Abstract
Central to most forms of autophagy are two ubiquitin-like proteins (UBLs), Atg8 and Atg12, which play important roles in autophagosome biogenesis, substrate recruitment to autophagosomes, and other aspects of autophagy. Typically, UBLs are activated by an E1 enzyme that (1) catalyzes adenylation of the UBL C terminus, (2) transiently covalently captures the UBL through a reactive thioester bond between the E1 active site cysteine and the UBL C terminus, and (3) promotes transfer of the UBL C terminus to the catalytic cysteine of an E2 conjugating enzyme. The E2, and often an E3 ligase enzyme, catalyzes attachment of the UBL C terminus to a primary amine group on a substrate. Here, we summarize our recent work reporting the structural and mechanistic basis for E1-E2 protein interactions in autophagy.
Keywords: Atg10, Atg12, Atg3, Atg7, Atg8, E1 enzyme, E2 enzyme, ubiquitin-like protein
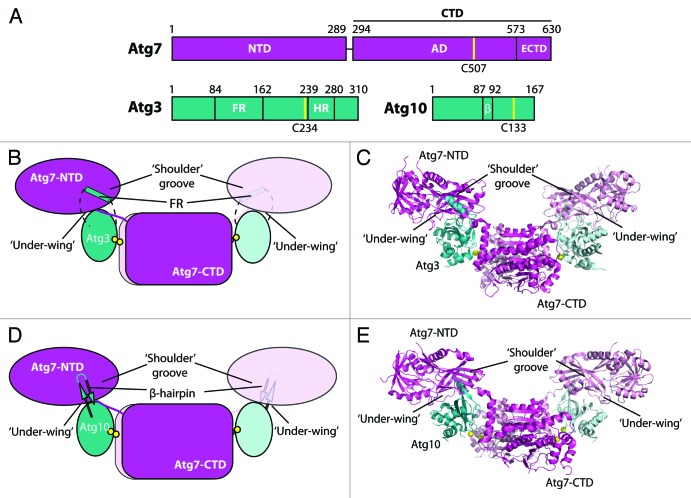
Atg8 and Atg12 are activated by the same E1 enzyme, Atg7 (Fig. 1), which in vivo specifically promotes their transfer to the catalytic cysteine residue of their specific E2 enzyme (Atg3 and Atg10, respectively). Ultimately, Atg8 and Atg12 are ligated to phosphatidylethanolamine and Atg5, respectively.
Figure 1. Structural analysis of proteins involved in the autophagy ubiquitin-like protein conjugation systems. (A) Domain diagrams for Atg7, Atg3 and Atg10. NTD, N-terminal domain; CTD, C-terminal domain; AD, adenylation domain; ECTD, extreme C-terminal domain; FR, flexible region; HR, handle region; β, β-hairpin. Catalytic cysteine residues are indicated with yellow lines. (B and C) Schematic and structure of the Atg7-Atg3 complex indicating key structural features involved in binding. Catalytic-cysteine thiols are shown as yellow spheres. (D and E) Schematic and structure of the Atg7-Atg10 complex indicating key structural features involved in binding. Yellow spheres represent the catalytic-cysteine thiols of Atg7 and His131 or Pro132 of Atg10, in which the catalytic cysteine was not visible.
Atg7, Atg3, and Atg10 are called “noncanonical” because they differ in many respects from so-called “canonical” E1 and E2 enzymes such as those for ubiquitin, SUMO, and NEDD8. Atg3 and Atg10 display a unique constellation of residues around the catalytic cysteine, and they have additional secondary structures inserted within the E2 fold. Atg7 is a symmetric homodimer with two catalytic cysteine residues, and it activates two different UBL proteins, Atg8 and Atg12, that share little sequence identity. Atg7 has a combined adenylation/catalytic cysteine-containing/homodimerization C-terminal domain (CTD), and a unique N-terminal domain (NTD) that previous work in our lab as well as the Inagaki and Song labs has shown to be important for recruiting E2 enzymes. In contrast, canonical E1 enzymes have a pseudosymmetric UBL adenylation domain, and a separate domain that houses the catalytic cysteine residue that forms a thioester bond with a specific UBL and passes it to the catalytic cysteine of a cognate E2 that binds to an E1 C-terminal ubiquitin-fold domain (UFD). Thus, although Atg7, Atg3, and Atg10 have long been known to play essential roles in autophagy, detailed knowledge of how Atg7 orchestrates the activation of two UBLs for transfer to two different E2s has remained elusive.
In our recent study, we used a chemical crosslinking approach to enable crystallization and structure determination for S. cerevisiae Atg7-Atg3 and Atg7-Atg10 complexes with E1 and E2 enzyme active site cysteine residues juxtaposed—as required for catalysis of UBL transfer. The use of nearly full-length proteins enabled us to observe multiple surfaces Atg7 uses to recruit its E2s (Fig. 1).
The structure of Atg7 is bird-like, with the CTD dimer resembling the body, and NTDs resembling wings, as indicated in Figure 1. Atg7 under-wing and NTD-CTD junction regions interact with the E2 core domain backsides and edges of Atg3 and Atg10, with unique insertions extending from Atg3 (flexible region) and Atg10 (β-hairpin) interacting with the Atg7 shoulder groove. These interactions, combined with Atg7 flexibility, result in the close apposition of the enzymes’ catalytic cysteine residues distal from the interaction surfaces (Fig. 1). Recent work from the Inagaki lab describes structures of complexes of the Atg7-NTD with Atg3 and Atg10, and although the proteins were derived from different organisms, the observed interactions are remarkably similar.
We evaluated the importance of the structurally observed interactions by using in vitro (crosslinking, pulse-chase UBL transfer, Atg8 lipidation) and in vivo (GFP-Atg8 localization, Pho8Δ60, GFP-Atg8 processing, Atg8 lipidation) assays. On the basis of these assays, we conclude that the interactions of the E2 insertions with the Atg7 shoulder groove are crucial, and that E2 interactions with the NTD-CTD junction region of Atg7 are also important. Atg7 under-wing interactions with the backside of Atg3 appear to be essential, whereas the observed loose interactions between the NTD and backside of Atg10 appear to not be required for Atg10 function. Notably, the structures explain our previous finding that a mutation in the Atg7 binding site for Atg3 impairs lipidation of the murine Atg8 homolog, LC3, in cells, without significantly decreasing Atg10-mediated Atg12 ligation to Atg5.
In addition to ascertaining the importance of the observed interactions, we used in vitro and in vivo assays to better understand the mechanism by which Atg7 catalyzes transfer of UBLs. Cross-linking experiments using engineered mixed dimer versions of Atg7 confirmed the structurally observed configurations and demonstrate that Atg7 transfers UBLs via a noncanonical trans mechanism, where an E2 receives the UBL from the E1 monomer opposite to that with which it is bound. In complex with Atg7, both Atg3 and Atg10 have conserved Tyr and His residues in equivalent positions flanking the E2 catalytic Cys residues. Consistent with the hypothesis that these residues are poised to contribute to catalysis, UBL transfer and Pho8Δ60 assays examining the effects of mutations of these residues demonstrate that they are required for Atg3 and Atg10 function. Interestingly, in a crystal structure of Atg3 alone, the catalytic cysteine is buried. Thus, binding to Atg7 may induce an activating conformational change around the active site of Atg3.
Combined with the recent data from the Inagaki lab, our work establishes a new paradigm for how E2 enzymes recognize flexibly tethered domains in E1 enzymes to achieve UBL conjugation. One important unanswered question is how Atg7 specifically recruits Atg3 and Atg10 for transfer of Atg8, and Atg12, respectively. We speculate that the activated UBL or other presently unknown factors may help achieve this specificity by influencing which E2 binds or contributing to catalysis of its own transfer. We anticipate that future work focused on E1-E2-UBL interactions may explain how specificity arises in this important pathway.
Acknowledgments
This work was supported by ALSAC, the St. Jude Cancer Center grant (5P30CA021765), NIH R01GM077053 to BAS and R01GM053396 to D.J.K. B.A.S. is an Investigator of the Howard Hughes Medical Institute.
Glossary
Abbreviations:
- UBLs
ubiquitin-like proteins
- UFD
ubiquitin-fold domain
- NTD
N-terminal domain
- CTD
C-terminal domain
- AD
adenylation domain
- ECTD
extreme C-terminal domain
- FR
flexible region
- HR
handle region
- β
beta-hairpin
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/23644