophagy is connected with inhibition of cell proliferation. The antigrowth effect of IFNA2c was evaluated using a Trypan Blue viability assay, and propidium iodide (PI) cell cycle analysis. Results of PI cell cycle analysis indicated changes in cell cycle phase fractions for most of the tested cell lines (Table 1). Based on the effect of IFNA2c treatment on the cell cycle, tested cell lines can be divided into three groups: (1) Daudi, T98G and MDA-MB-231 (increase in G1 fraction, decrease in S and/or G2+M fractions); (2) HeLa S3, B-JAB, and A549 (decrease in G1 fraction, increase in S and/or G2+M fractions); (3) U937 (no effect on cell cycle). The viability of all cell lines was greater than 90% (data not show). Inhibition of proliferation greater than 30% was observed in Daudi, B-JAB, HeLa S3 and T98G cell lines (Table 1). No increase in apoptosis between treated and untreated samples was detected in cell cycle analysis for any of the tested cell lines. Interestingly, no inhibition of cell proliferation was observed in U937. In this cell line, STAT2 was below detection level in WB, and we did not detect an increase of MAP1LC3-II (Fig. 3; Fig. S2). Thus, these results support our evidence that STAT2 may play a role in type I IFN-induced autophagy. In conclusion, our results showed that type I IFN has the ability to induce autophagy in a number of cancer cell lines and in general is correlated with cell cycle arrest.
Induction of autophagy and correlation with inhibition of MTORC1 activity following IFN treatment
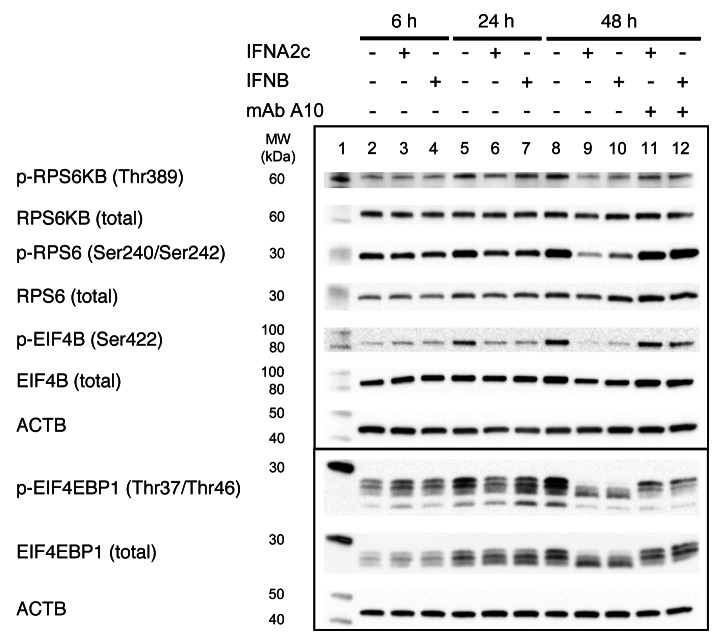
Previous studies have shown that global protein synthesis is downregulated in response to IFN treatment.34 One of the master regulators of protein synthesis and negative regulator of autophagy is MTORC1.19 The activity of MTORC1 can be monitored by phosphorylation changes in downstream target proteins like RPS6KB and EIF4EBP1.35 To examine the effect of type I IFN on MTORC1 we treated Daudi cells with 3.6 ng/mL of either IFNA2c or IFNB for 6, 24 and 48 h. We found that treatment of Daudi cells with type I IFNs decreased phosphorylation of RPS6KB (Thr389) and EIF4EBP1 (Thr37/Thr46), indicating that IFNs attenuate MTORC1 signaling (Fig. 8). To further examine activity of RPS6KB, we studied phosphorylation changes of two RPS6KB-dependent proteins: RPS6 and EIF4B, which both play a role in translational regulation. We observed decreased phosphorylation of RPS6 (Ser240/Ser242) and EIF4B (Ser422) proteins at 24, and 48 h post-IFN-treatment (Fig. 8). Neutralizing anti-IFNAR2 mAb A10 reversed the phosphorylation profile of all tested proteins (Fig. 8). Results of the concentration-dependent experiments confirmed that treatment of Daudi cells for 48 h with IFNA2c decreased MTORC1 activity and induced autophagy (Fig. S1). No changes in phospohorylation of RPS6 were detected in the STAT2-deficient Daudi cell line.
Figure 8. Detection of MTORC1 activity in Daudi cells after IFN treatment by western blot. Lanes: (1) molecular weight marker; (2) negative control (NC), untreated cells, 6 h; (3) IFNA2c (3.6 ng/mL), 6 h; (4) IFNB (3.6 ng/mL), 6 h; (5) NC 24 h; 6, IFNA2c (3.6 ng/mL), 24 h; (7) IFNB (3.6 ng/mL), 24 h; (8) NC 48 h; (9) IFNA2c (3.6 ng/mL) 48 h; (10) IFNB (3.6 ng/mL), 48 h; (11) IFNA2c (3.6 ng/mL) 48 h plus anti-IFNAR2 – mAb A10; (12) IFNB (3.6 ng/mL) 48 h plus anti-IFNAR2 – mAb A10. Data are representative of three individual experiments.
Direct inhibition of MTORC1 activity can lead to induction of autophagy. To further elucidate the role of MTORC1 in IFNA2c-induced autophagy, we treated Daudi cells with a nonsaturating dose of rapamycin, an MTORC1 inhibitor / autophagy inducer. We observed increased MAP1LC3-II (Fig. 9) in Daudi cells treated for 48 h with a combination of IFNA2c and rapamycin in comparison to treatment with inhibitor or IFN only (Fig. 9, lanes 5 and 6). Efficiency of MTORC1 inhibition by rapamycin was monitored by measuring phosphorylation of the downstream effector protein RPS6. Together, these results suggest that greater inhibition of MTORC1 activity through rapamycin treatment further enhances IFN-induced autophagy in Daudi cells.
Figure 9. Detection of MAP1LC3-I and MAP1LC3-II, p-RPS6 and p-MAPK1/3 after 48 h treatment of Daudi cells with chemical inhibitors and IFNA2c. Lanes: (1) molecular weight marker; (2) negative control, untreated cells; (3) IFNA2c (0.036 ng/mL); (4) rapamycin (0.27 nM); (5) IFNA2c (0.036 ng/mL) plus rapamycin (0.27 nM); (6) LY294002 (10 µM); (7) IFNA2c (0.036 ng/mL) plus LY294002 (10 µM); (8) PD98059 (46.7 µM); (9) IFNA2c (0.036 ng/mL) plus PD98059 (46.7 µM). Data are representative of three individual experiments. Ratios of MAP1LC3 were calculated as the division of the ratio of induced MAP1LC3-I to induced MAP1LC3-II by the ratio of basal MAP1LC3-I to basal MAP1LC3-II, and the numbers are shown below the MAP1LC3 lanes.
In addition to Daudi cells, we examined phosphorylation changes of RPS6 in HeLa S3, MDA-MB-231, T98G, A549, U937 and BJAB cells. We did not observe decreased RPS6 phosphorylation in those cell lines 48 h post-IFN treatment when compared with untreated cells (Fig. 3). To further explore the role of MTORC1, we performed siRNA knockdown experiments. T98G cells transfected with MTOR siRNA showed significantly more IFNA2c-induced MAP1LC3-II generation compared with cells transfected with a nonspecific siRNA (Fig. 10A). Efficiency of MTOR knockdown was monitored by measuring phosphorylation of downstream effector protein RPS6. Treatment of MTOR siRNA-transfected cells with IFNA2c had an additive effect on growth inhibition when compared with either as a single treatment, supporting a role of MTOR in cell proliferation (Table 2). In addition, combinatory treatment of T98G cells with nonsaturating doses of rapamycin or LY294002 in addition to IFN increased the level of MAP1LC3-II in comparison to treatment with IFN alone (Fig. 10B). Thus, these results suggest that MTOR and PI3K inactivation enhances IFN-induced autophagy.

Figure 10. Role of the MTORC1 activity in IFN-induced autophagy. (A) siRNA-mediated RNA silencing of MTOR. T98G cells were transfected for 48 h with 100 nM SignalSilenceRMTOR siRNA or SignalSilenceR control siRNA follow by IFNA2c (3.6 ng/mL) treatment for 48 h. The effect of MTOR-siRNA on MAP1LC3-II level was examined by western blot. Lanes: (1) molecular weight marker; (2) negative siRNA control; (3) negative siRNA control plus IFNA2c (3.6 ng/mL); (4) specific siRNA for MTOR; (5) specific siRNA for MTOR plus IFNA2c (3.6 ng/mL). Data are representative of three individual experiments. Ratios of MAP1LC3 were calculated as the division of the ratio of induced MAP1LC3-I to induced MAP1LC3-II by the ratio of basal MAP1LC3-I to basal MAP1LC3-II, and the numbers are shown below the MAP1LC3 lanes. (B) Detection of MAP1LC3-I, MAP1LC3-II, and p-RPS6 upon treatment with inhibitors rapamycin, LY294002 and IFNA2c. Lanes: (1) molecular weight marker; (2) negative control, untreated cells; (3) IFNA2c (3.6 ng/mL); (4) rapamycin (2.7 nM); (5) IFNA2c (3.6 ng/mL) + rapamycin (2.7 nM); (6) LY294002 (10 µM); (7) IFNA2c (3.6 ng/mL) + LY294002 (10 µM) Data are representative of two individual experiments. Ratios of MAP1LC3 were calculated as the division of the ratio of induced MAP1LC3-I to induced MAP1LC3-II by the ratio of basal MAP1LC3-I to basal MAP1LC3-II, and the numbers are shown below the MAP1LC3 lanes.
Table 2.MTOR siRNA and IFNA2c inhibit cell growth.
| Growth Inhibition (%; mean ± SD) | |
|---|---|
| Negative control |
0 |
| IFNA2c |
17 ± 7* |
|
MTOR siRNA |
13 ± 9 ns |
| MTOR siRNA + IFNA2c | 30 ± 11* |
T98G cells were transfected for 48 h with 100 nM SignalSilenceR MTOR siRNA or SignalSilenceR control siRNA followed by IFNA2c (3.6 ng/mL) treatment for 48 h. The effect of MTOR siRNA, IFN, or their combination on growth inhibition was examined using Cellometer in combination with Trypan Blue staining. Results shown are average of three individual experiments, ± SD of experimental replicates. We determined two-tailed p values by using a paired t-test that compared each treatment group relative to untreated control. Statistical significance was reported as follows: *p < 0.05 (significant); ns: p > 0.05 (not significant).
Evaluation of upstream regulators of MTORC1 activity
To determine the mechanism by which IFNA2c modulates MTORC1 activity in Daudi cells, we investigated the phosphorylation profile of three families of MAP kinases upstream of MTORC1: MAPK1/3, MAPK14 and MAPK8/9. At early time points (15 min, 1 and 4 h post IFNA2c treatment), we only observed an increase in phosphorylation of MAPK1/3 at 4 h. This phosphorylation was not accompanied by changes in the level of MAP1LC3-II (data not shown). Twenty-four h treatment with IFNA2c resulted in a significant decrease in phosphorylation of MAPK1/3, and a minimal decrease in the level of MAPK14 phosphorylation in comparison with untreated cells (Fig. 11A). Phosphorylation of MAPK8/9 was unobserved in untreated or IFNA2c-treated Daudi cells (data not shown). Similar results were observed at 48 h (data not shown). Because significant changes were observed in the phosphorylation profile of MAPK1/3, we further investigated the significance of in MAPK1/3 phosphorylation in IFNA2c-induced autophagy by culturing Daudi cells for 48 h in the presence of IFNA2c with or without a known MAPK1/3 inhibitor, PD98059. PD98059 inhibited phosphorylation of MAPK1/3 at 48 h in IFN-treated and control cells. Interestingly, combinatory treatment of PD98059 and IFNA2c did not increase cleavage of MAP1LC3-I to MAP1LC3-II in comparison to single treatments with inhibitor or IFN only (Fig. 9, lanes 8 and 9). These results suggest that downregulation of MAPK1/3 activity did not sensitize Daudi cells to IFN-induced autophagy.
Figure 11. Dose-dependent effects of IFNA2c on (A) MAP and (B) AKT kinases. Daudi cells were incubated with the designated amounts of IFN for 48 h. Lanes: (1) molecular weight marker; (2) untreated cells; (3) IFNA2c (3.6 ng/mL); (4) IFNA2c (0.36 ng/mL); (5) IFNA2c (0.036 ng/mL); (6) IFNA2c (0.0036 ng/mL); (7) negative control for AKT (cell extract from serum starved Jurkat cells, followed by LY294002-treatment); (8) positive control for AKT (cell extract from serum starved Jurkat cells, followed by Calyculin A-treatment). Data are representative of two individual experiments.
Multiple studies have demonstrated that type I IFNs activate the PI3K-AKT pathway, starting as early as 15 min post IFN treatment.10,11 AKT is activated by phosphorylation of Threonine 308 (Thr 308) and Serine 473 (Ser 473). The PI3K-AKT signaling pathway is directly involved in the activation of MTORC1.19 To determine the role of this signaling cascade in IFN-induced autophagy, we studied phosphorylation changes of AKT at 48 h post-IFNA2c treatment. We found that AKT (Ser 473) was constitutively phosphorylated in control cells, and that treatment of Daudi cells with IFNA2c minimally altered phosphorylation of AKT (Ser 473) at 48 h. Phosphorylation of AKT (Thr 308) in control cells or IFN-treated cells was not detected at 48 h (Fig. 11B). Similar results were observed at 24 h (data not shown). At early time points (15 min, 1 and 4 h post IFNA2c treatment), we observed a modest increase in AKT phosphorylation only at Ser 473 at 4 h (data not shown). This phosphorylation had no effect on the level of MAP1LC3-II (data not shown). To further explore the role of PI3K-AKT signaling pathway in IFN-induced autophagy, we treated Daudi cells with LY294002, a PI3K-dependent AKT phosphorylation inhibitor. Treatment of cells with both LY294002 and IFNA2c increased the level of MAP1LC3-II compared with cells treated with inhibitor or IFN only (Fig. 9, lanes 6 and 7), suggesting that PI3K-AKT signaling may inhibit IFN-induced autophagy.
Discussion
Induction of autophagy by type I IFN has not been previously reported. In this study, we demonstrate that type I IFN can induce autophagy in human cancer cell lines. Induction of autophagy after IFN treatment was observed in cells that display growth inhibition in response to IFN treatment, suggesting a correlation between those activities.
Autophagy is upregulated under conditions of stress or starvation.36 The presence of several other cytokines and pathogens can also promote increased autophagy.14,37,38 Type II IFN uses autophagy to clear mycobacteria and chlamydia.21,39 Type I IFN is produced by cells in response to viral infection and/or TLR activation, and induces an antiviral state through regulation of protein synthesis and induction of ISGs.40 Additionally, type I IFN can induce soluble factors associated with cytotoxic and inflammatory effects.41 Type I IFN can also inhibit cellular proliferation and has been shown to upregulate of MHC I.1,2,42 It has been reported that autophagy is required for production of type I IFN in plasmacytoid dendritic cells.43,44 Our data demonstrated a direct effect of type I IFN on induction of autophagy. Type I IFN-induced autophagy may play a role in viral clearance or antigen presentation, as well as represent a positive feedback loop for greater IFN production during viral infection. Moreover, our findings provided evidence for a potential mechanism to explain the antiproliferative effects of type I IFN in certain cell types, considering that autophagy is often concomitant with cell cycle arrest and senescence.45,46
It is well established that type I IFN mediates signaling primarily through the JAK-STAT pathway.47 IFN receptor binding stimulates gene transcription through the JAK1-TYK2-mediated phosphorylation of STAT1 and STAT2, facilitating interaction with IRF9 to form ISGF3 complex.8,48 This complex translocates to the nucleus and binds to IFN-stimulated response elements (ISREs), resulting in transcriptional activation of ISGs.8,49,50 Interestingly, treatment of STAT2-defective mutant Daudi cells did not increase levels of MAP1LC3-II and a decrease in SQSTM1, suggesting that IFN-induced autophagy is STAT2 dependent. Future biochemical studies are required to link STAT2 activation with type I IFN-induced autophagy.
Type I IFN activates several other signaling pathways, including PI3K through an insulin receptor substrate (IRS)-dependent, STAT-independent mechanism.51 Importantly, PI3K may play a role in nuclear translocation of IRF7 and type I IFN production after activation of TLR7 and 9 in predendritic cells.52 AKT is a downstream effector of PI3K and mediates antiapoptotic and prosurvival signals.53-56 Recent published evidence demonstrates a role for AKT kinases in regulating the MTOR-RPS6KB pathway for IFN-dependent translational regulation.10,57,58 We observed that treatment of Daudi cells with a combination of the PI3K inhibitor LY294002 and IFNA2c led to increased autophagy. Given that PI3K-AKT-MTOR signaling is constitutively active in this context prior to IFN exposure, our results suggest that modulating this signaling axis may enhance IFN-induced autophagy. In the present study, we observed less phosphorylation of RPS6KB at threonine 389 in Daudi cells after 24 to 48 h of IFN treatment, suggesting that type I IFN-induced autophagy correlates with decreased MTORC1 activity. Indeed, IFN treatment also resulted in decreased phosphorylation of two RPS6KB-dependent proteins, RPS6 and EIF4B, which are involved in translational regulation. These data suggest that prolonged type I IFN signaling may promote autophagy when MTORC1-mediated stimulation of protein translation is inhibited.
The multiprotein complex MTORC1 can directly control protein synthesis and integrate myriad signals from the environment (e.g., nutrients, growth factors, energy and stress) to regulate cellular homeostasis, growth and proliferation.19 The ability of MTORC1 to regulate so many different events suggests multiple signaling mechanisms may be involved in different cell lines. In Daudi cells, we detected phosphorylation changes of MTORC1 substrates that play a role in mRNA translation and thus in protein synthesis. In other cell lines, decreased phosphorylation of selected marker RPS6 was not observed, suggesting IFN treatment did not directly modulate this specific MTORC1-regulated event. In those cell lines where IFN-induced autophagy was readily observed, it is possible that other signaling events downstream of MTORC1 were inhibited independently of RPS6KB activity. It is also possible that IFN-induced autophagy may proceed independently of MTORC1 modulation, perhaps by influencing intracellular Ca2+ or cAMP levels that dictate MTOR-independent autophagy.59 More studies are needed, utilizing an expanding toolbox of specific autophagy inhibitors, to parse out specific IFN-controlled signaling outcomes for autophagy in different cell types.
The lipophilic macrolide rapamycin is an inhibitor of MTORC1 signaling.60-62 We observed enhanced, IFN-induced MAP1LC3-II processing and autophagy in Daudi and T98G cells treated with IFNA2c and rapamycin. In addition, siRNA-mediated knockdown of MTOR augmented IFN-induced growth inhibition and autophagy in T98G cells. Hence our results support the notion that a decrease of MTORC1 activity generally enhances the magnitude of IFN-induced autophagy, even if type I IFN itself does directly down modulate MTORC1 signaling in certain sensitive cell lines (e.g., T98G, HeLa). Again, further studies will determine exactly how the balance of JAK-STAT activation and PI3K-AKT-MTORC1 inhibition dictates autophagy induction in certain tumor cells exposed to type I IFN.
In conclusion, our findings demonstrated that type I IFN can induce autophagy in certain human cancer cell lines, which is a function not previously ascribed to type I IFN treatment. Importantly, our findings provide some evidence that type I IFN-induced autophagy is STAT2-dependent. Additionally we showed that decreased MTOR activation increases IFN-induced autophagy. Given the broad usage of type I IFN in treating viral infections, autoimmune disorders and certain cancers, the ability of type I IFN to induce autophagy illuminates a new cellular effect that should be considered. For example, it is possible that pharmacological modulation of IFN-induced autophagy could increase the therapeutic index of type I IFNs. Future studies are necessary to understand the detailed mechanism of type I IFN-mediated autophagy in different cell types, its biological significance and impact on therapeutic applications.
Materials and Methods
Cell culture
Daudi cells derived from a Burkitt’s lymphoma were obtained from Dr. P. Grimley (Department of Pathology, Uniformed Services University of the Health Sciences, Bethesda, MD). Human cervical carcinoma HeLa S3 cells (ATCC, CCL-2.2), human monocyte-like cell line derived from histiocytic lymphoma U937 (ATCC, CRL-1593.2) and human lung carcinoma A549 (CCL-185) were obtained from the American Type Cell Culture Collection. Human fibroblast glioblastoma cell line T98G and human epithelial breast adenocarcinoma cell line MDA-MB-231 were obtained from Dr. Raj Puri (FDA, CBER, Bethesda, MD) and human B cell lymphoma BJAB were obtained from Dr. Michael Lenardo (NIAID, NIH, Bethesda, MD). STAT2-defective mutant Daudi cells were purchased from KeraFAST (EH0001). All cells were propagated using RPMI 1640 media (Invitrogen, 2189) supplemented with 10% FBS (Gibco, 2614), 1% penicillin-streptomycin (Sigma, P4458) and 1% L-glutamine (Gibco, 25030). To study the kinetics of autophagy induction, Daudi cells (3 × 106) were mock-treated or treated with 3.6 ng/mL (approximately 700 IU/mL) of IFNA2c or IFNB for 6, 24 or 48 h. To study IFN concentration dependence of induction of autophagy, Daudi cells (3 × 106) were mock-treated or treated with various concentrations of IFNA2c for 24 or 48 h. To study autophagy flux, Daudi cells (3 × 106) were incubated with 3.6 ng/mL IFNA2c for 48 h followed by a 4 h incubation with 0.161 µM bafilomycin A1 (Enzo Life Sciences, BML-CM110) under nutrient-rich (RPMI 1640 media supplemented with 10% FBS) or starvation conditions (Hank’s balanced salt solution, Sigma, H9269). To study the influence of PD98059, a MAPK inhibitor (EMD/Calbiochem, 513001), LY294002, a PI3K inhibitor (Cell Signaling, 9901S) or rapamycin, a MTOR inhibitor (Enzo Life Sciences, BML-A275) on autophagy, Daudi cells were incubated with 0.036 ng/mL (approximately 7 IU/mL) of IFNA2c for 48 h in the presence or absence of inhibitors. To study the influence of rapamycin on autophagy in the adherent cells, T98G cells were incubated with 3.6 ng/mL of IFNA2c for 48 h in the presence or absence of rapamycin. Concentrations of the aforementioned inhibitors or inducer (rapamycin) were selected based on dose-dependent experiments in which Daudi (3 × 106) or T98G cells (1 × 105) were treated with 10-fold serial dilutions of chemical compounds or IFNA2c for 48 h. Concentrations that did not saturate MAP1LC3-II turnover (as detected by western blot) were selected to investigate a potential additive effect when combined with IFNA2c in treatment of cells. Additionally, we evaluated the effect of treatments on cell viability (assay described below). Concentrations selected did not affect cell viability (data not shown). For neutralizing experiments, anti-IFNAR2 monoclonal antibody A10 was used at a concentration of 10 µg/mL for 48 h. To show that induced autophagy is a direct effect of IFNA2c treatment, culture media was collected from mock-treated Daudi cells (3 × 106) or cells treated with 0.36 ng/ml IFNA2c for 24 h. Cell-free supernatants were subsequently transferred onto fresh Daudi cells for incubation in the presence or absence of anti-IFNAR2-mAb A10. Samples were collected at 24 h post-treatment, and analyzed by western blot. To study induction of autophagy in HeLa S3, MDA-MB-231, T98G, A549, 1 × 105 cells, and in U937, BJAB, 3 × 106 cells were treated with 3.6 ng/mL IFNA2c for 48 h. To evaluate induction of autophagy in STAT2-defective mutant Daudi cells, 3 × 106 cells were treated with 3.6 ng/mL IFNA2c for 48 h.
IFN
IFNA2c was prepared and analyzed as previously described.63,64 IFNB1a/IFN-β1a (Avonex) was obtained from Biogen Idec, Inc. The specific AV activities of IFNs on A549 cells are 2 × 108 IU/ mg. All material was tested for endotoxin levels, and the concentration of endotoxin was determined to be less than 0.07 EU/mg.
Western blot analysis
At the desired time points, cells were harvested, counted, and an equal number of cells per sample was used for cell lysate preparation. Cell lysates were prepared using lysis buffer (20 mM TRIS, 400 mM NaCl, 1mM EDTA, 1% TRITON X-100, 1 mM DTT) with protease (Pierce, 78425) and HaltTM phosphatase inhibitor (Pierce, 1861277) cocktails. Ten microliters of each sample were analyzed by SDS-PAGE using 10–20% Tris-Glycine gels (Invitrogen, EC61352) under reducing conditions, followed by transfer onto nitrocellulose membrane (Invitrogen, IB3010-02) using iBlotTM gel transfer system (Invitrogen, IB 1001), except for MAP1LC3, where a PVDF membrane (Bio-Rad, 162-174) was used in conjunction with a XCell SureLock® Mini-Cell (Invitrogen, EI0001). Primary antibodies against ACTB/Beta-Actin (Abcam, 4967L), MAP1LC3 (3868S), RPS6KB (2708P), phospho-RPS6KB (Thr389, 9205S), RPS6 (2317S), phospho-RPS6 (Ser235/236, 4858S and Ser240/244, 2215S), MAPK1/3 (4695P), phospho-MAPK1/3 (Thr202/204, 4370S), EIF4B (3592P), phospho-EIF4B (Ser422, 3591), EIF4EBP1 (53H11), phospho-EIF4EBP1 (Thr37/Thr46, 9459S), MAPK14 (9212P), phospho-MAPK14 (Thr180/Tyr182, 9215S), AKT (9272), phospho-AKT (Ser473 4060, Thr308 2965S), MAPK8/9 (9258P), phospho-MAPK8/9 (Thr183/Tyr185, 9255S), as well as MAPK8/9 control cell extracts (9253) and AKT control cell extracts (9273) were obtained from Cell Signaling Technology, Inc. Antibodies for detection of SQSTM1 (M153-3) and ATG12–ATG5 conjugates (M162-3) were obtained from MBL International Corp., antibody for detection of p-STAT1 (Tyr701, 612232) was obtained from BD Biosciences. Secondary goat anti-mouse (sc-2005) and anti-rabbit (sc-2004) antibodies were obtained from Santa Cruz Biotechnology. Biotinylated protein ladder (10 to 200 kDa, Cell Signaling Technology, 7727S) was used as molecular weight marker. The membranes were developed by using a SuperSignal West Femto Maximum Sensitivity Kit (Pierce, 34096), and images were visualized using an LAS-3000 Imaging system (GE Healthcare Biosciences). Post-exposure image processing was applied to the whole image. Adjustments were restricted to linear changes in brightness, contrast and color balance. Ratios of MAP1LC3 were calculated using MultiGauge 3.0 (GE Healthcare Biosciences) as the division of the ratio of induced MAP1LC3-I to induced MAP1LC3-II by the ratio of basal MAP1LC3-I to basal MAP1LC3-II {(MAP1LC3-Iinduced/MAP1LC3-IIinduced)/(MAP1LC3-Ibasal/MAP1LC3-IIbasal)}. Numbers are shown below MAP1LC3 lanes in western blots.
Analysis of cell growth and viability
Cell counts and viability for different cell lines at different time points were determined using Trypan Blue Stain (Lonza, 17-942E) exclusion and a hemocytometer (Nexcelom, CP2-002) and Cellometer Vision CBA (Nexcelom Bioscience LLC). Inhibition of cell growth by IFN was calculated and results were analyzed for statistical significance using a paired t-test.
Cell cycle analysis
Cell cycle analysis was performed using a Cellometer image-based cytometer in combination with PI kit (Nexcelom Bioscience, CSK-0122), following the manufacturer’s instructions. Results were analyzed using FCS Express 4 software (De Novo Flow Cytometry Software).
Detection of autophagy in living cells
To measure levels of autophagy in living cells, Cellometer image-based cytometer in combination with Cyto-ID autophagy detection kit (Enzo, ENZ-51031-K200) was used. Samples were prepared following the kit manufacturer’s instructions and guidance by the manufacturer of the instrument, Nexcelom Bioscience.65,66 Briefly, suspension cells (3 × 106) or adherent cells (1 × 105) were incubated with IFNA2c (3.6 ng/mL) for 48 h. After treatment, cells were harvested and labeled with Cyto-IDR Green autophagy dye, which is a component of Enzo autophagy detection kit. Instead of Hoechst 33342 reagent, propidium iodide (Nexcelom Bioscience, CS1-0109-5ML) was used to stain nonviable cells, in order to exclude these cells from data analysis. Labeling kit assay buffer was supplemented with 2% FBS. To measure fluorescence signal, appropriate optic modules were selected.66 Data were analyzed using FCS Express 4 software, and Autophagy Activity Factor (AAF) values were calculated using the following equation: AAF = 100 x (MFI treated – MFI control)/ MFI treated. MFI is mean fluorescence intensity, and AAF expresses the level of autophagy in live cells as the difference between the amount of Cyto-IDR Green autophagy dye gathered within cells in the presence and absence of an autophagy inducer.66
Silencing of MTOR using siRNA
HeLa S3 cells were seeded at 1 × 105 cells/well in 6-well plates and immediately transfected for 48 h with 100 nM SignalSilenceR MTOR siRNA (Cell Signaling 6381), or SignalSilenceR control siRNA (Cell Signaling 6568), using 5 µg LipofectamineTM 2000 (Invitrogen, 11668019) diluted in Opti-MEM (Gibco, 31985070). Cells were then treated for 48 h with 3.6 ng/mL IFNA2c. After IFN treatment, cells were harvested and the effect of MTOR-siRNA on MAP1LC3-II level was examined by western blot.
Transmission electron microscopy
Specimens were fixed overnight at 4°C with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4. Samples were post-fixed for 30 min with 0.5% osmium tetroxide/0.8% potassium ferricyanide, transferred to 1% tannic acid for 1 h, and then to 1% uranyl acetate overnight at 4°C. Samples were dehydrated with a graded ethanol series, and embedded in Spurr’s resin. Thin sections were cut with a Leica EM UC6 ultramicrotome (Leica), and stained with 1% uranyl acetate and Reynold’s lead citrate prior to viewing at 120 kV on a Tecnai BT Spirit transmission electron microscope (FEI, Eindhoven, The Netherlands). Digital images were acquired with a Hammamatsu XR-100 side mount digital camera system (Advanced Microscopy Techniques) and processed using Adobe Photoshop CS5 (Adobe Systems Inc.).
Supplementary Material
Acknowledgments
We thank Drs. J. Hartley, D. Esposito and W. Gillette (NCI/SAIC) for expression and purification of IFNA2c, Drs. H. Young (NCI), M. Lotze (University of Pittsburgh), M. Lenardo (NIAID), Li Yu (NIAID), F. Schmeisser (FDA), D. Jankovic (NIAID), G. Fabozzi (FDA), N. Lai and L.L. Chan (Nexcelom Bioscience LLC) for reviewing the manuscript and valuable discussions. This research was supported by the Intramural Research Program of the NIH (NIAID).
Glossary
Abbreviations:
- ACTB
actin, beta
- AKT
v-akt murine thymoma viral oncogene homolog (also known as protein kinase B)
- IFN
interferon
- TGFB/TGFβ
transforming growth factor beta
- TEM
transmission electron microscopy
- MTOR
mechanistic target of rapamycin, (serine/threonine kinase)
- MTORC1
MTOR complex 1 (composed of MTOR, RPTOR (regulatory-associated protein of MTOR, complex 1)
- MLST8
MTOR-associated protein, LST8 homolog (S. cerevisiae)
- AKT1S1
AKT1 substrate 1 (proline-rich)/PRAS40, proline-rich AKT1 substrate 1, 40 kDa, and DEPTOR, DEP domain containing MTOR-interacting protein
- MAP1LC3 (LC3)
microtubule-associated protein 1 light chain 3
- ATG
autophagy-related
- SQSTM1 (p62)
sequestosome 1 (a ubiquitin-binding scaffold protein)
- EIF4EBP1
eukaryotic translation initiation factor 4E binding protein 1 (a translation repressor protein)
- MAPK1 (ERK2)
mitogen-activated protein kinase 1 (also known as p42 MAPK)
- MAPK3 (ERK1)
mitogen-activated protein kinase 3 (also known as p44 MAPK)
- MAPK8
mitogen-activated protein kinase 8 (also known as JNK1)
- MAPK9
mitogen-activated protein kinase 9 (also known as JNK2)
- MAPK14
mitogen-activated protein kinase 14 (also known as p38 MAP kinase alpha)
- RPS6 (S6)
ribosomal protein S6
- RPS6KB
ribosomal protein S6 kinase, 70kDa
- EIF4B
eukaryotic translation initiation factor 4B
- ER
endoplasmic reticulum
- MAP
mitogen activated protein
- PI3K
phosphatidylinositol-4,5-bisphosphate 3-kinase
- ISGs
IFN-stimulated genes
- JAK1
Janus kinase 1
- TYK2
tyrosine kinase 2
- STAT
signal transducer and activator of transcription
- IFNAR2
interferon (alpha, beta and omega) receptor 2, AAF, autophagy activity factor
- TLR
toll-like receptor
- MHC
major histocompatibility complex
- ISGF3
IFN-stimulated gene factor 3, heterotrimeric complex
- IRF7
IFN regulatory factor 7
- IRF9
IFN regulatory factor 9
- ISREs
IFN-stimulated response elements
- IRS
insulin receptor substrate
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/autophagy/article/23921
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/23921
References
- 1.Bekisz J, Baron S, Balinsky C, Morrow A, Zoon KC. Antiproliferative Properties of Type I and Type II Interferon. Pharmaceuticals (Basel) 2010;3:994–1015. doi: 10.3390/ph3040994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bekisz J, Schmeisser H, Hernandez J, Goldman ND, Zoon KC. Human interferons alpha, beta and omega. Growth Factors. 2004;22:243–51. doi: 10.1080/08977190400000833. [DOI] [PubMed] [Google Scholar]
- 3.Hu R, Gan Y, Liu J, Miller D, Zoon KC. Evidence for multiple binding sites for several components of human lymphoblastoid interferon-α. J Biol Chem. 1993;268:12591–5. [PubMed] [Google Scholar]
- 4.Einat M, Resnitzky D, Kimchi A. Close link between reduction of c-myc expression by interferon and, G0/G1 arrest. Nature. 1985;313:597–600. doi: 10.1038/313597a0. [DOI] [PubMed] [Google Scholar]
- 5.Schmeisser H, Mejido J, Balinsky CA, Morrow AN, Clark CR, Zhao T, et al. Identification of alpha interferon-induced genes associated with antiviral activity in Daudi cells and characterization of IFIT3 as a novel antiviral gene. J Virol. 2010;84:10671–80. doi: 10.1128/JVI.00818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akiyama M, Iwase S, Horiguchi-Yamada J, Saito S, Furukawa Y, Yamada O, et al. Interferon-α repressed telomerase along with G1-accumulation of Daudi cells. Cancer Lett. 1999;142:23–30. doi: 10.1016/S0304-3835(99)00109-3. [DOI] [PubMed] [Google Scholar]
- 7.Lee CK, Bluyssen HAR, Levy DE. Regulation of interferon-α responsiveness by the duration of Janus kinase activity. J Biol Chem. 1997;272:21872–7. doi: 10.1074/jbc.272.35.21872. [DOI] [PubMed] [Google Scholar]
- 8.Kessler DS, Levy DE, Darnell JE., Jr. Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc Natl Acad Sci U S A. 1988;85:8521–5. doi: 10.1073/pnas.85.22.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Platanias LC. The p38 mitogen-activated protein kinase pathway and its role in interferon signaling. Pharmacol Ther. 2003;98:129–42. doi: 10.1016/S0163-7258(03)00016-0. [DOI] [PubMed] [Google Scholar]
- 10.Kaur S, Katsoulidis E, Platanias LC. Akt and mRNA translation by interferons. Cell Cycle. 2008;7:2112–6. doi: 10.4161/cc.7.14.6258. [DOI] [PubMed] [Google Scholar]
- 11.Kaur S, Sassano A, Dolniak B, Joshi S, Majchrzak-Kita B, Baker DP, et al. Role of the Akt pathway in mRNA translation of interferon-stimulated genes. Proc Natl Acad Sci U S A. 2008;105:4808–13. doi: 10.1073/pnas.0710907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–70. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deretic V. Autophagy as an immune defense mechanism. Curr Opin Immunol. 2006;18:375–82. doi: 10.1016/j.coi.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 14.Schmid D, Münz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroemer G, Levine B. Autophagic cell death: the story of a misnomer. Nat Rev Mol Cell Biol. 2008;9:1004–10. doi: 10.1038/nrm2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–92. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 18.Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–14. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiczer BM, Thomas G. Phospholipase D and mTORC1: nutrients are what bring them together. Sci Signal. 2012;5:pe13. doi: 10.1126/scisignal.2003019. [DOI] [PubMed] [Google Scholar]
- 21.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 22.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002;157:455–68. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pyo JO, Jang MH, Kwon YK, Lee HJ, Jun JI, Woo HN, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death. J Biol Chem. 2005;280:20722–9. doi: 10.1074/jbc.M413934200. [DOI] [PubMed] [Google Scholar]
- 24.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–55. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 25.Du Z, Fan M, Kim J-G, Eckerle D, Lothstein L, Wei L, et al. Interferon-resistant Daudi cell line with a Stat2 defect is resistant to apoptosis induced by chemotherapeutic agents. J Biol Chem. 2009;284:27808–15. doi: 10.1074/jbc.M109.028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, et al. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–54. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Yin X-M, Ding W-X, Gao W. Autophagy in the liver. Hepatology. 2008;47:1773–85. doi: 10.1002/hep.22146. [DOI] [PubMed] [Google Scholar]
- 28.Mijaljica D, Prescott M, Devenish RJ. Endoplasmic reticulum and Golgi complex: Contributions to, and turnover by, autophagy. Traffic. 2006;7:1590–5. doi: 10.1111/j.1600-0854.2006.00495.x. [DOI] [PubMed] [Google Scholar]
- 29.Eskelinen EL. To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy. 2008;4:257–60. doi: 10.4161/auto.5179. [DOI] [PubMed] [Google Scholar]
- 30.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- 32.Klionsky DJ, Elazar Z, Seglen PO, Rubinsztein DC. Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy. 2008;4:849–950. doi: 10.4161/auto.6845. [DOI] [PubMed] [Google Scholar]
- 33.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–5. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 34.Johnson TC, Lerner MP, Lancz GJ. Inhibition of protein synthesis in noninfected L cells by partially purified interferon preparations. J Cell Biol. 1968;36:617–24. doi: 10.1083/jcb.36.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavieu G, Scarlatti F, Sala G, Carpentier S, Levade T, Ghidoni R, et al. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem. 2006;281:8518–27. doi: 10.1074/jbc.M506182200. [DOI] [PubMed] [Google Scholar]
- 36.Amaravadi RK, Lippincott-Schwartz J, Yin X-M, Weiss WA, Takebe N, Timmer W, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17:654–66. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang YP, Tsai CC, Huang WC, Wang CY, Chen CL, Lin YS, et al. Autophagy facilitates IFN-gamma-induced Jak2-STAT1 activation and cellular inflammation. J Biol Chem. 2010;285:28715–22. doi: 10.1074/jbc.M110.133355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–32. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Zeer MA, Al-Younes HM, Braun PR, Zerrahn J, Meyer TF. IFN-gamma-inducible Irga6 mediates host resistance against Chlamydia trachomatis via autophagy. PLoS One. 2009;4:e4588. doi: 10.1371/journal.pone.0004588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008;8:559–68. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wichers MC, Koek GH, Robaeys G, Verkerk R, Scharpé S, Maes M. IDO and interferon-alpha-induced depressive symptoms: a shift in hypothesis from tryptophan depletion to neurotoxicity. Mol Psychiatry. 2005;10:538–44. doi: 10.1038/sj.mp.4001600. [DOI] [PubMed] [Google Scholar]
- 42.Herberman RB, Holden MT, Djeu JY, Jerrells TR, Varesio L, Tagliabue A, et al. Macrophages as regulators of immune response against tumors. Adv Exp Med Biol. 1979;121B:361–79. doi: 10.1007/978-1-4684-8914-9_35. [DOI] [PubMed] [Google Scholar]
- 43.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 44.Zhou D, Kang KH, Spector SA. Production of interferon α by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J Infect Dis. 2012;205:1258–67. doi: 10.1093/infdis/jis187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Filippi-Chiela EC, Villodre ES, Zamin LL, Lenz G. Autophagy interplay with apoptosis and cell cycle regulation in the growth inhibiting effect of resveratrol in glioma cells. PLoS One. 2011;6:e20849. doi: 10.1371/journal.pone.0020849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young ARJ, Narita M, Ferreira M, Kirschner K, Sadaie M, Darot JFJ, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:784–7. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr. Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–13. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 48.Kessler DS, Veals SA, Fu XY, Levy DE. Interferon-alpha regulates nuclear translocation and DNA-binding affinity of ISGF3, a multimeric transcriptional activator. Genes Dev. 1990;4:1753–65. doi: 10.1101/gad.4.10.1753. [DOI] [PubMed] [Google Scholar]
- 49.Levy D, Darnell JE., Jr. Interferon-dependent transcriptional activation: signal transduction without second messenger involvement? New Biol. 1990;2:923–8. [PubMed] [Google Scholar]
- 50.Fu X-Y, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr. ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci U S A. 1990;87:8555–9. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uddin S, Yenush L, Sun XJ, Sweet ME, White MF, Platanias LC. Interferon-alpha engages the insulin receptor substrate-1 to associate with the phosphatidylinositol 3′-kinase. J Biol Chem. 1995;270:15938–41. doi: 10.1074/jbc.270.27.15938. [DOI] [PubMed] [Google Scholar]
- 52.Guiducci C, Ghirelli C, Marloie-Provost MA, Matray T, Coffman RL, Liu YJ, et al. PI3K is critical for the nuclear translocation of IRF-7 and type I IFN production by human plasmacytoid predendritic cells in response to TLR activation. J Exp Med. 2008;205:315–22. doi: 10.1084/jem.20070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 54.Barca O, Ferré S, Seoane M, Prieto JM, Lema M, Señarís R, et al. Interferon beta promotes survival in primary astrocytes through phosphatidylinositol 3-kinase. J Neuroimmunol. 2003;139:155–9. doi: 10.1016/S0165-5728(03)00160-7. [DOI] [PubMed] [Google Scholar]
- 55.Ruuth K, Carlsson L, Hallberg B, Lundgren E. Interferon-alpha promotes survival of human primary B-lymphocytes via phosphatidylinositol 3-kinase. Biochem Biophys Res Commun. 2001;284:583–6. doi: 10.1006/bbrc.2001.5025. [DOI] [PubMed] [Google Scholar]
- 56.Yang CH, Murti A, Pfeffer SR, Kim JG, Donner DB, Pfeffer LM. Interferon alpha /beta promotes cell survival by activating nuclear factor kappa B through phosphatidylinositol 3-kinase and Akt. J Biol Chem. 2001;276:13756–61. doi: 10.1074/jbc.M011006200. [DOI] [PubMed] [Google Scholar]
- 57.Lekmine F, Uddin S, Sassano A, Parmar S, Brachmann SM, Majchrzak B, et al. Activation of the p70 S6 kinase and phosphorylation of the 4E-BP1 repressor of mRNA translation by type I interferons. J Biol Chem. 2003;278:27772–80. doi: 10.1074/jbc.M301364200. [DOI] [PubMed] [Google Scholar]
- 58.Lekmine F, Sassano A, Uddin S, Smith J, Majchrzak B, Brachmann SM, et al. Interferon-gamma engages the p70 S6 kinase to regulate phosphorylation of the 40S S6 ribosomal protein. Exp Cell Res. 2004;295:173–82. doi: 10.1016/j.yexcr.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 59.Ravikumar B, Futter M, Jahreiss L, Korolchuk VI, Lichtenberg M, Luo S, et al. Mammalian macroautophagy at a glance. J Cell Sci. 2009;122:1707–11. doi: 10.1242/jcs.031773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raught B, Gingras A-C, Sonenberg N. The target of rapamycin (TOR) proteins. Proc Natl Acad Sci U S A. 2001;98:7037–44. doi: 10.1073/pnas.121145898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/S0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 62.Oshiro N, Yoshino K, Hidayat S, Tokunaga C, Hara K, Eguchi S, et al. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells. 2004;9:359–66. doi: 10.1111/j.1356-9597.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- 63.Schmeisser H, Kontsek P, Esposito D, Gillette W, Schreiber G, Zoon KC. Binding Characteristics of IFN-alpha Subvariants to IFNAR2-EC and Influence of the 6-Histidine Tag. J Interferon Cytokine Res. 2006;26:866–76. doi: 10.1089/jir.2006.26.866. [DOI] [PubMed] [Google Scholar]
- 64.Hu R, Bekisz J, Hayes M, Audet S, Beeler J, Petricoin E, et al. Divergence of binding, signaling, and biological responses to recombinant human hybrid IFN. J Immunol. 1999;163:854–60. [PubMed] [Google Scholar]
- 65.Chan LL, Zhong X, Qiu J, Li PY, Lin B. Cellometer vision as an alternative to flow cytometry for cell cycle analysis, mitochondrial potential, and immunophenotyping. Cytometry A. 2011;79:507–17. doi: 10.1002/cyto.a.21071. [DOI] [PubMed] [Google Scholar]
- 66.Chan LL, Shen D, Wilkinson AR, Patton W, Lai N, Chan E, et al. A novel image-based cytometry method for autophagy detection in living cells. Autophagy. 2012;8:1371–82. doi: 10.4161/auto.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.