Abstract
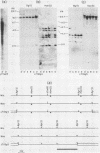
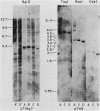
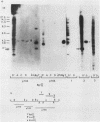
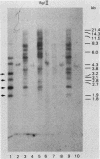
We analyzed the extent, reproducibility, and developmental control of genomic rearrangements in the somatic macronucleus of the ciliate Tetrahymena thermophila. To exclude differences caused by genetic polymorphisms, we constructed whole-genome homozygotes, and we compared the homozygous progeny derived from single macronuclear differentiation events. This strategy enabled us to identify a novel form of variable rearrangement and to confirm previous findings that rearranged sequences occur at a high frequency in the Tetrahymena genome. Rearrangements studied here were deletions of both unique and interchromosomally dispersed repetitive DNA sequences involving DNA rejoining of internal, nontelomeric regions of macronuclear DNAs. We showed that although rearrangements of some sequence classes are reproducible among independently developed macronuclei, other specific sequence classes are variably rearranged in macronuclear development. The variable somatic genomes so produced may be the source of phenotypically variant cell lines.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen S. L. Cytogenetics of genomic exclusion in Tetrahymena. Genetics. 1967 Apr;55(4):797–822. doi: 10.1093/genetics/55.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerberry C. F., Allis C. D., Yao M. C. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7383–7387. doi: 10.1073/pnas.81.23.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannon G. A., Bowen J. K., Yao M. C., Gorovsky M. A. Tetrahymena H4 genes: structure, evolution and organization in macro- and micronuclei. Nucleic Acids Res. 1984 Feb 24;12(4):1961–1975. doi: 10.1093/nar/12.4.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beermann S. The diminution of Heterochromatic chromosomal segments in Cyclops (Crustacea, Copepoda). Chromosoma. 1977 Apr 20;60(4):297–344. doi: 10.1007/BF00292858. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H., Budarf M. L., Challoner P. B., Cherry J. M., Howard E. A., Katzen A. L., Pan W. C., Ryan T. DNA termini in ciliate macronuclei. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1195–1207. doi: 10.1101/sqb.1983.047.01.135. [DOI] [PubMed] [Google Scholar]
- Bromberg S., Pratt K., Hattman S. Sequence specificity of DNA adenine methylase in the protozoan Tetrahymena thermophila. J Bacteriol. 1982 May;150(2):993–996. doi: 10.1128/jb.150.2.993-996.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunk C. F., Tsao S. G., Diamond C. H., Ohashi P. S., Tsao N. N., Pearlman R. E. Reorganization of unique and repetitive sequences during nuclear development in Tetrahymena thermophila. Can J Biochem. 1982 Sep;60(9):847–853. doi: 10.1139/o82-107. [DOI] [PubMed] [Google Scholar]
- Bruns P. J., Brussard T. B., Merriam E. V. Nullisomic Tetrahymena. II. a Set of Nullisomics Define the Germinal Chromosomes. Genetics. 1983 Jun;104(2):257–270. doi: 10.1093/genetics/104.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns P. J., Katzen A. L., Martin L., Blackburn E. H. A drug-resistant mutation in the ribosomal DNA of Tetrahymena. Proc Natl Acad Sci U S A. 1985 May;82(9):2844–2846. doi: 10.1073/pnas.82.9.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan R. C., Shalke G., Gorovsky M. A. Developmental rearrangements associated with a single type of expressed alpha-tubulin gene in Tetrahymena. Cell. 1984 Feb;36(2):441–445. doi: 10.1016/0092-8674(84)90237-x. [DOI] [PubMed] [Google Scholar]
- Emery H. S., Weiner A. M. An irregular satellite sequence is found at the termini of the linear extrachromosomal rDNA in Dictyostelium discoideum. Cell. 1981 Nov;26(3 Pt 1):411–419. doi: 10.1016/0092-8674(81)90210-5. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A. Genome organization and reorganization in Tetrahymena. Annu Rev Genet. 1980;14:203–239. doi: 10.1146/annurev.ge.14.120180.001223. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Hattman S., Pleger G. L. ( 6 N)methyl adenine in the nuclear DNA of a eucaryote, Tetrahymena pyriformis. J Cell Biol. 1973 Mar;56(3):697–701. doi: 10.1083/jcb.56.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovsky M. A. Macro- and micronuclei of Tetrahymena pyriformis: a model system for studying the structure and function of eukaryotic nuclei. J Protozool. 1973 Feb;20(1):19–25. doi: 10.1111/j.1550-7408.1973.tb05995.x. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Ostrander D. A., Hodnett J. L., Legerski R. J., Robberson D. L. Extracellular nucleases of Pseudomonas BAL 31. I. Characterization of single strand-specific deoxyriboendonuclease and double-strand deoxyriboexonuclease activities. Nucleic Acids Res. 1975 Sep;2(9):1459–1492. doi: 10.1093/nar/2.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan C. E., Warren G. J. Viability of palindromic DNA is restored by deletions occurring at low but variable frequency in plasmids of Escherichia coli. Gene. 1983 Oct;24(2-3):317–326. doi: 10.1016/0378-1119(83)90092-6. [DOI] [PubMed] [Google Scholar]
- Iwamura Y., Sakai M., Muramatsu M. Rearrangement of repeated DNA sequences during development of macronucleus in Tetrahymena thermophila. Nucleic Acids Res. 1982 Jul 24;10(14):4279–4291. doi: 10.1093/nar/10.14.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer K. M. Germ line-specific DNA sequences are present on all five micronuclear chromosomes in Tetrahymena thermophila. Mol Cell Biol. 1983 Nov;3(11):1909–1919. doi: 10.1128/mcb.3.11.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel A. R., Gorovsky M. A. Organization of the 5S RNA genes in macro- and micronuclei of Tetrahymena pyriformis. Chromosoma. 1978 Jun 23;67(1):1–20. doi: 10.1007/BF00285644. [DOI] [PubMed] [Google Scholar]
- King B. O., Yao M. C. Tandemly repeated hexanucleotide at Tetrahymena rDNA free end is generated from a single copy during development. Cell. 1982 Nov;31(1):177–182. doi: 10.1016/0092-8674(82)90417-2. [DOI] [PubMed] [Google Scholar]
- Klobutcher L. A., Jahn C. L., Prescott D. M. Internal sequences are eliminated from genes during macronuclear development in the ciliated protozoan Oxytricha nova. Cell. 1984 Apr;36(4):1045–1055. doi: 10.1016/0092-8674(84)90054-0. [DOI] [PubMed] [Google Scholar]
- Lawn R. M., Heumann J. M., Herrick G., Prescott D. M. The gene-size DNA molecules in Oxytricha. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):483–492. doi: 10.1101/sqb.1978.042.01.051. [DOI] [PubMed] [Google Scholar]
- Orias E., Bruns P. J. Induction and isolation of mutants in Tetrahymena. Methods Cell Biol. 1976;13:247–282. [PubMed] [Google Scholar]
- Pan W. C., Orias E., Flacks M., Blackburn E. H. Allele-specific, selective amplification of a ribosomal RNA gene in Tetrahymena thermophila. Cell. 1982 Mar;28(3):595–604. doi: 10.1016/0092-8674(82)90214-8. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Yao M. C., Kimmel A. R., Gorovsky M. A. Sequence organization within and flanking clusters of 5S ribosomal RNA genes in Tetrahymena. Nucleic Acids Res. 1984 Mar 26;12(6):3003–3021. doi: 10.1093/nar/12.6.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampay J., Szostak J. W., Blackburn E. H. DNA sequences of telomeres maintained in yeast. Nature. 1984 Jul 12;310(5973):154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Choi J., Yokoyama S., Austerberry C. F., Yao C. H. DNA elimination in Tetrahymena: a developmental process involving extensive breakage and rejoining of DNA at defined sites. Cell. 1984 Feb;36(2):433–440. doi: 10.1016/0092-8674(84)90236-8. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Gorovsky M. A. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48(1):1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Yao C. H. Repeated hexanucleotide C-C-C-C-A-A is present near free ends of macronuclear DNA of Tetrahymena. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7436–7439. doi: 10.1073/pnas.78.12.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama R. W., Yao M. C. Elimination of DNA sequences during macronuclear differentiation in Tetrahymena thermophila, as detected by in situ hybridization. Chromosoma. 1982;85(1):11–22. doi: 10.1007/BF00344591. [DOI] [PubMed] [Google Scholar]