Summary
Mineral nitrogen in nature is often found in the form of nitrate (NO3-). Numerous microorganisms evolved to assimilate nitrate and use it as a major source of mineral nitrogen uptake1. Nitrate, which is central in nitrogen metabolism, is first reduced to nitrite (NO2-) through a two-electron reduction reaction2,3. The accumulation of cellular nitrite can be harmful because nitrite can be reduced to the cytotoxic nitric oxide. Instead, nitrite is rapidly removed from the cell by channels and transporters, or reduced to ammonium or dinitrogen through the action of assimilatory enzymes3. Despite decades of effort no structure is currently available for any nitrate transport protein and the mechanism by which nitrate is transported remains largely obscure. Here we report the structure of a bacterial nitrate/nitrite transport protein, NarK, from Escherichia coli, with and without substrate. The structures reveal a positively charged substrate-translocation pathway lacking protonatable residues, suggesting that NarK functions as a nitrate/nitrite exchanger and that H+s are unlikely to be co-transported. Conserved arginine residues form the substrate-binding pocket, which is formed by association of helices from the two halves of NarK. Key residues that are important for substrate recognition and transport are identified and related to extensive mutagenesis and functional studies. We propose that NarK exchanges nitrate for nitrite by a rocker-switch mechanism facilitated by inter-domain H-bond networks.
Keywords: Nitrate, Nitrite, X-ray crystallography, nitrate/nitrite porter (NNP), Major Facilitator Superfamily (MFS), membrane protein, exchanger
The nitrate/nitrite porter (NNP) family of membrane proteins evolved to efficiently translocate the ionic molecules NO3- and NO2- across the membrane4,5. Two nitrate/nitrite transport protein NarK and NarU were identified in Escherichia coli 6-8,9,10. NarK proteins have been shown to catalyze either nitrate/nitrite exchange or nitrate uptake, presumably by symport with a proton 9,1,10. The former activity would be associated with respiration whereas the latter could be associated either with respiration of the net assimilation of nitrite into cell material. (An additional membrane protein, NirC, functions as a H+/nitrite channel in E. coli but is not a member of the NNP10). Nitrate is the preferred source of nitrogen for plans and at least 16 nitrate/nitrite transport proteins have been identified 11. In plants the function of NarK proteins is likely related solely to the net assimilation of nitrogen.
The NNP family belongs to the Major Facilitator Superfamily (MFS) of secondary transporters. MFS members exhibit specificity to a wide range of molecules12. Although more than 58 distinct families of transporters make up the major facilitator superfamily, representatives of only 6 such families have been crystallized and their structure determined13-19. These 6 representative transporters require H+s for their function. All MFS members are postulated to function through the rocker switch mechanism12. They all share a common structural topology but share little or no sequence homology.
Here we report the first crystal structure of a nitrate transport protein that also transports nitrite. The structure of the E. coli NarK with and without substrate was determined by X-ray crystallography. Functionally important residues that form the substrate-binding pocket are identified and related to previously described mutagenesis and functional studies. We provide the first evidence that NarK functions as a nitrate/nitrite exchanger, and that H+s are likely not co-transported with substrate.
We overexpressed NarK, and purified the protein to homogeneity as described in the Methods section. Well-ordered high quality crystals were obtained when NarK was co-crystallized with Fab fragment of a monoclonal antibody we developed. The data extended to 2.6 Å were phased using molecular replacement with Fab as a search model.
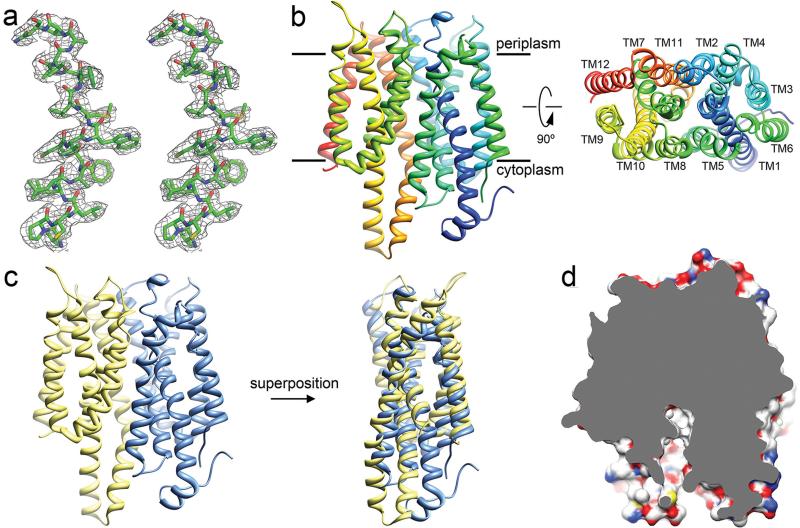
The resulting density map was of high enough quality allowing us to build and refine the NarK structure (Fig. 1a). The asymmetric unit contains one molecule of NarK forming a complex with one molecule of Fab (Fig. S1). NarK, as other MFS proteins, is structurally divided into two domains, the N- terminal half and the C-terminal half each consisting of 6 transmembrane helices (TM1-6 and TM7-12, respectively) (Fig. 1b-c). The two domains are connected by a long loop between TM6 and TM7 (disordered in our structure) and it is thought that the substrate transport pathway is localized at the interface between these two domains. NarK appears to be in the inward-facing conformation as the hydrophilic central cavity is exposed to the cytosolic side (Fig. 1d). A detailed description of the crystal packing and the overall architecture of NarK can be found in the Supplementary Text.
Fig. 1. The crystal structure of NarK.
a. Part of TM2 of NarK is shown in stereo view with a sigma-weighted 2Fo-Fc map at 2.6Å resolution, contoured at 1.0 σ. b. Left, NarK structure viewed from the plane of membrane with the putative location of the lipid bilayer as indicated. NarK is colored in rainbow with the N-terminus in blue. Right, NarK viewed from the periplasmic side. The identity of the 12 transmembrane helices is indicated. c. The N-terminal domain (TM1-6) of NarK (blue) is psudo-symmetric to the C-terminal domain (TM7-12) (yellow) and can be superimposed with an r.m.s.d of 2.9Å. d. Cut-away surface representation of the inward-facing NarK shows the central cavity exposed to the cytosol.
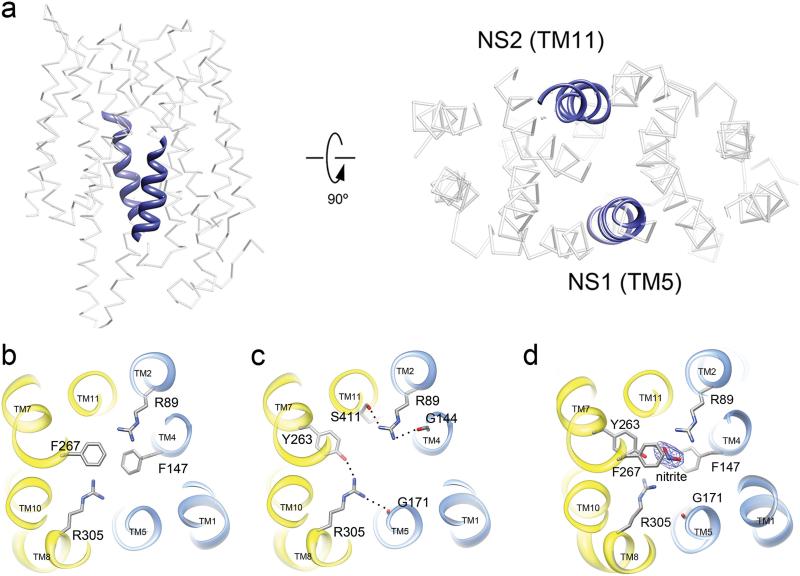
All members of the nitrate/nitrite porter (NNP) family contain two stretches of conserved residues called the nitrate signature (NS) motifs (Fig. S2). The NS motifs are not found in other MFS members but are a unique feature of the NNP family20. In NarK, the NS1 motif is formed by residues 164 – 175 (GGALGLNGGLGN) located on TM5. The NS2 motif of NarK is formed by residues 408 – 420 (GFISAIGAIGGFF) located on TM11 (Fig. 2a, blue). The NS motifs in NarK are located at the center of the protein, lining part of the substrate transport pathway (Fig 2a, right). Both of the NS motifs are glycine rich which ensures a tight fit among the surrounding helices. The result is a significantly more compact structure for NarK when compared to other known structures of MFS members (Fig. S3).
Fig. 2. The substrate-binding site in NarK.
a. Two highly conserved NS motifs in TM5 and TM11 (blue helices) at the center of NarK form the nitrate/nitrite transport pathway. b. The substrate-binding pocket is defined by two evolutionarily conserved and functionally important arginine residues R89 and R305. The binding site is capped above and below by F267 and F147, respectively. c. R89 and R305 are stabilized by inter-domain H-bonds as depicted. The two halves of NarK are indicated as blue (N-terminal domain) and yellow (C-terminal domain). d. Nitrite bound structure of NarK. The density for nitrite is observed at the substrate-binding site after soaking the NarK crystals with sodium nitrite. R305 undergoes a conformational change upon substrate binding. This conformational change affects the inter-domain network of H-bonds between Y263, G171 and R305. The displayed map is a sigmaA-weighted Fo-Fc at 3σ.
In order to transport anions like NO3- and NO2-, polar residues lining the central pore are most likely to form the substrate-binding pocket and be involved in substrate recognition and transport. Two arginine residues, R89 from TM2 and R305 from TM8 are absolutely conserved among all nitrate/nitrite transporters in both prokaryotes and eukaryotes (Fig. S2). Structurally, R89 and R305 are in plane and appose one another at the very center of NarK, with their side chains extending well into the central cavity of the transporter (Fig. 2b). These arginines are capped by two phenylalanine residues: F267 above and F147 below. Together the arginines and the phenylalanines form the substrate-binding pocket. The only bulky side chain in plane with the arginines is Y263, which hydrogen bonded with R305 (Fig. 2c). The two arginine side chains are stabilized by an intricate system of inter-domain H-bonds that link the two halves of NarK (Fig. 2c). Mutation of the residues described above led to a complete loss of function in NarK and its close homologues (Fig. S2 and Table 1). A detailed discussion of the relevant mutational and functional studies can be found in Supplementary Text.
Table 1.
Mutagenesis and functional study of key residues important for nitrate/nitrite exchange.
Soaking our NarK crystals with sodium nitrate deteriorated the crystal packing and did not yield meaningful data. In sharp contrast, soaking the crystals with sodium nitrite did not significantly affect crystallinity and yielded data to 2.8Å resolution (Table S1) allowing us to visualize the nitrite bound in the substrate binding pocket (Fig. 2d and S4). Overall the structure of substrate-free NarK and nitrite-bound NarK are very similar having an all Cα-atom r.m.s.d of 0.6Å (Fig. S5). This is not surprising because NarK is likely stabilized in the inward facing conformation by crystal contacts and the Fab could further restrict protein movement (Fig. S1). Nevertheless, clear densities were observed for nitrite in the substrate-binding pocket (Fig. 2d and S4). Nitrite was observed in-plane with R89 and R305 at the substrate binding pocket where it is capped above and below by F267 and F147, respectively (Fig. 2d). This binding configuration stabilizes the substrate via the π-electron delocalization among the arginine and phenylalanine side chains. Arginine 305 changed its conformation so that the inter-domain H-bond network involving Y263 and G171 was disrupted upon nitrite binding.
While it is clear that NarK and other NNP members are capable of NO3- uptake and NO2- export, it is not clear what the mechanism is or whether the process is proton coupled. Three distinct modes of action have been proposed: H+/NO3- symport, H+/NO2- antiport, or a NO3-/NO2- exchange without H+ translocation1,9,10.
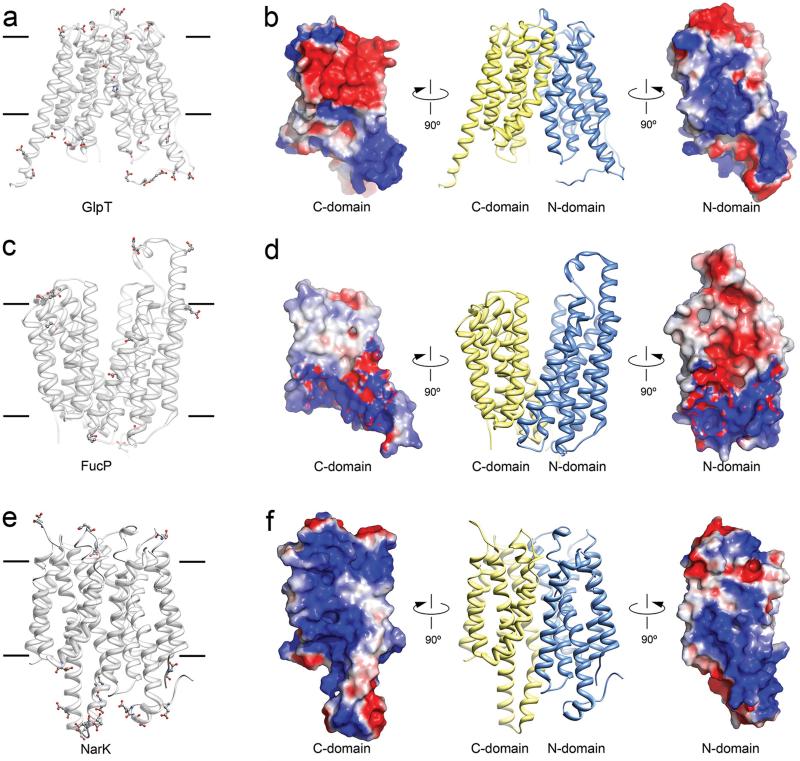
Typically, channels and transporters that translocate protons employ residues that are capable of protonation or deprotonation along the pore or substrate pathway12. For example, the lactose permease LacY that co-transports lactose with protons, uses glutamate and histidine residues to translocate the H+ 21. The glycerol-3-phosphate/phosphate antiporter GlpT uses a protonated histidine to facilitate substrate binding18 (Fig. 3a and b). The fucose transporter FucP uses glutamates and aspartates to translocate protons together with substrate16 (Fig. 3c and d). NirC, which is a H+ coupled nitrite channel has a functionally important histidine residue at the center of the channel22. In these four examples, glutamates, aspartates and/or histidines are found on transmembrane helices with their side chains extending into the substrate translocation pathway.
Fig. 3. Protons are likely excluded from the substrate translocation pathway of NarK.
a. Location of histidine, aspartate and glutamate residues in the anion transporter GlpT. Acidic residues line the substrate translocation pathway. b. Electrostatic surface representation for each domain of GlpT showing a relatively even distribution of positive and negative charges in the substrate translocation pathway. c. Location of histidine, aspartate and glutamate residues in the fucose transporter FucP. Acidic residues line the substrate translocation pathway. d. Electrostatic surface representation for each domain of FucP. e. Location of histidine, aspartate and glutamate residues in NarK. No acidic residues are found in the substrate translocation pathway of NarK. f. Electrostatic surface representation for NarK showing a dominantly positively charged substrate translocation pathway. It represents a formidable barrier for the translocation of H+ but could attract negatively charged molecules like nitrate and nitrite.
NarK does not contain glutamates, aspartates or histidines on its transmembrane helices. Instead, all glutamates and aspartates are found on either cytoplasmic or periplasmic soluble domains of NarK, well away from the substrate translocation pathway (Fig. 3e, balls and sticks). Moreover, NarK only contains three histidine residues and these are also found on soluble loops. Therefore NarK has no candidate residue for proton translocation or deprotonation in its substrate translocation pathway.
Consistent with the above postulate, surface electrostatic potential calculations in NarK indicate that the substrate translocation pathway is highly positively charged. The electrostatic potentials for the N- and C-terminal halves of NarK are presented in Fig 3f. The positively charged pathway can facilitate the transport of the negatively charged nitrate and nitrite anions but at the same time it would represent a formidable barrier for the translocation of protons. In sharp contrast, MFS members that couple the movement of substrate to the movement of protons, have a much more balanced electrostatic distribution in their translocation pathway (Fig 3). We note that it is possible that protons could be required for NarK activation, but our data suggests that NarK is a nitrate/nitrite exchanger in which protons are not co-transported with substrate. Moreover, it is still not clear whether NarK could function in nitrite transport alone and if so, whether this could be bi-directional. Further functional studies would have to be performed to answer these questions.
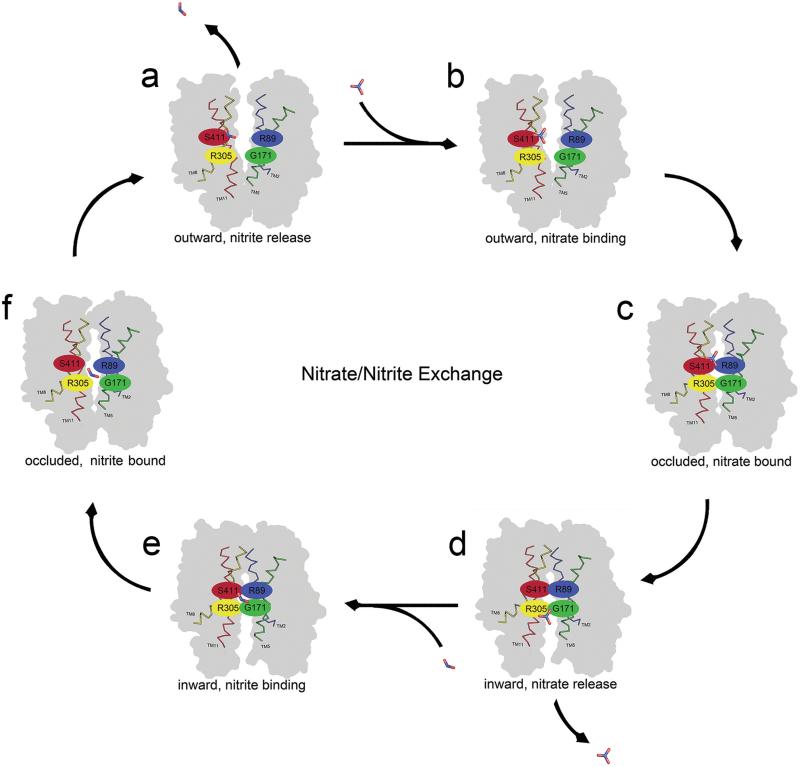
Our data argues that NarK functions as a nitrate/nitrite exchanger. We propose the following mechanism for nitrate/nitrite exchange. The mechanism is based on the structural analysis that is presented above, and the previously proposed rocker switch12 (Fig. 4). Our proposed mechanism of action begins with NarK in the outward conformation in which the substrate translocation pathway is open to the periplasm (Fig. 4a). The positively charged substrate translocation pathway can attract a nitrate molecule, which can enter the pore and bind at directly above the two arginines at the substrate-binding site (Fig. 4b and S4). There the nitrate forms H-bonds with R305 and N175 (Fig. 4b). The binding event could then trigger a conformational change in NarK into the transient occluded state where the pore is closed both at the periplasm and cytoplasm (Fig. 4c). The conformational change could push the nitrate from above R89 and R305 to directly below, and as the transporter adopts the inward conformation, its substrate translocation pathway opens to the cytoplasm and nitrate can then be released (Fig. 4d). As nitrate is released it is exchanged with nitrite. Nitrite enters the substrate translocation pathway and binds in plane with R89 and R305 (Fig. 2d and 4e). The binding of nitrite at the substrate-binding site triggers the conformational change of NarK from its inward facing conformation into the outward facing conformation via the transient occluded state (Fig. 4e and f). During this process nitrite is pushed directly above R89 and R305 and once NarK is outward facing the nitrite is released into the periplasm (Fig. 4a). The cycle of exchange can then continue.
Fig. 4. Proposed mechanism of nitrate/nitrite exchange.
Six conformations of NarK are depicted as a. outward facing, b. outward facing with nitrate bound, c. occluded with nitrate bound, d. inward facing with nitrate release, e. inward facing with nitrite bound, f. occluded with nitrite bound. Once NarK completes the cycle and returns to the outward facing conformation nitrite is released to the periplasm. The proposed mechanism is based on the rocker switch12. For NarK the rocker switch is facilitated by breaking and reforming inter-domain H-bonds involving R89 and R305 as described in Fig 2.
Such a rocker switch mechanism would require a hinge to allow the two halves of NarK to rock against one another as described above. We propose that the hinge is formed by the inter-domain H-bonds involving the conserved arginine residues R89 and R305. As discussed above, these two residues are stabilized by inter-domain H-bonding: Y263 from the C-terminal domain and G171 from the N-terminal domain of NarK make H-bonds with R305; and G144 of the N-terminal domain and S411 of the C-terminal domain of NarK make H-bonds with R89 (Fig. 2c). As the substrate interacts with R89 and R305, it would disrupt the H-bond networks between the two domains that are mediated by the arginines. Our structure of nitrite-bound NarK indicates that R305 undergoes a conformational change upon substrate binding consistent with a break in the inter-domain H-bonding network (Fig. 2d). We propose that it is the breaking and reforming of these inter domain H-bonds through the arginines that allow the two halves of NarK to rock against one another as depicted in Fig. 4. Consistent with this postulate, G171, G144, S411 and Y263 are residues that are conserved in NNP members (Fig. S2).
The rocker switch mechanism proposed in other MFS members whose structures are known involves the breaking and the formation of salt bridges and H-bonds between various protein residues 15-19,23. As discussed above, NarK contains no acidic residues in its pore so salt bridges could only form between protein residues and the substrate, but salt bridges between various protein residues are unlikely to be involved in its mechanism of action. Instead, the pore of NarK is highly positively charged, probably to exclude protons and to attract anions like nitrate and nitrite, while the rocking appears to involve the breaking and formation of inter domain H-bonds at the substrate binding pocket. It remains to be seen as more structures of MFS members from various families are determined if other members use a similar pattern of H-bond breaking and formation for their function.
Methods Summary
NarK from E. coli strain K12 was overexpressed in E. coli BL21 (DE3) C41. Fab antibody fragments were generated as described in Methods. NarK-Fab complex was purified in the presence of 0.2 % (w/v) n-decyl β-D-maltoside and crystallized in the following condition: 0.1M citric acid (pH 3.5), 0.1M NaCl, 0.1M Li2SO4 and 28% PEG400. Nitrite-bound NarK-Fab crystal was obtained by soaking the NarK-Fab crystal in the buffer containing 50mM sodium nitrite. Diffraction data sets of both crystals were collected at the Advanced Light Source (beamline 8.2.2). Data processing and structure determination were performed using the HKL2000, COOT and CCP4 programs.
Supplementary Material
Acknowledgements
We thank Edwin McCleskey (Howard Hughes Medical Institute) for critically reading this manuscript and for fruitful scientific discussions. We thank Daniel Cawley (Monoclonal antibody core facility, Oregon Health Sciences University) for development and production of monoclonal antibodies, and staff at the Advanced Light Source, Lawrence Berkeley National Laboratory for assistance with X-ray data collection. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Research in the Gonen laboratory is funded by the Howard Hughes Medical Institute.
Footnotes
Author contributions
H.Z and T.G designed the project. H.Z. performed all biochemical experiments including cloning, expression, purification, antibody production and binding assays, crystallization and X-ray data collection for both apo- and nitrite-bound NarK. H.Z. and G.W. built and refined the structures. All authors participated in data analysis and figure preparation. H.Z. and T.G. wrote the manuscript.
Author Information
Structures of substrate free and nitrite bound NarK have been deposited in PDB under accession numbers 4JR9 and 4JRE, respectively. Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no financial conflict related to this work.
Full Methods and associated references are available in the supplementary material.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
References
- 1.Wood NJ, Alizadeh T, Richardson DJ, Ferguson SJ, Moir JW. Two domains of a dual-function NarK protein are required for nitrate uptake, the first step of denitrification in Paracoccus pantotrophus. Mol Microbiol. 2002;44:157–170. doi: 10.1046/j.1365-2958.2002.02859.x. [DOI] [PubMed] [Google Scholar]
- 2.Martinez-Espinosa RM, Cole JA, Richardson DJ, Watmough NJ. Enzymology and ecology of the nitrogen cycle. Biochem Soc Trans. 2011;39:175–178. doi: 10.1042/BST0390175. doi:10.1042/BST0390175. [DOI] [PubMed] [Google Scholar]
- 3.Einsle O, Kroneck PM. Structural basis of denitrification. Biol Chem. 2004;385:875–883. doi: 10.1515/BC.2004.115. doi:10.1515/BC.2004.115. [DOI] [PubMed] [Google Scholar]
- 4.Saier MH, Jr., et al. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim Biophys Acta. 1999;1422:1–56. doi: 10.1016/s0304-4157(98)00023-9. [DOI] [PubMed] [Google Scholar]
- 5.Pao SS, Paulsen IT, Saier MH., Jr. Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jia W, Cole JA. Nitrate and nitrite transport in Escherichia coli. Biochem Soc Trans. 2005;33:159–161. doi: 10.1042/BST0330159. doi:10.1042/BST0330159. [DOI] [PubMed] [Google Scholar]
- 7.DeMoss JA, Hsu PY. NarK enhances nitrate uptake and nitrite excretion in Escherichia coli. J Bacteriol. 1991;173:3303–3310. doi: 10.1128/jb.173.11.3303-3310.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowe JJ, Ubbink-Kok T, Molenaar D, Konings WN, Driessen AJ. NarK is a nitrite-extrusion system involved in anaerobic nitrate respiration by Escherichia coli. Mol Microbiol. 1994;12:579–586. doi: 10.1111/j.1365-2958.1994.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 9.Moir JW, Wood NJ. Nitrate and nitrite transport in bacteria. Cell Mol Life Sci. 2001;58:215–224. doi: 10.1007/PL00000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia W, Tovell N, Clegg S, Trimmer M, Cole J. A single channel for nitrate uptake, nitrite export and nitrite uptake by Escherichia coli NarU and a role for NirC in nitrite export and uptake. Biochem J. 2009;417:297–304. doi: 10.1042/BJ20080746. doi:10.1042/BJ20080746. [DOI] [PubMed] [Google Scholar]
- 11.Wang YY, Hsu PK, Tsay YF. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012;17:458–467. doi: 10.1016/j.tplants.2012.04.006. doi:10.1016/j.tplants.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. doi:10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature. 2012;490:361–366. doi: 10.1038/nature11524. doi:10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 14.Solcan N, et al. Alternating access mechanism in the POT family of oligopeptide transporters. Embo J. 2012;31:3411–3421. doi: 10.1038/emboj.2012.157. doi:10.1038/emboj.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newstead S, et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. Embo J. 2011;30:417–426. doi: 10.1038/emboj.2010.309. doi:10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dang S, et al. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467:734–738. doi: 10.1038/nature09406. doi:10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 17.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–744. doi: 10.1126/science.1125629. doi:10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–620. doi: 10.1126/science.1087619. doi:10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 19.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. doi:10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 20.Trueman LJ, Richardson A, Forde BG. Molecular cloning of higher plant homologues of the high-affinity nitrate transporters of Chlamydomonas reinhardtii and Aspergillus nidulans. Gene. 1996;175:223–231. doi: 10.1016/0378-1119(96)00154-0. [DOI] [PubMed] [Google Scholar]
- 21.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. Embo J. 2006;25:1177–1183. doi: 10.1038/sj.emboj.7601028. doi:10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu W, et al. Structural and functional characterization of the nitrite channel NirC from Salmonella typhimurium. Proc Natl Acad Sci U S A. 2012;109:18395–18400. doi: 10.1073/pnas.1210793109. doi:10.1073/pnas.1210793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin L, et al. Sialin (SLC17A5) functions as a nitrate transporter in the plasma membrane. Proc Natl Acad Sci U S A. 2012;109:13434–13439. doi: 10.1073/pnas.1116633109. doi:10.1073/pnas.1116633109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.