Abstract
Cytogenetic analysis is an informative classical approach to understanding the relationships among members in a group of closely related species of mosquitoes. Anopheles ovengensis is a recently discovered species of the An. nili group and is one of the important malaria vectors in the African equatorial forest. This study characterized polytene chromosomes of An. ovengensis and compared them with polytene chromosomes of An. nili. Using fluorescent in situ hybridization and chromosome banding pattern comparison we have established correspondence between chromosomal arms of An. ovengensis and An. nili. Analysis of chromosome morphology in the two species revealed a limited similarity in the banding patterns. The most extensive reorganization occurs in pericentromeric and intercalary heterochromatin. Chromosomes of An. ovengensis are joined together by a diffuse chromocenter and they have two large regions of intercalary heterochromatin in arms 2L and 3R. In contrast, the chromocenter and intercalary heterochromatin are not seen in An. nili chromosomes. Comparative analysis of the arm association suggests the occurrence of a whole-arm translocation between the two members of the group. The observed, substantial reorganizations of chromosome structure implies either a rapid rate of chromosome evolution in the An. nili group, or that the two species belong to different taxonomic groups within subgenus Cellia.
Keywords: malaria mosquito, Anopheles ovengensis, Anopheles nili, polytene chromosomes, fluorescent in situ hybridization
1. Introduction
Malaria has a devastating impact on public health and welfare on the African continent. It is now becoming clear that the end point of all efforts to reduce the current malaria impact must be the eventual elimination of the disease (Enayati and Hemingway, 2010). Vector control is seen as a cornerstone of the malaria control strategy. Because of economic and practical reasons, vector control mainly relies on the use of synthetic insecticides (Takken and Knols, 2009). However, this strategy is inefficient if all major vector species are not targeted. Many malaria vectors belong to species complexes or groups, and members within these complexes or groups can vary significantly in their vectorial capacity. Moreover, species can be further subdivided into populations adapted to different environments. Some malaria control initiatives have failed because they targeted the wrong species or population (Coluzzi, 1992; Van Bortel et al., 2001). Understanding and targeting the heterogeneity and complexity of all major vector species and populations is necessary for effective vector control and malaria elimination (Enayati and Hemingway, 2010). Anopheles gambiae, An. arabiensis, An. funestus, An. moucheti, and An. nili are the major malaria vectors in sub-Saharan Africa because they are anthropophilic and susceptible to Plasmodium falciparum (Fontenille and Simard, 2004). Most studies of African malaria vectors have involved the An. gambiae complex and, to a lesser extent, the An. funestus group, in part, because molecular and cytogenetic tools for characterizing population structure, ecological adaptation, and taxonomic status of species are available for these species. Similar tools for the An. nili group have been lacking until recently (Berthomieu et al., 2003; Kengne et al., 2003; Peery et al., 2011; Sharakhova et al., 2011). This represented a critical barrier to progress in the field of vector biology and control because members of the An. nili group contribute substantially to malaria transmission in African humid savannah and forested areas. The recent findings of circulation of P. falciparum along with other Plasmodium species in great apes and monkeys (Duval et al., 2010; Prugnolle et al., 2010; Prugnolle et al., 2011) raise concerns about pathogen transfer between humans and primates and highlight the need to improve our knowledge of forest malaria vectors.
Although correct species identification is crucial for successful vector control, the taxonomic status of members of the An. nili group remains unclear. Because of this knowledge gap, the distribution, behavior, adaptation, and role in malaria transmission that can be attributed to each member of this group is also largely unknown (Fontenille and Simard, 2004). Analysis of sequence variation in the ribosomal DNA second internal transcribed spacer (ITS2) and D3 28S region allowed identification of four species within the An. nili group (namely, An. nili s.s., An. somalicus, An. carnevalei, and An. ovengensis) (Kengne et al., 2003). A comprehensive study in Cameroon confirmed that An. nili s.s. (hereafter An. nili) is the major malaria vector of the group and is widespread in humid savannah and degraded forest environments (Antonio-Nkondjio et al., 2006). This study has emphasized the exophagic behavior of An. ovengensis and An. carnevalei. It has also demonstrated that An. ovengensis is abundant in deep intact forests of Central Africa, where it substantially contributes to malaria transmission (Antonio-Nkondjio et al., 2006). In Equatorial Guinea, sporozoite rates in An. ovengensis can reach 4.1% (n=74), which is higher than that of An. gambiae in the same area (3.3%, n=603) (Ridl et al., 2008), confirming a major yet overlooked role for An. ovengensis in malaria epidemiology in these settings.
A recent study used a combination of nuclear (microsatellite and ribosomal DNA) and mitochondrial DNA markers to explore the levels of genetic polymorphism and divergence among species of the An. nili group in the savannah and forested areas of Cameroon (Ndo et al. PLoS ONE, in press). The study detected a large number of fixed mutations between An. nili and An. ovengensis, as well as among other members of the group. The genetic distance has been estimated 4 to 8 fold higher than that commonly reported among cryptic Anopheles species. This high genetic divergence within the An. nili group suggests that its members might belong to different species groups. The aim of our study was to perform the first cytogenetic analysis of An. ovengensis and to compare structural organization of polytene chromosomes in An. ovengensis and An. nili.
2. Materials and methods
2.1. Wild mosquito collection, preservation, and species identification
Anopheles ovengensis and An. nili adult females were collected by pyrethrum spraying and bednet traps in the villages of Nyabessan in Cameroon (2°80’N; 10°25’E) and Dinderesso (11°14'N; 4°23'W) in Burkina Faso, respectively. Specimens were identified in the field as members of the An. nili group by using morphological identification keys (Awono-Ambene et al., 2004; Gillies and Coetzee, 1987) and were further characterized by molecular assays as An. ovengensis and An. nili (Kengne et al., 2003). Semi-gravid females were dissected under a microscope, and their ovaries, at the appropriate stage, were preserved in Carnoy's fixative solution (3 methanol: 1 glacial acetic acid by volume). Tissues of both species were stored under identical conditions and were processed using the same protocol.
2.2. Chromosome preparation and imaging
Ovaries from half-gravid females stored in Carnoy's fixative solution were dissected in 50% propionic acid under a Leica MZ6 dissection microscope (Leica Microsystems GmbH, Wetzlar, Germany). A cover slip was placed on the follicles and tapped with a pencil to squash the cells. Preparations of semi-squashed nuclei were obtained by placing a coverslip on the follicles in a drop of 50% propionic acid followed by very gentle tapping. Hard tapping on the coverslip obtained preparations of fully squashed nuclei. The banding pattern of polytene chromosomes was examined using an Olympus CX-41 phase-contrast microscope (×1000) (Olympus America Inc., Melville, NY, USA). Slides with good chromosomal preparations were dipped in liquid nitrogen, then cover slips were removed and slides were dehydrated in 50%, 70%, 95%, and 100% ethanol. Chromosomes that showed a suitable level of polytenization were imaged by an Olympus BX-41 with an attached Olympus Q-Color 5 camera and Q-Imaging software (Olympus America Inc., Melville, NY, USA). The six best chromosomal slides were utilized to develop a preliminary map of An. ovengensis. Images of the chromosomes were combined, straightened, shaped, and cropped using Adobe® Photoshop. Chromosome preparations from 60 An. ovengensis females and from 100 An. nili females were analyzed for this study.
2.3. Fluorescence in situ hybridization
Primers were designed using the Primer3 program (Rozen and Skaletsky, 2000) and gene sequences from the An. gambiae genome assembly. PCR products ranged from 400–600 bp in size. The genomic DNA of single An. nili mosquitoes was extracted using the Wizard SV Genomic Purification System (Promega Corporation, Madison, WI, USA) and was used as a template for PCR. PCR products were gel purified using the Geneclean kit (Qbiogene, Inc., Irvine, CA). The fluorescence in situ hybridization (FISH) procedure was conducted as previously described (Sharakhova et al., 2006). The DNA was labeled with Cy3-dUTP and Cy5-dUTP (GE Healthcare UK Ltd., Buckinghamshire, England) using Random Primers DNA Labeling System (Invitrogen Corporation, Carlsbad, CA, USA). DNA probes were hybridized to the chromosomes of An. ovengensis at 39°C overnight in hybridization solution (Invitrogen Corporation, Carlsbad, CA, USA). Then the chromosomes were washed in 0.2XSSC (Saline-Sodium Citrate: 0.03M Sodium Chloride, 0.003M Sodium Citrate), counterstained with YOYO-1, and mounted in DABCO. Fluorescent signals were detected and recorded using a Zeiss LSM 510 Laser Scanning Microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY, USA).
3. Results
3.1. Structure of polytene chromosomes in An. ovengensis
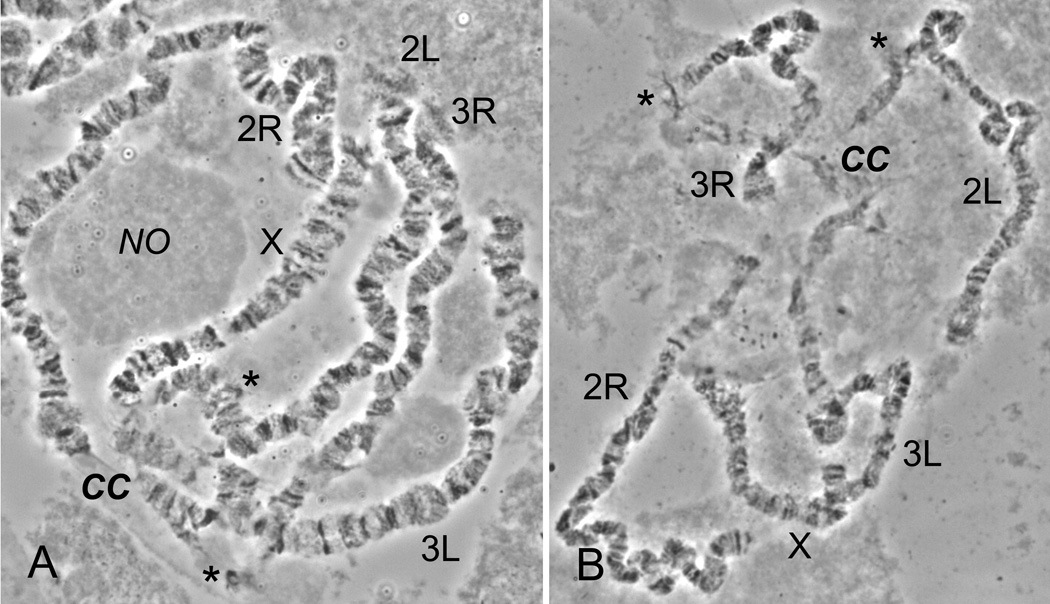
We analyzed readable polytene chromosomes from the ovarian nurse cells of 44 An. ovengensis females obtained from a total of 60 chromosome preparations. The polytene chromosome complement consists of chromosome X and four autosomal arms. Pericentromeric regions of the polytene chromosomes in An. ovengensis form a diffuse chromocenter (Fig. 1A). Chromosome X can be easily distinguished from other chromosomes by having the shortest length. Arm 2R is the longest among the five arms and can be recognized by the presence of several distinct bands in the telomere. The major landmarks for arm 2L are a light telomere and a region of intercalary heterochromatin located approximately six bands apart from the centromere. Arm 3R has a large region of intercalary heterochromatin also located approximately six bands distally from the centromere. The regions of intercalary heterochromatin are indicated by asterisks in Fig. 1. Unlike arm 2L, arm 3R often displays asynapsis of homologous chromosomes in the region of intercalary heterochromatin and the chromosome can be easily broken in this region by squashing (Fig. 1B). Arm 3L has a lightly flared telomeric region and several large puffs. We detected no polymorphic inversions among our samples of An. ovengensis.
Fig. 1.
Polytene chromosomes from ovarian nurse cells of An. ovengensis. Semi-squashed (A) and fully squashed (B) unstained nuclei are imaged with a phase-contrast microscopy. The chromocenter located at the nuclear periphery, CC, and nucleolus, NO, are shown in (A). The spread-out chromocenter is shown in (B). Regions of intercalary heterochromatin are marked by asterisks.
3.2. Chromosome arm associations in An. ovengensis
The assignment of chromosomal arms was done based on their relative length and associations. Because pericentromeric regions of the polytene chromosomes in An. nili do not form a chromocenter, it was easy to determine arm association in our previous study (Sharakhova et al., 2011). We found only one type of arm association in An. nili: 2R + 2L, 3R + 3L. In contrast, pericentromeric heterochromatin of the polytene chromosomes in An. ovengensis forms a diffuse chromocenter, which makes it more difficult to determine arm association. A chromocenter is a structure where heterochromatic pericentromeric regions of all polytene arms join together by ectopic contacts. In contrast, arms of mitotic chromosomes do not join with each other in a chromocenter, but they are associated as the following: 2R + 2L, 3R + 3L; the X chromosome is not associated with the autosomes. The organization of pericentromeric regions in a chromocenter is not uncommon feature of polytene chromosomes from ovarian nurse cell nuclei of mosquitoes. For example, the compact chromocenter has been observed in ovarian nurse cell nuclei of An. funestus (Sharakhov et al., 2001). When cell nuclei are squashed during chromosome preparation, the chromocenter loses its integrity and breaks up. Although this breakage may result in all possible arm associations, the real (not ectopic) arm associations are expected to be more frequent. To identify autosomal arm pairs with prevalent association we have analyzed 43 chromosomes spreads with a broken chromocenter in 10 squashed chromosome preparations (Fig. 1B). Although the analysis revealed all six possible variants of pair-wise associations in An. ovengensis (2R + 2L, 2R + 3R, 2R + 3L, 3R + 2L, 2L + 3L, and 3R + 3L) the associations 2R + 2L and 3R + 3L were more prevalent (Table 1). Pearson’s χ2 test indicates that the observed arm associations are significantly different than expected by random chance (χ2 = 12.6, d.f. = 5, P < 0.026).
Table 1.
Frequencies of chromosome arm associations in ovarian nurse cells of An. ovengensis.
| Arm association type | |||||||
|---|---|---|---|---|---|---|---|
| 2R+3L | 2R+2L | 2R+3R | 3L+2L | 3R+3L | 3R+2L | Total | |
| Number of cases | 3 | 13 | 6 | 4 | 12 | 5 | 43 |
| Frequency (%) | 7.0 | 30.2 | 14.0 | 9.3 | 27.9 | 11.6 | 100 |
3.3. Chromosome arm homology among An. ovengensis, An. nili and An. gambiae
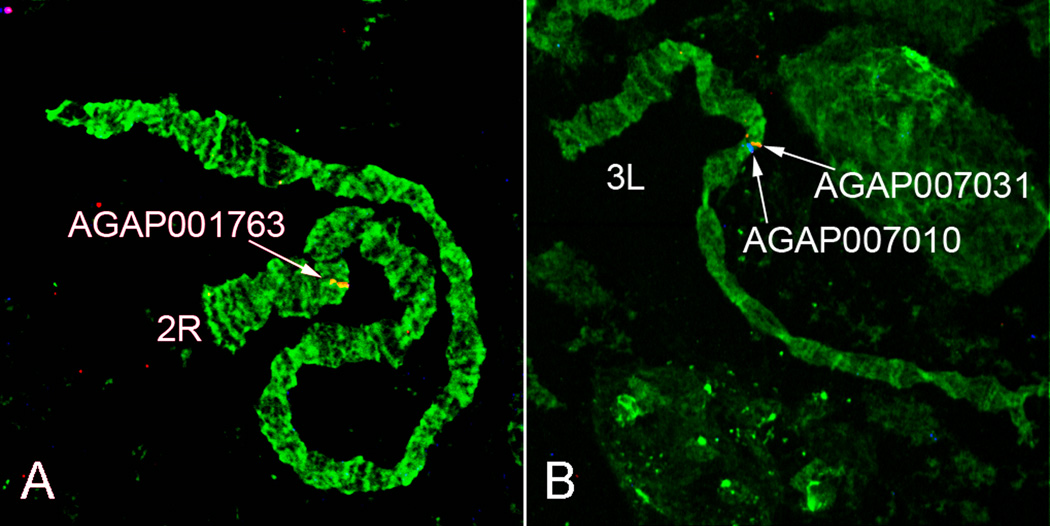
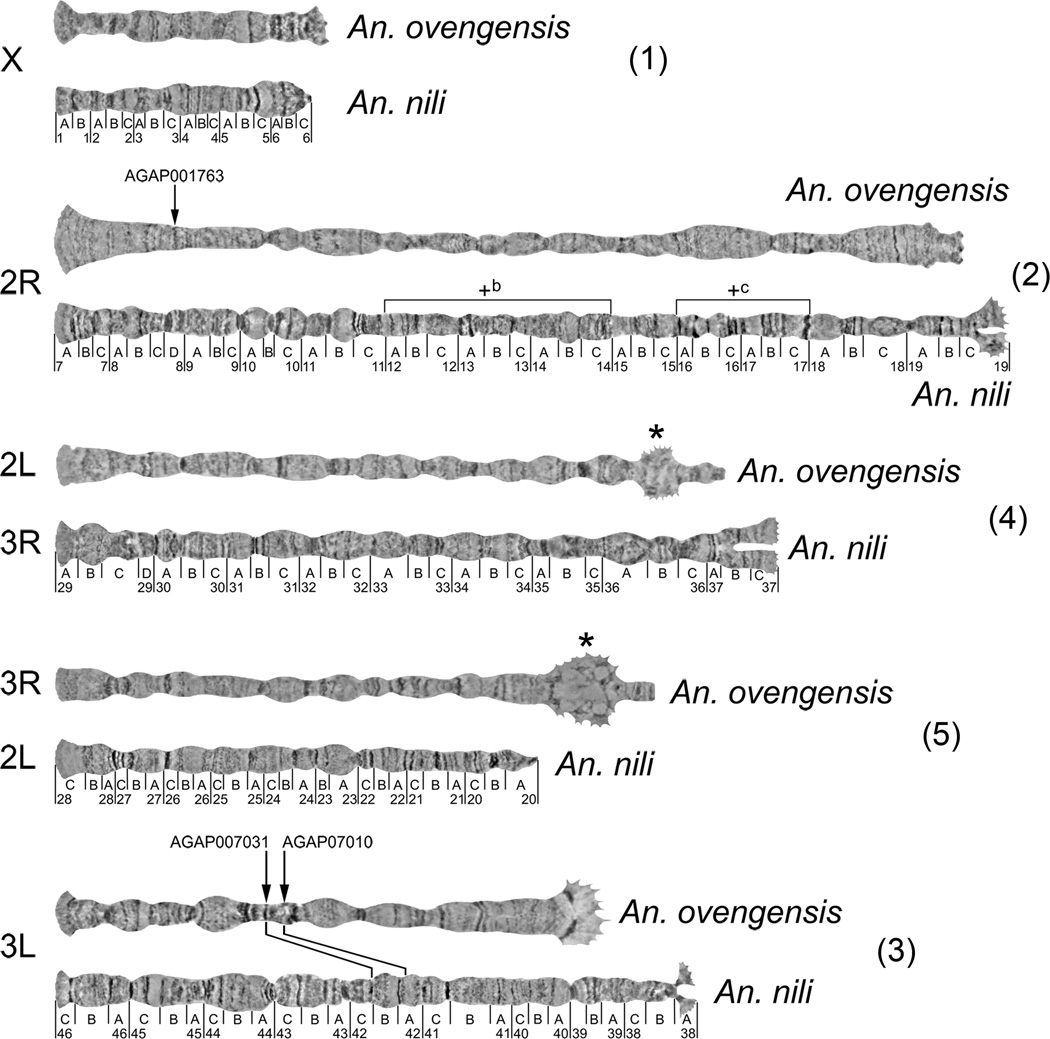
The appearance of An. ovengensis chromosomes is quite different from that of An. nili or An. gambiae chromosomes. The sex chromosome X, as the shortest, is obviously homologous among the three species. We established autosomal arm homology by successful FISH of three DNA probes derived from An. gambiae genes. Probe AGAP001763 was from arm 2R of An. gambiae, probes AGAP007010 and AGAP007031 were from arm 2L of An. gambiae. We mapped AGAP001763 to arm 2R while AGAP007010 and AGAP007031 mapped to arm 3L in An. ovengensis (Fig. 2). In our previous study, we labeled 25 gene fragments derived from arms 2R and 2L in An. gambiae and mapped them to chromosomes 2R and 3L in An. nili, including AGAP007010 and AGAP007031 (Sharakhova et al., 2011). Thus, we demonstrated that the 2R arms are homologous among the three species and the 2L arm of An. gambiae corresponds to the 3L arms in An. ovengensis and An. nili. We also established homology between polytene chromosomes 3R of An. ovengensis and 2L of An. nili based on the similarity in the banding pattern. The apparently identical banding pattern starts in subtelomeric regions and extends to subdivision 22C on 2L on the An. nili map (Sharakhova et al., 2011) (Fig. 3). For comparative purposes we adopted the system of autosomal arm notation proposed by (Green and Hunt, 1980). Accordingly, the autosomal arms in An. gambiae are named as the following: 2R = 2, 2L = 4, 3R = 3, and 3L = 5. We propose to name the sex chromosome X as element 1 in agreement with nomenclature of Anopheles autosomes (Green and Hunt, 1980). Likewise, H. J. Muller named chromosomal elements of Drosophila as A (chromosome X) and B, C, D, E, F (autosomes) (Muller, 1940).
Fig. 2.
FISH of DNA probes homologous to the An. gambiae gene sequences with polytene chromosomes 2R (A) and 3L (B) of An. ovengensis. Probes were labeled with Cy3, red, and Cy5, blue, fluorochromes. Arrows indicate signals of hybridization.
Fig. 3.
Comparison of the banding pattern of polytene chromosomes between An. ovengensis and An. nili. Chromosomal arm homology between the species is shown by numbers in parentheses. Arrows indicate localization of DNA probes homologous to the An. gambiae gene sequences on chromosomes of An. ovengensis. Intercalary heterochromatin in An. ovengensis is shown by asterisks. Lines between chromosomes 3L connect homologous markers mapped in both species. Polymorphic inversions in An. nili are shown by brackets above the 2R arm. Pericentromeric regions of chromosomes are oriented to the right. Unstained chromosome images were taken with a phase-contrast microscopy. The chromosomes are ordered according to the arm notation in An. ovengensis.
Therefore, elements 1 and 2 correspond to the chromosomes X and 2R in An. ovengensis, An. nili and An. gambiae. Element 3 corresponds to arm 3L in An. ovengensis and An. nili (Fig. 2, 3) but to arm 2L in An. gambiae (Sharakhova et al., 2011). In contrast, element 4 has the same notation in An. nili and An. gambiae (arm 3R) but a different notation in An. ovengensis (arm 2L). Finally, element 5 corresponds to arm 3R in An. ovengensis, arm 2L in An. nili and arm 3L in An. gambiae. Thus, our data suggest a whole-arm translocation between chromosome elements 2 and 3 in An. nili and An. ovengensis.
3.4. Structural divergence of chromosomes in the An. nili group
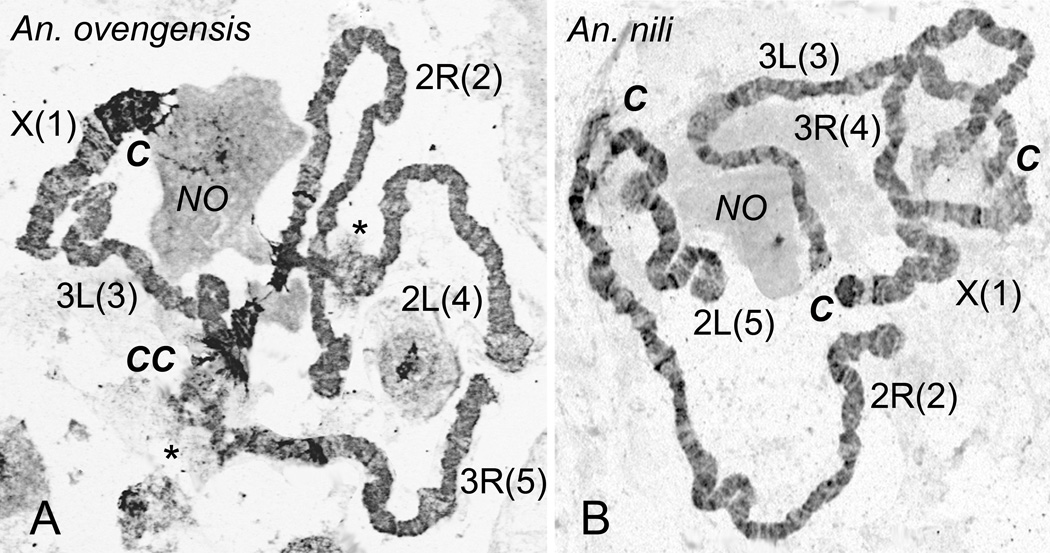
Our analysis of chromosomes stained with a fluorescent dye, YOYO-1, revealed remarkable difference in pericentromeric heterochromatin between An. ovengensis and An. nili (Fig. 4). Based on chromosome morphology, we identified two types of heterochromatin: dark compact, and light diffuse. There was more heterochromatin in An. ovengensis chromosomes than in An. nili chromosomes. Anopheles ovengensis has large blocks of dark compact heterochromatin in pericentomeric regions of all chromosomes. While only the X chromosome has a large block of dark compact heterochromatin in An. nili. The two species were also different with respect to the morphology of the pericentromeric regions of autosomes (Fig. 3). The pericentromeric regions of chromosome elements 2, 3 and 4 were typically asynaptic in An. nili but not in An. ovengensis. In addition, chromosome arms 2L and 3R of An. ovengensis displayed large regions of light diffuse intercalary heterochromatin. This type of heterochromatin (both pericentomeric and intercalary) forms attachments to the nuclear periphery (Fig. 1, 4). Because the chromosomal location of nuclear envelope contacts differs between An. ovengensis and An. nili and because the pericentromeric regions of different arms stay closer to each other in An. ovengensis than in An. nili, the spatial organization of chromosomes is expected to be different between the two species. The regions of intercalary heterochromatin and the occurrence of asynapsis were consistent between nuclei and between specimens.
Fig. 4.
Differences in organization of polytene chromosomes between An. ovengensis and An. nili. A) Chromosomes of An. ovengensis form a chromocenter, CC, and display a large amount of heterochromatin—dark and granular areas of chromosomes. Asterisks indicate regions of intercalary heterochromatin. B) Pericentromeric regions, C, of An. nili do not form a chromocenter and display a small amount of heterochromatin. Chromosomal arm homologous between the species are shown with numbers in parentheses. NO—nucleolus. The preparations were stained with YOYO-1. Chromosome images were taken with a fluorescent microscope and converted into grayscale inverted images.
The euchromatic portion of polytene chromosomes was also found to be significantly divergent between An. ovengensis and An. nili (Fig. 3). We determined only limited similarity in banding patterns between all homologous chromosome elements of the two species except in element 5. The likely reason for this poor homology between the two maps is a large number of fixed chromosomal inversions between An. ovengensis and An. nili. However, the number and locations of the fixed inversions could not be precisely determined because of the limited similarity of the banding pattern. Extensive physical mapping on chromosomes in both species can be performed to identify the fixed inversions.
4. Discussion
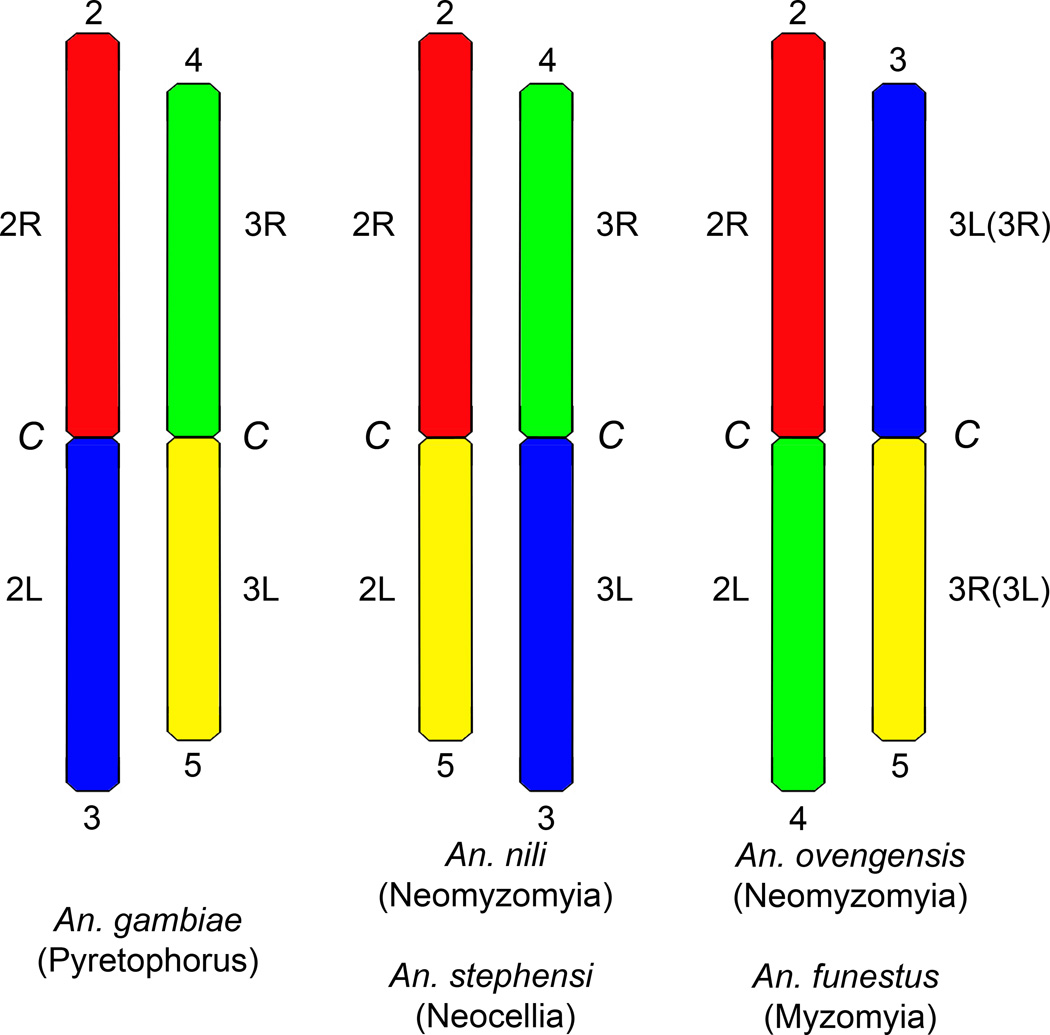
Cytogenetic studies of polytene chromosomes have been useful for understanding population genetics, taxonomy and systematics of various groups of malaria mosquitoes (Chandra et al., 2010; Coluzzi et al., 2002; Rafael et al., 2004; Somboon et al., 2008; Spillings et al., 2009). The major goal of this study was to conduct the cytogenetic analysis of a neglected malaria vector, An. ovengensis, and to evaluate karyotypic divergence in the An. nili group. Our comparative cytogenetic study of An. nili and An. ovengensis has yielded surprising results. First, we detected very few similarities in banding pattern between polytene chromosomes of An. nili and An. ovengensis. Usually, polytene chromosomes of species of the same group or complex have very similar banding patterns and differ only by few fixed inversions with no other apparent chromosomal differences (Coluzzi et al., 2002; Green and Hunt, 1980). Second, we discovered dramatic differences in the location and morphology of heterochromatic regions between An. nili and An. ovengensis. In addition to the differences in morphology of the pericentric heterochromatin, An. ovengensis has intercalary heterochromatin that could not be identified in An. nili. Finally, we determined the difference in arm association between An. nili and An. ovengensis. This finding indicates that a whole-arm translocation might have occurred during the evolution of the An. nili group. Whole-arm translocations and paracentric inversions are the common types of rearrangements in anopheline mosquitoes of subgenus Cellia (Green and Hunt, 1980; Sharakhov et al., 2001). Partial arm translocations and pericentric inversions have not been described in Anopheles even when species from different subgenera were compared (Cornel and Collins, 2000). Therefore, chromosomal arms are expected to preserve their integrity even across large evolutionary distances. However, whole-arm translocations have never been documented within a group of closely related species but is normally found among much more distantly related species that belong to different series (e.g., among An. gambiae, An. funestus, and An. stephensi (Xia et al., 2010) (Fig. 5). Therefore, these data point to a high chromosomal divergence between two species of the An. nili group.
Fig. 5.
A scheme showing proposed chromosomal arm homology and arm translocations among members of subgenus Cellia. Homologous arms are indicated by the numbers above and below chromosomes. Names of series are shown in parentheses below the species names. Notations for 3R and 3L arms of An. funestus are in parentheses. C—pericentromeric regions.
Two hypotheses could explain the profound differences in the karyotypes of An. nili and An. ovengensis. First, members of the An. nili group could have unusually high rates of chromosome evolution. Second, An. nili and An. ovengensis could actually belong to different taxonomic groups within subgenus Cellia. Nonuniform speeds of chromosomal rearrangements have been documented in different organisms. Comparison of vertebrate genomes demonstrated a slow rate of chromosomal evolution in fish and birds and an accelerated rate of genome rearrangements in mammals (Ellegren, 2010; Murphy et al., 2005; Postlethwait et al., 2000). Within mammals, rodent lineages have undergone 3.2–3.5 chromosome rearrangements per million years (Myr) while primates have accumulated only 1.6 rearrangements per Myr since the two lineages diverged. Within carnivores, the rate of chromosome evolution in Canidae is much higher than in other lineages (Yang et al., 1999). Comparison of genomic sequences of 12 species of Drosophila revealed that inversions have been fixed at different rates in different lineages (Bhutkar et al., 2008). For example, 29 fixed inversions are located between D. melanogaster and D. yakuba. All but one of these inversions occurred in the D. yakuba lineage (Ranz et al., 2007). Likewise, the distribution of polymorphic rearrangements varies dramatically among lineages. More than 500 polymorphic inversions are known for D. melanogaster while only 14 inversions have been described for its close relative, D. simulans (Aulard et al., 2004). Within the An. gambiae species complex, An. gambiae s.s. and An. arabiensis are highly polymorphic for chromosomal inversions while their sibling species An. merus is chromosomally monomorphic (Coluzzi et al., 2002). Two highly polymorphic inversions have been found in An. nili from Burkina Faso and one of them has been recorded at low frequency in An. nili from Cameroon (Peery et al., 2011; Sharakhova et al., 2011). The lack of inversion polymorphism in An. ovengensis reported here could be due to a limited sample size and distribution. Thus, the level of inversion polymorphism in the An. nili group remains to be investigated.
Variations in heterochromatin structure and location have been observed within the An. maculipennis subgroup (Sharakhova et al., 1997; Stegnii, 1987), albeit to a lesser extent than within the An. nili group. Evolutionary transformations of heterochromatin with respect to chromosomal location and structure have been demonstrated between An. gambiae and An. funestus, which belong to different series within subgenus Cellia (Sharakhov et al., 2002; Sharakhov et al., 2001). Another study has found a cluster of genes within the centric heterochromatin in D. melanogaster but within euchromatin in D. ananassae, D. pseudoobscura, and D. virilis (Yasuhara et al., 2005). The unique features of heterochromatic genes in D. melanogaster include the accumulation of transposable elements, increased AT richness, longer introns, and association with H3K9me2-enriched domains (a heterochromatin-specific histone modification) (Yasuhara et al., 2005; Yasuhara and Wakimoto, 2006, 2008). A study of the An. gambiae genome showed that heterochromatin accumulates genes important for regulation of gene expression and chromatin organization and harbors genes encoding for odorant receptors, cuticular proteins, and serin-type endopeptidases (Sharakhova et al., 2010). It is possible that changes in heterochromatin have been a mechanism of genetic reinforcement during speciation in the An. nili group. A number of studies have demonstrated direct associations between diffuse heterochromatin and the nuclear envelope in fruit flies and mosquitoes (Baricheva et al., 1996; Hochstrasser and Sedat, 1987; Sharakhov et al., 2001). Closely related species within the An. maculipennis and the D. melanogaster subgroups can be discriminated on the basis of the spatial localization and morphology of the chromosomal regions to which the nuclear envelope is attached in germ-line cells (Stegnii, 1987; Stegnii and Vasserlauf, 1994). The differences in organization of heterochromatin between An. nili and An. ovengensis suggest spatial reorganization of nuclear architecture during speciation in the An. nili group. Reorganizations of heterochromatin and nuclear architecture may lead to substantial changes in global gene expression patterns (Jost et al., 2012; Pezer et al., 2010; Van de Vosse et al., 2011).
The first cytogenetic study of An. nili included 4 specimens collected in the bushes around a cattle kraal near Popa Falls of Okavango River (18º03’ S, 21º39’ E) in Namibia and 68 specimens from the M’Poka village area (3º55’ S, 14º29’ E) in Congo. We investigated the correspondence between the first published An. nili cytogenetic map (Miles et al., 1984) and photomaps of An. nili (Sharakhova et al., 2011) and An. ovengensis (Fig. 3). Our analysis revealed that the originally published chromosomes of An. nili (Miles et al., 1984) were more similar to chromosomes of An. ovengensis than to the more recently published chromosomes of An. nili (Sharakhova et al., 2011). In addition to the similarity of the banding patterns, we noticed the same position of diffuse intercalary heterochromatin in chromosomes of An. nili (Miles et al., 1984) and in chromosomes of An. ovengensis (Fig. 3). Regions of intercalary heterochromatin appear as puffy areas separated from the centromeres by regions of euchromatin. We consider these regions of intercalary heterochromatin as the landmarks for recognition of chromosome arms in An. ovengensis. Specifically, intercalary heterochromatin is seen on arms 4 and 5 of the An. nili map (Miles et al., 1984) as well as on arms 2L and 3R of An. ovengensis, which correspond to arms 3R and 2L of An. nili, respectively (Fig. 3). Intercalary heterochromatin has not been detected in chromosomes of An. nili in our studies (Fig. 3, 4) (Sharakhova et al., 2011). Importantly, the autosomal arms 2, 3, 4, and 5 of the An. ovengensis map correspond to the arms with the same names of the An. nili map (Miles et al., 1984). Distinction between members of the An. nili group is difficult because the morphological differences are subtle (Awono-Ambene et al., 2004; Gillies and Coetzee, 1987) and molecular diagnostics assays have not been available until 2003 (Kengne et al., 2003). A recent study has demonstrated that the equatorial forest might harbor many more species of the An. nili group than were previously described (Ndo et al. PLoS ONE, in press). Therefore, we believe that the first published An. nili chromosomes (Miles et al., 1984) belong an An. ovengensis-like species from the An. nili group.
Anopheles nili and An. ovengensis might belong to different taxonomic groups within subgenus Cellia. According to the accepted rates of molecular evolution of 2.2% per Myr for ITS2 (Schlotterer et al., 1994) and 2% per Myr for mtDNA (DeSalle et al., 1987), a recent study estimated the divergence time among members of the An. nili group at about 0.8 to 6 Myr or 0.2 to 3 Myr, respectively (Ndo et al. PLoS ONE, in press). In addition, ribosomal DNA haplotypes found in populations and species from the An. nili group differed from one another by a large number of fixed mutations and insertion/deletions leading to genetic distance estimates 4 to 8 fold higher than those commonly reported among cryptic Anopheles species (Collins and Paskewitz, 1996; Kengne et al., 2003). This high genetic divergence within the An. nili group suggests that its members may not belong to the same species group. Therefore, the diversity of Anopheles species in the African equatorial forest could be much higher than expected and needs to be thoroughly assessed to improve current vector control measures.
5. Conclusion
This cytogenetic study of An. ovengensis is one of the first steps toward detailed characterization of genome sequences for this important but neglected malaria vector. The observed, substantial reorganizations of chromosome structure between An. ovengensis and An. nili suggest either a rapid rate of chromosome evolution in the An. nili group, or that the two species belong to different taxonomic groups. These hypotheses can be tested by a combination of extensive physical mapping and whole-genome molecular analyses. Cytogenetic and physical mapping, coupled with advances in genome sequencing are the major approaches to understanding mosquito taxonomy, systematics, evolution, ecology, and population genetics. Future studies will highlight genetic features associated with ecological adaptation, population differentiation, and speciation of malaria vectors in equatorial forest. We hypothesize that other species, which currently belong to the An. nili group, could differ from each other by fixed chromosomal inversions and heterochromatin structure. Cytogenetic and physical mapping can identify fixed inversions and reorganizations of heterochromatin. Because of the high readability of polytene chromosomes in species of the An. nili group, this study will be both feasible and informative.
Highlights.
We report on the first cytogenetic analysis of Anopheles ovengensis.
We found a limited similarity in the banding patterns between species.
We observed substantial reorganization of heterochromatin.
Our comparative analysis suggests a whole-arm translocation in the An. nili group.
Acknowledgements
This work was supported by National Institutes of Health grant 5R21AI079350 (to I.V.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributions
I.V.S. designed research; M.V.S., A.P., A.X., and I.V.S. performed cytogenetic research; C.N. and C.A-N. conducted field work and mosquito identification under supervision of P. A-A. and F.S., M.V.S., A.P. and I.V.S. analyzed data; I.V.S. and M.V.S. wrote the paper which was critically revised by A.P., C.A-N., and F.S.
Conflict of interests
The authors declare no conflict of interest.
References
- Aulard S, Monti L, Chaminade N, Lemeunier F. Mitotic and polytene chromosomes: comparisons between Drosophila melanogaster and Drosophila simulans. Genetica. 2004;120:137–150. doi: 10.1023/b:gene.0000017637.10230.c4. [DOI] [PubMed] [Google Scholar]
- Antonio-Nkondjio C, Kerah CH, Simard F, Awono-Ambene P, Chouaibou M, Tchuinkam T, Fontenille D. Complexity of the malaria vectorial system in Cameroon: contribution of secondary vectors to malaria transmission. J Med Entomol. 2006;43:1215–1221. doi: 10.1603/0022-2585(2006)43[1215:cotmvs]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Awono-Ambene HP, Kengne P, Simard F, Antonio-Nkondjio C, Fontenille D. Description and bionomics of Anopheles (Cellia) ovengensis (Diptera: Culicidae), a new malaria vector species of the Anopheles nili group from south Cameroon. J Med Entomol. 2004;41:561–568. doi: 10.1603/0022-2585-41.4.561. [DOI] [PubMed] [Google Scholar]
- Baricheva EA, Berrios M, Bogachev SS, Borisevich IV, Lapik ER, Sharakhov IV, Stuurman N, Fisher PA. DNA from Drosophila melanogaster beta-heterochromatin binds specifically to nuclear lamins in vitro and the nuclear envelope in situ. Gene. 1996;171:171–176. doi: 10.1016/0378-1119(96)00002-9. [DOI] [PubMed] [Google Scholar]
- Berthomieu A, Kengne P, Awono-Ambene P, Raymond M, Fontenille D, Weill M. Isolation and characterization of microsatellite DNA markers in the malaria vector Anopheles nili. Molecular Ecology Notes. 2003;3:394–396. [Google Scholar]
- Bhutkar A, Schaeffer SW, Russo SM, Xu M, Smith TF, Gelbart WM. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics. 2008;179:1657–1680. doi: 10.1534/genetics.107.086108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra G, Bhattacharjee I, Chatterjee S. A review on Anopheles subpictus Grassi--a biological vector. Acta Trop. 2010;115:142–154. doi: 10.1016/j.actatropica.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Collins FH, Paskewitz SM. A review of the use of ribosomal DNA (rDNA) to differentiate among cryptic Anopheles species. Insect Mol Biol. 1996;5:1–9. doi: 10.1111/j.1365-2583.1996.tb00034.x. [DOI] [PubMed] [Google Scholar]
- Coluzzi M. Malaria vector analysis and control. Parasitol Today. 1992;8:113–118. doi: 10.1016/0169-4758(92)90277-9. [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Cornel AJ, Collins FH. Maintenance of chromosome arm integrity between two Anopheles mosquito subgenera. J Hered. 2000;91:364–370. doi: 10.1093/jhered/91.5.364. [DOI] [PubMed] [Google Scholar]
- DeSalle R, Templeton A, Mori I, Pletscher S, Johnston JS. Temporal and spatial heterogeneity of mtDNA polymorphisms in natural populations of Drosophila mercatorum. Genetics. 1987;116:215–223. doi: 10.1093/genetics/116.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval L, Fourment M, Nerrienet E, Rousset D, Sadeuh SA, Goodman SM, Andriaholinirina NV, Randrianarivelojosia M, Paul RE, Robert V, Ayala FJ, Ariey F. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10561–10566. doi: 10.1073/pnas.1005435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Evolutionary stasis: the stable chromosomes of birds. Trends Ecol Evol. 2010;25:283–291. doi: 10.1016/j.tree.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Enayati A, Hemingway J. Malaria management: past, present, and future. Annual Review of Entomology. 2010;55:569–591. doi: 10.1146/annurev-ento-112408-085423. [DOI] [PubMed] [Google Scholar]
- Fontenille D, Simard F. Unravelling complexities in human malaria transmission dynamics in Africa through a comprehensive knowledge of vector populations. Comparative immunology, microbiology and infectious diseases. 2004;27:357–375. doi: 10.1016/j.cimid.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Gillies MT, Coetzee M. A supplement to the Anophelinae of Africa south of the Sahara. Publ. S. Afr. Inst. Med. Res. 1987;55:1–143. [Google Scholar]
- Green C, Hunt R. Interpretation of variation in ovarian polytene chromosomes of Anopheles funestus Giles, A. parensis Gillies, and A. aruni? Genetica. 1980;51:187–195. [Google Scholar]
- Hochstrasser M, Sedat JW. Three-dimensional organization of Drosophila melanogaster interphase nuclei. I. Tissue-specific aspects of polytene nuclear architecture. J Cell Biol. 1987;104:1455–1470. doi: 10.1083/jcb.104.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost KL, Bertulat B, Cardoso MC. Heterochromatin and gene positioning: inside, outside, any side? Chromosoma. 2012;121:555–563. doi: 10.1007/s00412-012-0389-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kengne P, Awono-Ambene P, Antonio-Nkondjio C, Simard F, Fontenille D. Molecular identification of the Anopheles nili group of African malaria vectors. Med Vet Entomol. 2003;17:67–74. doi: 10.1046/j.1365-2915.2003.00411.x. [DOI] [PubMed] [Google Scholar]
- Miles SJ, Carnevale P, Green CA. The ovarian polytene chromosomes of the taxon Anopheles (Cellia) nili Theobald. Coh. O.R.S.T.O.M., ser. Ent. med. et Parasitol. 1984;22:9–11. [Google Scholar]
- Muller HJ. Bearings of the Drosophila work on systematics. In: Huxley J, editor. The new systematics. Oxford: Clarendon Press; 1940. pp. 185–268. [Google Scholar]
- Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, Beever JE, Chowdhary BP, Galibert F, Gatzke L, Hitte C, Meyers SN, Milan D, Ostrander EA, Pape G, Parker HG, Raudsepp T, Rogatcheva MB, Schook LB, Skow LC, Welge M, Womack JE, O'Brien SJ, Pevzner PA, Lewin HA. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Peery A, Sharakhova MV, Antonio-Nkondjio C, Ndo C, Weill M, Simard F, Sharakhov IV. Improving the population genetics toolbox for the study of the African malaria vector Anopheles nili: microsatellite mapping to chromosomes. Parasit Vectors. 2011;4:202. doi: 10.1186/1756-3305-4-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezer Z, Brajkovic J, Feliciello I, Ugarkovc D. Satellite DNA-mediated effects on genome regulation. Genome dynamics. 2010;7:153–169. doi: 10.1159/000337116. [DOI] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Durand P, Neel C, Ollomo B, Ayala FJ, Arnathau C, Etienne L, Mpoudi-Ngole E, Nkoghe D, Leroy E, Delaporte E, Peeters M, Renaud F. African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proc Natl Acad Sci U S A. 2010;107:1458–1463. doi: 10.1073/pnas.0914440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prugnolle F, Ollomo B, Durand P, Yalcindag E, Arnathau C, Elguero E, Berry A, Pourrut X, Gonzalez JP, Nkoghe D, Akiana J, Verrier D, Leroy E, Ayala FJ, Renaud F. African monkeys are infected by Plasmodium falciparum nonhuman primate-specific strains. Proc Natl Acad Sci U S A. 2011;108:11948–11953. doi: 10.1073/pnas.1109368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafael MS, Tadei WP, Hunter FF. The physical gene Hsp70 map on polytene chromosomes of Anopheles darlingi from the Brazilian Amazon. Genetica. 2004;121:89–94. doi: 10.1023/b:gene.0000019959.45267.d7. [DOI] [PubMed] [Google Scholar]
- Ranz JM, Maurin D, Chan YS, von Grotthuss M, Hillier LW, Roote J, Ashburner M, Bergman CM. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biol. 2007;5:e152. doi: 10.1371/journal.pbio.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridl FC, Bass C, Torrez M, Govender D, Ramdeen V, Yellot L, Edu AE, Schwabe C, Mohloai P, Maharaj R, Kleinschmidt I. A pre-intervention study of malaria vector abundance in Rio Muni, Equatorial Guinea: their role in malaria transmission and the incidence of insecticide resistance alleles. Malar J. 2008;7:194. doi: 10.1186/1475-2875-7-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Schlotterer C, Hauser MT, von Haeseler A, Tautz D. Comparative evolutionary analysis of rDNA ITS regions in Drosophila. Mol Biol Evol. 1994;11:513–522. doi: 10.1093/oxfordjournals.molbev.a040131. [DOI] [PubMed] [Google Scholar]
- Sharakhov IV, Serazin AC, Grushko OG, Dana A, Lobo N, Hillenmeyer ME, Westerman R, Romero-Severson J, Costantini C, Sagnon N, Collins FH, Besansky NJ. Inversions and gene order shuffling in Anopheles gambiae and A. funestus. Science. 2002;298:182–185. doi: 10.1126/science.1076803. [DOI] [PubMed] [Google Scholar]
- Sharakhov IV, Sharakhova MV, Mbogo CM, Koekemoer LL, Yan G. Linear and spatial organization of polytene chromosomes of the African malaria mosquito Anopheles funestus. Genetics. 2001;159:211–218. doi: 10.1093/genetics/159.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, Antonio-Nkondjio C, Xia A, Ndo C, Awono-Ambene P, Simard F, Sharakhov IV. Cytogenetic map for Anopheles nili: Application for population genetics and comparative physical mapping. Infect Genet Evol. 2011;11:746–754. doi: 10.1016/j.meegid.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, George P, Brusentsova IV, Leman SC, Bailey JA, Smith CD, Sharakhov IV. Genome mapping and characterization of the Anopheles gambiae heterochromatin. BMC Genomics. 2010;11:459. doi: 10.1186/1471-2164-11-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, Stegnii VN, Braginets OP. Interspecies differences in the ovarian trophocyte precentromere heterochromatin structure and evolution of the malaria mosquito complex Anopheles maculipennis. Genetika. 1997;33:1640–1648. [PubMed] [Google Scholar]
- Sharakhova MV, Xia A, McAlister SI, Sharakhov IV. A standard cytogenetic photomap for the mosquito Anopheles stephensi (Diptera: Culicidae): application for physical mapping. J Med Entomol. 2006;43:861–866. doi: 10.1603/0022-2585(2006)43[861:ascpft]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Somboon P, Thongwat D, Morgan K, Walton C. Crossing experiment of Anopheles maculatus form K and Anopheles willmori (James) (Diptera: Culicidae) Parasitol Res. 2008;103:1317–1322. doi: 10.1007/s00436-008-1135-9. [DOI] [PubMed] [Google Scholar]
- Spillings BL, Brooke BD, Koekemoer LL, Chiphwanya J, Coetzee M, Hunt RH. A new species concealed by Anopheles funestus Giles, a major malaria vector in Africa. Am J Trop Med Hyg. 2009;81:510–515. [PubMed] [Google Scholar]
- Stegnii VN. Systemic reorganization of the architectonics of polytene chromosomes in the onto- and phylogenesis of malarial mosquitoes. II. Species specificity in the pattern of chromosome relations with the nuclear envelope of nutrient ovarian cells. Genetika. 1987;23:1194–1199. [PubMed] [Google Scholar]
- Stegnii VN, Vasserlauf IE. Species architecture of generative tissue chromosomes and problems of phylogenetic relationships in the melanogaster subgroup of the Drosophila genus (Sophophora) Genetika. 1994;30:478–483. [PubMed] [Google Scholar]
- Takken W, Knols BGJ. Malaria vector control: current and future strategies. Trends in parasitology. 2009;25:101–104. doi: 10.1016/j.pt.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Van Bortel W, Harbach RE, Trung HD, Roelants P, Backeljau T, Coosemans M. Confirmation of Anopheles varuna in Vietnam, previously misidentified and mistargeted as the malaria vector Anopheles minimus. Am J Trop Med Hyg. 2001;65:729–732. doi: 10.4269/ajtmh.2001.65.729. [DOI] [PubMed] [Google Scholar]
- Van de Vosse DW, Wan Y, Wozniak RW, Aitchison JD. Role of the nuclear envelope in genome organization and gene expression. Wiley interdisciplinary reviews. Systems biology and medicine. 2011;3:147–166. doi: 10.1002/wsbm.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia A, Sharakhova M, Leman S, Tu Z, Bailey J, Smith C, Sharakhov IV. Genome landscape and evolutionary plasticity of chromosomes in malaria mosquitoes. PLoS ONE. 2010;5:e10592. doi: 10.1371/journal.pone.0010592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, O'Brien PC, Milne BS, Graphodatsky AS, Solanky N, Trifonov V, Rens W, Sargan D, Ferguson-Smith MA. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics. 1999;62:189–202. doi: 10.1006/geno.1999.5989. [DOI] [PubMed] [Google Scholar]
- Yasuhara JC, DeCrease CH, Wakimoto BT. Evolution of heterochromatic genes of Drosophila. Proc Natl Acad Sci U S A. 2005;102:10958–10963. doi: 10.1073/pnas.0503424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuhara JC, Wakimoto BT. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 2006;22:330–338. doi: 10.1016/j.tig.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Yasuhara JC, Wakimoto BT. Molecular landscape of modified histones in Drosophila heterochromatic genes and euchromatin-heterochromatin transition zones. PLoS Genet. 2008;4:e16. doi: 10.1371/journal.pgen.0040016. [DOI] [PMC free article] [PubMed] [Google Scholar]