Abstract
Aim
To explore the role of the ACE gene polymorphisms in the risk of essential hypertension in Mexican Mestizo individuals and evaluate the correlation between these polymorphisms and the serum ACE levels.
Methods
Nine ACE gene polymorphisms were genotyped by 5′ exonuclease TaqMan genotyping assays and polymerase chain reaction (PCR) in 239 hypertensive and 371 non- hypertensive Mexican individuals. Haplotypes were constructed after linkage disequilibrium analysis. ACE serum levels were determined in selected individuals according to different haplotypes.
Results
Under a dominant model, rs4291 rs4335, rs4344, rs4353, rs4362, and rs4363 polymorphisms were associated with an increased risk of hypertension after adjusting for age, gender, BMI, triglycerides, alcohol consumption, and smoking. Five polymorphisms (rs4335, rs4344, rs4353, rs4362 and rs4363) were in strong linkage disequilibrium and were included in four haplotypes: H1 (AAGCA), H2 (GGATG), H3 (AGATG), and H4 (AGACA). Haplotype H1 was associated with decreased risk of hypertension, while haplotype H2 was associated with an increased risk of hypertension (OR = 0.77, P = 0.023 and OR = 1.41, P = 0.004 respectively). According to the codominant model, the H2/H2 and H1/H2 haplotype combinations were significantly associated with risk of hypertension after adjusted by age, gender, BMI, triglycerides, alcohol consumption, and smoking (OR = 2.0; P = 0.002 and OR = 2.09; P = 0.011, respectively). Significant elevations in serum ACE concentrations were found in individuals with the H2 haplotype (H2/H2 and H2/H1) as compared to H1/H1 individuals (P = 0.0048).
Conclusion
The results suggest that single nucleotide polymorphisms and the “GGATG” haplotype of the ACE gene are associated with the development of hypertension and with increased ACE enzyme levels.
Introduction
Hypertension plays a major etiologic role in the development of cerebrovascular disease, ischemic heart disease, cardiac and renal failure. [1]. Essential hypertension is a complex disease where environmental, demographic, and genetic factors are involved [2], [3]: It has been estimated that approximately 30% of the interindividual variability in blood pressure is genetically determined [4]. Special attention has been given to the role of genetic variation in genes implicated in the renin-angiotensin system (RAS), particularly the angiotensin-converting enzyme (ACE) gene [5], [6]. ACE is a key enzyme in the renin-angiotensin-aldosterone (RAAS) and kalikrein-kinin systems [7], [8] playing a crucial role in blood pressure (BP) regulation and electrolyte balance [9]. The most studied polymorphism in the RAAS system is an insertion or deletion (I/D) of a 287 bp sequence of DNA in intron 16 of the ACE gene. It has been suggested that this polymorphism could explain up to 47% of total phenotypic variation in ACE serum levels and determines ACE enzyme activity [10]. ACE serum levels of D/D homozygous individuals are reported to be twice as high as those of I/I homozygous individuals, while I/D heterozygous individuals have intermediate ACE levels [11]. It is currently believed that the I/D polymorphism is not directly responsible for inherited ACE serum level variation in humans [10], [12], [13]. Tiret et al. demonstrated that this polymorphism is in close linkage disequilibrium to at least one, and perhaps more functional polymorphisms determining the phenotypic variations of enzyme levels [11]. Still, other studies have shown loci with variants of this sequence in complete linkage disequilibrium with the I/D polymorphism [14]. The role of ACE gene polymorphisms in hypertension has not been studied in the Mexican population. We selected 9 polymorphisms considering a previous study in which ACE haplotypes were described in African-American and European-American populations [15]. Population LD block differences were observed, as haplotypes were shorter and more diverse in Africans, while Europeans had longer and fewer haplotypes. This suggests that different linkage disequilibrium of the polymorphisms located in the ACE gene can be observed in other populations. On the other hand, the length of the genomic segment covered by the polymorphisms included in our study is 21 kb, increasing the possibility to detect some association with the disease. Thus, the objective of the present study was to analyze whether these polymorphisms or given haplotypes are associated with essential hypertension and ACE serum levels in Mexican Mestizo individuals.
Materials and Methods
Subjects
All participants provided written informed consent, and the study complies with the Declaration of Helsinki and was approved by the Ethics Committee of the Instituto Nacional de Cardiología ‘‘Ignacio Cháávez’’ (INCICH). This study included a group of 239 hypertensive individuals (120 men and 119 women) referred to the National Institute of Cardiology (INCICH) in Mexico City. Hypertension was defined as systolic blood pressure (BP) ≥140 mmHg, diastolic BP ≥90 mmHg, or the use of at least one class of antihypertensive drugs. One hundred hypertensive subjects (41.8%) taking antihypertensive drugs (64 patients one drug, 26 patients two drugs and 10 patients three drugs). The drugs used were ACE inhibitors in 37 patients, beta-blockers in 38, angiotensin II type-1 receptor antagonist in 33, diuretics in 28 and calcium channel antagonists in 18. Secondary hypertension was minimized using detailed health questionnaire and clinical evaluation, and none patient had evidence of cardiac or renal failure. A group of 371 non-hypertensive individuals (168 men and 203 women) were recruited among blood donors at the INCICH with systolic and diastolic blood pressures below 140 and 90 mmHg respectively. All participants were ethnically matched, defining Mexican Mestizos as individuals born in Mexico for three generations, including their own. All participants provided informed consent.
Genetic analysis
Genomic DNA from whole blood containing EDTA was isolated by standard techniques [16]. The A-239T (rs4291), A7941G (rs4318), A10539G (rs4335), A11599G (rs4343), A12292G (rs4344), A15990G (rs4353), C19329T (rs4362), and A20069G (rs4363) single nucleotide polymorphisms (SNPs) were genotyped using 5’ exonuclease TaqMan genotyping assays on an ABI Prism 7900HT Fast Real-Time PCR system, according to manufacturer’s instructions (Applied Biosystems, Foster City, CA, USA) (Table S1). The ACE polymorphism on intron 16 (I/D) was determined by PCR using the following primers: sense 5′-CTGCAGACCACTCCCATCCTTTCT-3′ and antisense 5′-GATGTGGCCATCACATTCGTCAGAT-3′ previously described [10]. Each sample found to have the D/D genotype was subjected to a second PCR amplification with insertion-specific primers (5a: 5′-TGGGACCACAGCGCCCGCCACTAC-3′ and 5c: 5′-TCGCCAGCCCTCCCATGCCCATAA-3′) in order to avoid D/D mistyping [17].
Pairwise linkage disequilibrium (LD, D) estimations between polymorphisms and haplotype reconstruction were performed with Haploview version 4:1 (Broad Institute of Massachusetts Institute of Technology and Harvard University, Cambridge, MA, USA).
Serum ACE levels
Serum ACE levels were measured in 169 hypertensive individuals selected according to haplotype (33 H1/H1, 94 H1/H2 heterozygous and 42 H2/H2 homozygous). ACE concentrations were determined with Boster’s human ACE ELISA Kit that was based on standard sandwich enzyme-linked immune-sorbent assay technology; measurements were in picograms per milliliter (pg/ml.).
Statistical analysis
Demographic and clinical variables between hypertensive and non-hypertensive groups were analyzed with Stata 8.0 for Windows software. Numerical variables not normally distributed are presented as median and percentiles 25 and 75, and were compared using Mann Whitney U test. Hardy-Weinberg equilibrium (HWE) and comparisons between categorical variables were analyzed using Chi2. Logistic regression analyses were used to test for associations of polymorphisms or haplotypes with hypertension under four inheritance models: co-dominant, dominant, recessive, and heterozygous advantage. The most appropriate inheritance model was selected based on Akaike information criteria (AIC) and was adjusted by age, gender, BMI, triglycerides, alcohol consumption, and smoking. Statistical power to detect association of polymorphisms with hypertension exceeded 0.80 as estimated with QUANTO software (http://hydra.usc.edu/GxE/).
Correlation between serum ACE concentration and haplotypes was tested using the Mann-Whitney U test. Statistical significance was set at P≤0.05.
Functional prediction analysis
We predicted the potential effect of the ACE polymorphisms associated with hypertension in our population using bioinformatics Tools, including FastSNP [18], SNP Function Prediction (http://snpinfo.niehs.nih.gov/snpfunc.htm), Human-transcriptome DataBase for Alternative Splicing (http://www.h-invitational.jp/h-dbas/), SplicePort (http://www.spliceport.cs.umd.edu/SplicingAnalyser2.html), ESE finder (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi), HSF (http://www.umd.be/HSF/) and SNPs3D (http://www.snps3d.org/).
Results
Characteristic of the study sample
Demographic and clinical characteristics of patients and controls are shown in Table 1. Body mass index, triglyceride levels, glucose levels and alcohol consumption were significantly higher in hypertensive than in non-hypertensive individuals; while mean age, sex ratio and cholesterol levels were similar in both groups.
Table 1. Comparison of cardiovascular risk factors in hypertensive and non-hypertensive individuals.
| Hypertensive (n = 239) | Non-hypertensive (n = 371) | P value | ||||||
| P25 | median | P75 | P25 | median | P75 | |||
| Age (years) | 51 | 58 | 63 | 52 | 56 | 62 | 0.066 | |
| Body-mass Index (kg/m2) | 26.8 | 29.3 | 32.1 | 24.5 | 27.1 | 29.8 | <10−3 | |
| Triglycerides (mg/dl) | 117 | 160 | 215 | 105 | 142 | 191 | 0.003 | |
| Glucose (mg/dl) | 88 | 95 | 108 | 83 | 90 | 115 | <10−3 | |
| Cholesterol (mg/dl) | 175.3 | 196 | 215 | 166 | 193 | 211 | 0.083 | |
| HDL(mg/dl) | 36 | 44 | 54 | 37 | 45 | 56 | 0.161 | |
| LDL(mg/dl) | 99.6 | 121.4 | 138 | 98 | 118.8 | 137 | 0.319 | |
| Blood Pressure (mmHg) | Systolic | 127 | 137.5 | 151 | 103 | 111.5 | 121 | <10−3 |
| Diastolic | 74.3 | 81.5 | 87.5 | 65 | 69.5 | 76 | <10−3 | |
| Gender (n(%)) | Male | 120(50.2) | 168(45.3) | 0.134 | ||||
| Female | 119(49.8) | 203(54.7) | ||||||
| Tobacco smoking (n(%)) | Yes | 43(18.0) | 104(28.0) | 0.003 | ||||
| Use of alcohol (n(%)) | Never used | 94 (39) | 177 (48) | 0.025 | ||||
| > = 6 g/day use | 145 (61) | 194 (52) | ||||||
| Type 2 diabetes mellitus (n(%)) | Yes | 33 (14) | 71 (19) | 0.054 | ||||
| Family history of hypertension (n(%)) | Yes | 121 (51.0) | 231 (62.0) | 0.005 | ||||
Data are expressed as median and percentiles 25 and 75.
P values were estimated using Mann-Whitney U-test continuous variables and Chi-square or Fisher test for categorical values.
Allele and genotype frequencies
No deviation from HWE was observed for rs4291, rs4318, rs4335, rs4344, rs4353, rs4362, and rs4363 polymorphisms; however, the rs4343 was not in HWE in the non-hypertensive individuals. Allele frequencies of rs4291, rs4335, rs4344, rs4353, rs4362 and rs4363 differed significantly between hypertensive and non-hypertensive individuals. According to the dominant model, rs4291 (OR = 1.5, 95%CI: 1.07–2.15, P = 0.02,), rs4335 (OR = 2.05, 95%CI: 1.39–2.99, P<10−3), rs4344 (OR = 1.82, 95%CI: 1.24–2.68, P = 0.002), rs4353 (OR = 1.80, 95%CI: 1.22–2.64, P = 0.003), rs4362 (OR = 1.71, 95%CI: 1.17–2.51, P = 0.006), and rs4363 (OR = 1.87, 95%CI: 1.27–2.74, P = 0.001,) were significantly associated with hypertension adjusting for age, gender, BMI, triglycerides, alcohol consumption, and smoking (Table 2).
Table 2. Associations of ACE polymorphisms with hypertension.
| Polymorphism | Alleles* | MAFa | MAFa | Genotypes | Genotypes | OR(95%CI); |
| HT | NHT | HT | NHT | Pdom value | ||
| A-239T (rs4291) | T/A | 0.35 | 0.29 | 25/118/96 | 34/149/188 | 1.51 (1.07–2.15); 0.020 |
| 0.11/0.49/0.40 | 0.09/0.40/0.51 | |||||
| A7941G (rs4318) | G/A | 0.01 | 0.02 | 0/7/232 | 0/12/359 | NS |
| 0.0/0.03/0.97 | 0.0/0.03/0.97 | |||||
| A10539G (rs4335) | G/A | 0.47 | 0.38 | 47/130/62 | 55/169/147 | 2.05 (1.39–2.99); <10−3 |
| 0.20/0.54/0.26 | 0.15/0.45/0.40 | |||||
| I/D (rs4646994) | I/D | 0.22 | 0.25 | 6/95/138 | 18/147/206 | NS |
| 0.02/0.40/0.58 | 0.05/0.40/0.55 | |||||
| A11599G (rs4343) | G/A | 0.42 | 0.37 | 41/117/81 | 39/196/136 | NS |
| 0.17/0.49/0.34 | 0.10/0.53/0.37 | |||||
| A12292G (rs4344) | G/A | 0.49 | 0.42 | 55/124/60 | 69/171/131 | 1.82 (1.24–2.68); 0.002 |
| 0.23/0.52/0.25 | 0.19/0.46/0.35 | |||||
| A159990G (rs4353) | A/G | 0.48 | 0.41 | 49/129/61 | 67/171/133 | 1.80 (1.22–2.64); 0.003 |
| 0.20/0.54/0.26 | 0.18/0.46/0.36 | |||||
| C19329T (rs4362) | T/C | 0.46 | 0.40 | 44/133/62 | 65/170/136 | 1.71 (1.17–2.51); 0.006 |
| 0.18/0.56/0.26 | 0.17/0.46/0.37 | |||||
| A20060G (rs4363) | G/A | 0.47 | 0.40 | 47/129/63 | 62/170/139 | 1.87 (1.27–2.74); 0.001 |
| 0.20/0.54/0.26 | 0.17/0.46/0.37 |
Adjusted for age, gender, BMI, triglycerides, alcohol consumption and smoking.
: MAF, minor allele frequency.
HT: Hypertensive individuals.
NHT: Non-hypertensive individuals.
NS: Not significant.
Underlined letter denotes the minor allele in the control samples.
SNP function prediction
Based on SNP functional prediction software’s, only the rs4335 (FastSNP software), rs4353 (FastSNP software), and rs4363 (ESE finder and HSF software’s) polymorphisms seem to be functional. The variation in these polymorphisms affects the DNA binding of the transcription factors AML-1, GATA-1, and Srp40, respectively.
Haplotype frequencies
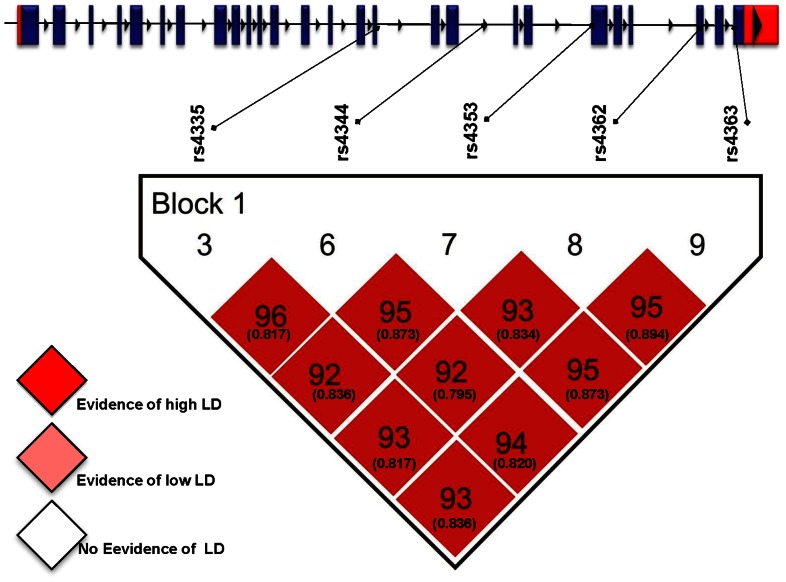
The linkage disequilibrium analysis was made considering the nine studied polymorphisms (Fig S1). Five polymorphism (rs4335, rs4344, rs4353, rs4362, rs4363) with high linkage disequilibrium (D’ values >0.9 and r2 values >0.8) were reanalyzed (Fig 1) and used to construct four haplotypes, H1 (AAGCA), H2 (GGATG), H3 (AGATG), and H4 (AGACA). The H1 haplotype was associated with decreased risk (OR = 0.77, 95%CI: 0.66–0.97, P = 0.023), while the H2 haplotype was associated with increased risk of hypertension (OR = 1.41, 95%CI: 1.11–1.80, P = 0.004) (Table 3). Afterwards, the most common haplotypes were tested for association with hypertension under different inheritance models. Under the codominant model H2/H2 and H1/H2 haplotype combinations were significantly associated with risk of hypertension after adjusted by age, gender, BMI, triglycerides, alcohol consumption, and smoking (Table 4).
Figure 1. Haploview linkage disequilibrium graph of five ACE gene polymorphisms (rs4335, rs4344, rs4353, rs4362, rs4363).
Pairwise linkage disequilibrium coefficients D ´× 100 are shown in each cell (D ´ values of 1.0 are not shown). Standard color scheme of Haploview was applied for linkage disequilibrium color display (logarithm of odds [LOD] score ≥2 and D ´ = 1, shown in bright red; LOD score ≥2 and D ´<1 shown in shades of pink/red; LOD score ≤2 and D ´<1 shown in white). D values of 1.0 are not shown. Numbers in boxes and parentheses represent D values and r2 values after the decimal point, respectively.
Table 3. ACE haplotype frequencies in hypertensive and non-hypertensive individuals.
| Haplotypes | Block 1 | HT | NHT | OR (95% CI) | P value |
| H1 | A-A-G-C-A | 0.489 | 0.556 | 0.77 (0.66–0.97) | 0.023 |
| H2 | G-G-A-T-G | 0.426 | 0.346 | 1.41 (1.11–1.80) | 0.004 |
| H3 | A-G-A-T-G | 0.011 | 0.022 | NS | — |
| H4 | A-G-A-C-A | 0.013 | 0.014 | NS | — |
HT, hypertensive; NHT, non-hypertensive; OR, odds ratio; CI, confidence interval; NS, not significant. The order of the polymorphisms in the haplotypes is according to the positions in the chromosome (rs4335, rs4344, rs4353, rs4362, rs4363).
Table 4. Risk assessment according to haplotypes using the codominant inherence model.
| Model | Haplotypes | OR | 95% CI | P value |
| H1/H1 | 1 | |||
| Co-dominant | H1/H2 | 2.00 | 1.294–3.118 | 0.002 |
| H2/H2 | 2.09 | 1.180–3.704 | 0.011 |
OR; odds ratio, CI; confidence interval.
Adjusted by age, gender, BMI, triglycerides, alcohol consumption, and smoking.
Correlation between haplotypes and serum ACE levels
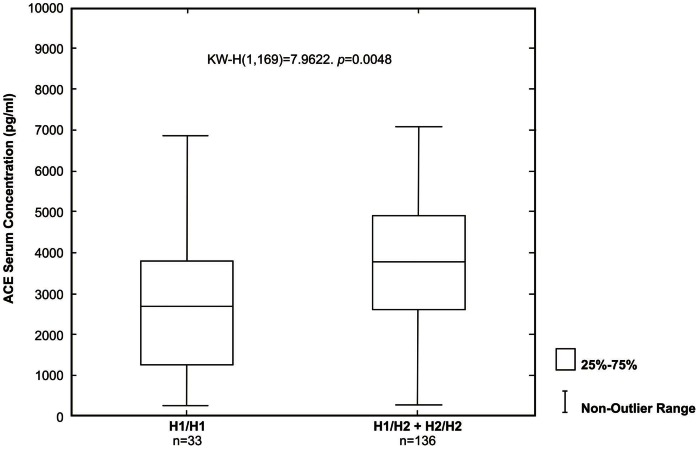
Serum ACE levels were analyzed in 33 hypertensive individuals homozygous for the H1 protective haplotype (H1/H1) and in 136 hypertensive individuals with at least one H2 risk haplotype (H1/H2 and H2/H2) (Fig. 2). Significant elevations in serum ACE concentrations were found in individuals with risk haplotypes (H2/H2 and H1/H2) (median 3802.37 pg/ml.) as compared with H1/H1 individuals (median 2692.73 pg/ml; P = 0.0048).
Figure 2. Graphic representation of serum ACE concentrations according to haplotypes.
The limits of the boxes represent the middle 50% of the data values; the extent of the lines encompass the interquartile range with extreme outlying data points shown as such. The central line within each box represents the median. H1/H1 = homozygous for protection, H1/H2 = heterozygous protection/risk and H2/H2 = homozygous for risk. Serum ACE concentrations were measured in patients without ACE inhibitors treatment.
Discussion
Essential hypertension involves interactions among genetic, environmental, demographic, vascular, and neuroendocrine factors. The etiologic role of genes implicated in the renin-angiotensin system (RAS) has been widely studied. Several clinical studies have reported an association between ACE polymorphisms and pathological phenotypes [19]–[28]. In the present study, we genotyped nine ACE gene polymorphisms in a group of Mexican individuals with and without essential hypertension. Six out of nine polymorphisms (rs4291, rs4335, rs4344, rs4353, rs4362 and rs4363) were associated with hypertension in our group of patients. These associations were independent from other hypertension-associated risk factors. The rs4343 polymorphism was not associated with hypertension perhaps because this polymorphism was not in HWE in the non-hypertensive individuals. Of the studied polymorphisms, only the rs4291 has been previously studied in patients with hypertension. Zhu et al. [29] reported that individuals with the rs4291 T allele showed increased blood pressure, in agreement with our observation in the Mexican population. On the other hand, the bioinformatics tools we used predicted that only the rs4335, rs4353 and rs4363 polymorphisms have a potential functional effect, and this variant is predicted to be in a binding site for the AML-1, GATA-1, and Srp40 transcription factors. The presence of specific alleles in these polymorphisms could result in loss (or reduction) of DNA binding of these transcriptional factors with important consequences in the expression of the enzyme. According to linkage disequilibrium analysis, five out of nine polymorphisms studied (rs4335, rs4344, rs4353, rs4362, rs4363) were in high linkage disequilibrium and four haplotypes were constructed, one of them associated with risk (H2: GGATG) and another with protection (H1: AAGCA) for hypertension. Zhu et al. [29] studied 10 polymorphisms in hypertensive patients from African-American and European-American populations. Our study included 8 of these 10 polymorphisms. In the study by Zhu et al. [29], a haplotype with three polymorphisms was associated with hypertension; however, the haplotype associated was different in African-American and European-American patients. In the African-Americans, the haplotype associated included the rs4343, rs4353, and rs4363 polymorphisms (AAA haplotype), whereas in European-Americans the haplotype included the rs4335, rs4343, and rs4344 polymorphisms (GGG haplotype). The risk haplotype in the Mexican Mestizo population includes the rs4335 G and rs4344 G alleles detected in European-American population and the rs4353 A allele detected in African-American individuals.
We also measured ACE serum levels to seek whether any haplotype was associated with this trait in hypertensive individuals. A limitation of our study is the fact that ACE levels were not measured in non-hypertensive individuals. Significant elevations in serum ACE concentrations were found in hypertensive individuals with the risk haplotypes when compared to individuals with the protective haplotype. This result supports the role of these haplotypes in the genetic susceptibility to developing essential hypertension. In a previous study, the relationship between the ACE levels and seven polymorphisms of the ACE gene was analyzed [30]. None of the polymorphisms studied by Zhu et al. [30] was included in our study. Zhu et al. [30] detected a haplotype associated with ACE levels. We suggest that these studies be extended to other ethnic groups to establish other regulatory regions in the ACE gene.
The most studied ACE gene polymorphism in this case is the insertion/deletion. In the study by Zhu et al. [12], two polymorphisms accounted for the variation of ACE concentration, A2350G had the most significant effect, accounting for 19% of the total variation in ACE, while rs4291 located in the 5′ section of the gene accounted for 6% of the variation. The effects of both these polymorphisms fitted best an additive model. After adjustment for the effect of A2350G, the I/D polymorphism was no longer associated with ACE, indicating that it is in LD with A2350G and unlikely to be a functional polymorphism. In this study, was established that the rs4291 A allele is associated with increased levels of ACE. This polymorphism was not associated with either hypertension or ACE levels in the Mexican Mestizo population. The association of other ACE polymorphisms with ACE levels is contradictory with positive and negative associations [31]–[34].
Conclusions
In summary, our results suggest that the rs4291, rs4335, rs4344, rs4353, rs4362, and rs4363 ACE gene polymorphisms might play an important role in the development of hypertension in the Mexican population. In our study, it was possible to identify a risk haplotype (H2: GGATG) and a protective haplotype (H1: AAGCA) for hypertension. Individuals with the risk haplotype showed increased ACE plasma levels as compared to individuals with the protective haplotype.
Supporting Information
Haploview linkage disequilibrium graph of nine ACE gene polymorphisms. Pairwise linkage disequilibrium coefficients D×100 are shown in each cell (D ´ values of 1.0 are not shown). Standard color scheme of Haploview was applied for linkage disequilibrium color display (logarithm of odds [LOD] score ≥2 and D = 1, shown in bright red; LOD score ≥2 and D ´<1 shown in shades of pink/red; LOD score ≤2 and D ´<1 shown in white). D values of 1.0 are not shown. Numbers in boxes and parentheses represent D values and r2 values after the decimal point, respectively.
(TIFF)
Genetic Polymorphisms studied in the ACE gene.
(DOC)
Acknowledgments
This work was submitted in partial fulfillment of the requirements for the Ph.D degree by Nancy L. Martínez-Rodríguez at the Graduate Studies in Biomedical Sciences of the Universidad Nacional Autónoma de México. The authors are grateful to the study participants.
Funding Statement
Nancy L. Martínez-Rodríguez was supported by a fellowship from the Consejo Nacional de Ciencia y Tecnología (CONACyT). This work was supported in part by grants from the Consejo Nacional de Ciencia y Tecnología (Project number 87356). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Collins R, Peto R, MacMahon S, Hebert P, Fiebach NH, et al. (1990) Blood pressure, stroke, and coronary heart disease. Part 2, short-term reductions in blood pressure: overview of randomized drug trials in their epidemiological context. Lancet 335: 827–838. [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization/International Society of Hypertension Guidelines for the management of hypertension (2003) J Hypertension. 17: 151–183. [PubMed] [Google Scholar]
- 3. Schork NJ (1997) Genetically complex cardiovascular traits. Origins, problems, and potential solutions. Hypertension 29: 145–149. [DOI] [PubMed] [Google Scholar]
- 4.Ward R (1995) Familial aggregation and genetic epidemiology of blood pressure. In: Laragh JH, Brenner BM, editors. Hypertension: Pathophysiology, Diagnosis and Management. New York: Raven Press. pp. 67–68.
- 5. Hubert C, Houot MA, Corvol P, Soubrier F (1991) Structure of the angiotensin I-converting enzyme gene. Two alternate promoters correspond to evolutionary steps of a duplicate gene. J Biol Chem 266: 15377–15383. [PubMed] [Google Scholar]
- 6. Mattei MG, Hubert C, Alhenc-Gelas F, Roeckel N, Corvol P, et al. (1989) Soubrier F. Angiotensin I converting enzyme gene is on chromosome 17. Cytogenet Cell Genet 51: 1041–1045. [Google Scholar]
- 7. Erdos EG, Skidgel RA (1987) The angiotensin I-converting enzyme. Lab Invest 56: 345–348. [PubMed] [Google Scholar]
- 8. Crisan D, Carr J (2000) Angiotensin I-converting enzyme, genotype and disease associations. J Mol Diagn 2: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Esther CR, Marino EM, Howard TE, Machaud A, Corvol P, et al. (1997) The critical role of tissue angiotensin-converting enzyme as revealed by gene targeting in mice. J Clin Invest 99: 2375–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rigat B, Hubert C, Alhenc-Gelas F, Cambien F, Corvol P, et al. (1990) An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest 86: 1343–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tiret L, Rigat B, Visvikis S, Breda C, Corvol P, et al. (1992) Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Genet 51: 197–205. [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu X, Bouzekri N, Southam L, Cooper RS, Adeyemo A, et al. (2001) Linkage and association analysis of angiotensin I- converting enzyme (ACE)-gene polymorphisms with ACE concentration and blood pressure. Am J Hum Genet 68: 1139–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davis GK, Roberts DH (1997) Molecular genetics of the renin-angiotensin system: implications for angiotensin II receptor blockade. Pharmacol Ther 75: 43–50. [DOI] [PubMed] [Google Scholar]
- 14. Rieder MJ, Taylor SL, Clark AG, Nikerson DA (1999) Sequence variation in the human angiotensin converting enzyme. Nature Genet 22: 59–62. [DOI] [PubMed] [Google Scholar]
- 15. Zhu X, Yan D, Cooper RS, Luke A, Ikeda MA, et al. (2003) Linkage disequilibrium and haplotype diversity in the genes of the rennin-angiotensin system. Findings from the family blood pressure program. Genome Res 13: 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller A (1993) A Single salting out procedure for extracting DNA from human nucleated cell. Nucleic Acid Res 16: 1215–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shanmugam V, Sell KW, Saha BK (1993) Mistyping ACE heterozygotes. PCR Methods Appl 3: 120–121. [DOI] [PubMed] [Google Scholar]
- 18.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, et al.. (2006) FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res 34 (Web Server issue): W635-41. Accessed 2012. [DOI] [PMC free article] [PubMed]
- 19. Kabadou IA, Soualmia H, Jemaa R, Feki M, Kallel A, et al. (2013) G protein beta3 subunit gene C825T and angiotensin converting enzyme gene insertion/deletion polymorphisms in hypertensive Tunisian population. Clin Lab 59: 85–92. [DOI] [PubMed] [Google Scholar]
- 20. Yadav S, Hasan N, Marjot T, Khan MS, Prasad K, et al. (2013) Detailed analysis of gene polymorphisms associated with ischemic stroke in South asians. PLoS One 8: e57305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sakai T, Shikishima K, Matsushima M, Tsuneoka H (2013) Genetic polymorphisms associated with endothelial function in nonarteritic anterior ischemic optic neuropathy. Mol Vis 19: 213–219. [PMC free article] [PubMed] [Google Scholar]
- 22. Yang SJ, Kim S, Park H, Kim SM, Choi KM, et al. (2013) Sex-dependent association between angiotensin-converting enzyme insertion/deletion polymorphism and obesity in relation to sodium intake in children. Nutrition 29: 525–530. [DOI] [PubMed] [Google Scholar]
- 23. Jing Q, Wang X, Ma Y, Yang M, Huang G, et al. (2013) Angiotensin-converting enzyme I/D polymorphism and the risk of thoracic aortic dissection in Chinese Han population. Mol Biol Rep 40: 1249–1254. [DOI] [PubMed] [Google Scholar]
- 24. Rasyid H, Bakri S, Yusuf I (2012) Angiotensin-converting enzyme gene polymorphisms, blood pressure and pulse pressure in subjects with essential hypertension in a South Sulawesi Indonesian population. Acta Med Indones 44: 280–283. [PubMed] [Google Scholar]
- 25. Dhar S, Ray S, Dutta A, Sengupta B, Chakrabarti S (2012) Polymorphism of ACE gene as the genetic predisposition of coronary artery disease in Eastern India. Indian Heart J 64: 576–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang W, Dong X, Li B, Zhang XQ, Zeng Y, et al. (2012) Correlation of angiotensin converting enzyme gene polymorphism with perioperative myocardial protection under extracorporeal circulation. Asian Pac J Trop Med 5: 995–999. [DOI] [PubMed] [Google Scholar]
- 27. Zhang XL, Wu LQ, Liu X, Yang YQ, Tan HW, et al. (2012) Association of angiotensin-converting enzyme gene I/D and CYP11B2 gene -344T/C polymorphisms with lone atrial fibrillation and its recurrence after catheter ablation. Exp Ther Med 4: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Z, Xu G, Liu D, Fan X, Zhu W, et al. (2012) Angiotensin-converting enzyme insertion/deletion polymorphism contributes to ischemic stroke risk: a meta-analysis of 50 case-control studies. PLoS One 7: e46495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu X, Chang YP, Yan D, Weder A, Cooper R, et al. (2003) Association between hypertension and genes in the rennin-angiotensin system. Hypertension 41: 1027–1034. [DOI] [PubMed] [Google Scholar]
- 30. Zhu X, McKenzie CA, Forrester T, Nickerson DA, Broeckel U, et al. (2000) Localization of a small genomic region associated with elevated ACE. Am J Hum Genet 67: 1144–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Villard E, Tiret L, Visvikis S, Rakotovao R, Cambien F, et al. (1996) Identification of new polymorphism of the angiotensin I-converting enzyme (ACE) gene, and study of their relationship to plasma ACE levels by two QTL segregation linkage analysis. Am J Hum Genet 58: 1268–1278. [PMC free article] [PubMed] [Google Scholar]
- 32. Keavney B, McKenzie CA, Connell JM, Julier C, Ratcliffe PJ, et al. (1998) Measured haplotype analysis of the angiotensin-I converting enzyme (ACE) gene. Hum Mol Genet 7: 1745–1751. [DOI] [PubMed] [Google Scholar]
- 33. Baghai TC, Binder EB, Schule C, Salyakina D, Eser D, et al. (2006) Polymorphisms in the angiotensin-converting enzyme gene are associated with unipolar depression, ACE activity and hypercortisolism. Mol Psychiatry 11: 1003–1015. [DOI] [PubMed] [Google Scholar]
- 34. Chung CM, Wang RY, Chen JW, Fann CS, Leu HB, et al. (2010) A genome-wide association study identifies new loci for ACE activity: potential implications for response to ACE inhibitor. Pharmacogenomics J 10: 537–544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Haploview linkage disequilibrium graph of nine ACE gene polymorphisms. Pairwise linkage disequilibrium coefficients D×100 are shown in each cell (D ´ values of 1.0 are not shown). Standard color scheme of Haploview was applied for linkage disequilibrium color display (logarithm of odds [LOD] score ≥2 and D = 1, shown in bright red; LOD score ≥2 and D ´<1 shown in shades of pink/red; LOD score ≤2 and D ´<1 shown in white). D values of 1.0 are not shown. Numbers in boxes and parentheses represent D values and r2 values after the decimal point, respectively.
(TIFF)
Genetic Polymorphisms studied in the ACE gene.
(DOC)