Abstract
MicroRNAs (miRNAs) form a class of ~21 nucleotide (nt) RNAs that post-transcriptionally repress partially complementary messenger RNAs. miRNA-mediated silencing is critical for control of many key biological processes such as tumorigenesis, neuronal synaptic plasticity and defense against bacteria and viruses. Thus, unsurprisingly, miRNA biogenesis, abundance and action are under refined feedback control that is only beginning to be experimentally uncovered. We recently discovered that DICER1 and EIF2C/AGO are targeted for degradation by autophagy as miRNA-free entities by the selective autophagy receptor CALCOCO2/NDP52 (calcium binding and coiled-coil domain 2/nuclear dot protein, 52 kDa). Strikingly, autophagy establishes a checkpoint for continued loading of miRNA, and this checkpoint is required for maintenance of miRNA abundance and proper miRNA activity. This newfound role for autophagy in miRNA biology suggests that human diseases exhibiting misregulated autophagy may be interdependent with defects in miRNA-mediated regulation of gene networks.
Keywords: autophagy, DICER, microRNA, NDP52
Pre-miRNA are 60–120 nt stem-loop structures that are cleaved in the cytoplasm by DICER1 into ~21-nt double-stranded miRNA-miRNA* duplexes (Fig. 1). The miRNA-miRNA* duplex is then transferred into the groove of EIF2C and, upon dissociation of the miRNA*-strand, a mature single-stranded miRNA remains loaded into EIF2C (Fig. 1). EIF2C can then repress miRNA-targeted mRNA by translation inhibition and accelerated mRNA decay. miRNA-mediated silencing is critical for control of cellular homeostasis. However, the regulation of miRNA biogenesis, abundance and action are not yet fully defined.
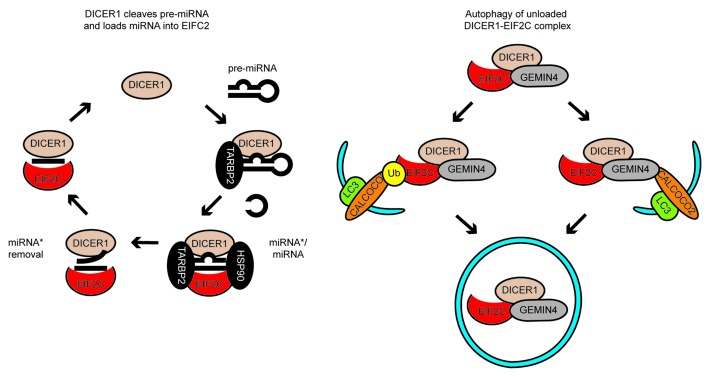
Figure 1. A model of miRNA biogenesis and loading into EIF2C and its regulation by autophagy. Left, miRNA biogenesis and loading into EIF2C. DICER1 cleaves stem-loop pre-miRNA into double-stranded miRNA-miRNA*complexes, which are loaded into EIF2C. TARBP2 and HSP90 facilitate loading of miRNA-miRNA* complexes into EIF2C. The miRNA* strand is removed and a complex of miRNA and EIF2C capable of repressing complementary mRNA is released. Right, targeting of DICER1 and EIF2C to autophagy. A complex of DICER1 and EIF2C not associated with a pre-miRNA-derived RNA associates with GEMIN4. EIF2C may associate with CALCOCO2 via two non-mutually exclusive means, ubiquitination of EIF2C, or via GEMIN4. This leads to degradation of complexes containing DICER1, GEMIN4 and EIF2C.
Autophagy is an intracellular process delivering cytosolic material to the lysosome for degradation, yet little is known regarding its impact on cellular RNA. Autophagy may indirectly modulate RNA processes by degrading RNA-binding proteins, or, considering that lysosomes contain multiple, potent RNases, it may alternatively degrade RNA itself. Previous work had shown that the miRNA-processing enzyme, DICER1, and the main miRNA effector, EIF2C, associate with intracytosolic membranes and that, at least in plants, the miRNA-binding protein, EIF2C1, is degraded in a proteasome-independent manner. Based on this evidence, we hypothesized that proteins in the miRNA pathway, or miRNA themselves, might be a target of autophagic degradation in mammalian cells.
When autophagy was discovered over 50 y ago it was considered a general, nonselective degradation pathway. However, it is increasingly recognized that autophagosomes can degrade cytosolic material in a selective manner. To enable selectivity, SQSTM1/p62-like receptors (SLRs) recognize substrates for autophagy and target them to nascent autophagosome membranes through their interaction with ATG8 family proteins. While ubiquitination is the best-characterized means of cargo recognition for autophagic degradation by SLRs, ubiquitin-independent mechanisms have additionally started to emerge.
We recently discovered that autophagy regulates miRNA biogenesis by degrading DICER1 and EIF2C, which are known to form a well-defined complex. Consistent with this, EIF2C1, EIF2C2 and DICER1 accumulate in cells lacking the critical autophagy components ATG5, ATG6 or ATG7, or the well-characterized SLR, CALCOCO2. DICER1 and EIF2C levels are also altered by several inhibitors (e.g., bafilomycin A1 and chloroquine, which inhibit autophagosome-lysosome fusion) or activators (e.g., MTOR inhibitors rapamycin and PP242) of autophagy. Immunofluorescence microscopy reveals that DICER1 accumulates with CALCOCO2 and HcRed-LC3, and electron microscopy shows its presence in membrane-bound structures bearing the structural hallmarks of autophagosomes. Moreover, both DICER1 and EIF2C2 cofractionate with autophagosomes in density gradients. Taken together, these data strongly suggest that a complex of DICER1 and EIF2C is degraded by autophagy
The accumulation of DICER1 and EIF2C2 in the absence of CALCOCO2, and the colocalization of DICER1 with CALCOCO2-containing autophagosomes suggested that these proteins might be degraded in a CALCOCO2-selective manner. Ubiquitinated EIF2C2 accumulates when CALCOCO2 is depleted, suggesting that ubiquitination is involved in autophagy-mediated EIF2C2 degradation. In addition, we discovered that ubiquitin-independent signals may also contribute to EIF2C degradation by autophagy. CALCOCO2 and EIF2C2 have been both previously shown to interact with the RNA-binding protein GEMIN4, albeit for unclear reasons. Using immunoprecipitation we confirmed that GEMIN4 and CALCOCO2 form a complex with DICER1 and EIF2C. Interestingly, in cells lacking GEMIN4, we observed an accumulation of EIF2C2, but not DICER1, suggesting that GEMIN4 links EIF2C2 to CALCOCO2 to engage its autophagic degradation. Together, these data highlight how ubiquitin-dependent and -independent recognition events direct the autophagic degradation of EIF2C in a CALCOCO2-dependent manner (Fig. 1). While DICER1 is degraded in a CALCOCO2-dependent manner that may redundantly involve GEMIN4, the critical recognition events in this process remain to be identified.
DICER1 binds pre-miRNA and mediates production of miRNA-miRNA* duplexes while EIF2C protects miRNA from cytoplasmic RNases. So how does autophagy affect levels of these RNAs in the cell? We observed that pre-miRNA, miRNA* and miRNA are not degraded by autophagy in parallel with DICER1 and EIF2C, suggesting that DICER1 and EIF2C are turned over as RNA-free entities (Fig. 1). Surprisingly, sustained inhibition of autophagy using siRNA targeting CALCOCO2, ATG5 or ATG7 decreases the cellular levels of the ubiquituous miRNAs MIRLET7A/let-7a and MIR16/miR-16. Inhibition of autophagy with bafilomycin A1 decreases the ability of EIF2C to bind miRNA-miRNA* duplexes, a step facilitated by DICER1, and causes accumulation of miRNA-free EIF2C2. Prevention of miRNA-loading onto EIF2C2 destabilizes miRNA in many circumstances, and likely results in the long-term decrease in miRNA levels observed when autophagy is inhibited. On the basis of these results, we propose that autophagy functions as a checkpoint to remove inactive DICER1–EIF2C complexes, i.e., complexes not bound to miRNA, which may compete with active complexes for limiting cofactors, such as TRBP [TAR (HIV-1) RNA binding protein 2] or HSP90. This would likely prevent EIF2C loading with miRNA and affect miRNA stability (Fig. 1).
As expected, decreased miRNA levels resulting from compromised autophagy, correlate with decreased miRNA activity, as measured using dual luciferase reporters. Levels of endogenous proteins whose mRNA is normally silenced by miRNA, such as RAS (a critical oncogene in humans), also increase following depletion of ATG7. It had been previously shown that the mRNA encoding DICER1 is itself targeted by miRNA. We showed that inhibition of autophagy relieves miRNA-mediated repression of DICER1 mRNA, activating production of new DICER1 protein to replenish the decreasing miRNA levels. Thus, autophagy regulates miRNA homeostasis by direct post-translational degradation of DICER1 and EIF2C proteins, and by gauging post-transcriptional regulation of DICER1 mRNA.
In summary, we have identified autophagy as a newfound regulator of miRNA homeostasis. The inhibition of autophagy leads to inhibited miRNA function. Autophagy is modulated in many stresses, diseases and infections. These changes in autophagy may modulate miRNA function, adding a new regulatory dimension to our understanding of these pathophysiological situations.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/23694