Abstract
Characterization of large numbers of single nucleotide polymorphisms (SNPs) throughout a genome has the power to refine the understanding of population demographic history and to identify genomic regions under selection in natural populations. To this end, population genomic approaches that harness the power of next-generation sequencing to understand the ecology and evolution of marine invertebrates represent a boon to test long-standing questions in marine biology and conservation. We employed restriction-site-associated DNA sequencing (RAD-seq) to identify SNPs in natural populations of the sea anemone Nematostella vectensis, an emerging cnidarian model with a broad geographic range in estuarine habitats in North and South America, and portions of England. We identified hundreds of SNP-containing tags in thousands of RAD loci from 30 barcoded individuals inhabiting four locations from Nova Scotia to South Carolina. Population genomic analyses using high-confidence SNPs resulted in a highly-resolved phylogeography, a result not achieved in previous studies using traditional markers. Plots of locus-specific FST against heterozygosity suggest that a majority of polymorphic sites are neutral, with a smaller proportion suggesting evidence for balancing selection. Loci inferred to be under balancing selection were mapped to the genome, where 90% were located in gene bodies, indicating potential targets of selection. Results from analyses with and without a reference genome supported similar conclusions, further supporting RAD-seq as a method that can be efficiently applied to species lacking existing genomic resources. We discuss the utility of RAD-seq approaches in burgeoning Nematostella research as well as in other cnidarian species, particularly corals, to determine phylogeographic relationships of populations and identify regions of the genome undergoing selection.
Keywords: balancing selection, estuarine, genome, Nematostella, next-generation sequencing, phylogeography
INTRODUCTION
Population genomic approaches offer revolutionary opportunities over traditional population genetic markers to characterize the history of species and populations, and the genetic mechanisms of adaptation by analyzing polymorphic markers dispersed throughout the entire genome (Luikart et al. 2003; Nadeau & Jiggins 2010). Historically, methods to identify large numbers of genetic markers and characterize their geographic distribution in natural populations were labor-intensive and cost-prohibitive for almost any species, particularly those lacking extensive sequence resources. However, advances in sequencing technology in recent years have opened new avenues for the generation of large numbers of molecular markers in a panel of individuals to better characterize the ecology and evolution of traditionally non-model species (Rowe et al. 2011). One of these methods is restriction-site-associated DNA sequencing (RAD-seq), which combines enzymatic fragmentation of the genome with high throughput sequencing for generation of large numbers of SNP markers (Baird et al. 2008).
Knowing the proportion of genetic exchange among populations and the spatial distribution of genetic diversity for particular species within aquatic ecosystems is critical in order to understand biodiversity and inform conservation and management decisions (Palumbi 2003; Palumbi 2004; Botsford et al. 2009). Marine and estuarine habitats are relatively poorly characterized ecosystems for which we know little about the population genetics of most resident species when compared to terrestrial systems. Current data support a spectrum of expectations for species’ dispersal, and the resulting population connectivity, from nearly open to a higher degree of population genetic structure over unexpectedly small geographic distances due to local recruitment (Hauser & Carvalho 2008; Ciannelli et al. 2010). Previous expectations for connectivity relied on the pelagic larval duration (PLD) to hypothesize relative dispersal distances, and thus the probability of gene flow in natural populations (Cowen et al. 2000; Bay et al. 2006; Cowen & Sponaugle 2009). However, recent studies have convincingly shown that PLD is at best weakly correlated with population genetic structure (Bradbury et al. 2008; Weersing & Toonen 2009), which may be driven by errors and uncertainties when calculating FST (Faurby & Barber 2012), making confident, accurate predictions about population connectivity in the marine environment difficult.
The attributes of genetic markers for making population genetic inferences can have substantial impacts on what hypotheses can be adequately tested. Previous reviews have discussed the relative merits and limitations of the diverse set of molecular markers available for studying population processes (Parker et al. 1998; Sunnucks 2000; Mariette et al. 2002; Brumfield 2003; Brito & Edwards 2008; Diniz-Filho 2008). To date, a large majority of studies that characterize the population genetics of marine or estuarine species have utilized allozymes, anonymous markers (e.g., AFLPs, RFLPs), a small number of microsatellites, or a handful of sequence-based markers (e.g., mitochondrial DNA, nuclear ribosomal DNA). These markers have trade-offs that frequently balance diversity (e.g., microsatellites, AFLPs) with the ease of interpretation and ability to compare among species (e.g., sequence markers). More recent surveys discussing the utility of genetic markers have emphasized the significant advantages of single nucleotide polymorphisms (SNPs) for population genetic studies (Brumfield 2003; Morin et al. 2004; Brito & Edwards 2008). Although SNPs have the limitation of lower diversity due to only four possible allelic states and a low mutation rate, they have clear advantages for accommodating diverse assumptions of linkage or independence of markers depending on the discovery strategy, explicit models of evolutionary change, and potential for roles in functional evolution (e.g., polymorphisms in coding or promoter regions). In addition, SNPs can be readily compared among genomes (nuclear, mitochondrial, chloroplast) to utilize the underlying mutational scales to characterize evolutionary processes (Morin et al. 2004; Petit et al. 2004).
Nematostella vectensis is an anthozoan cnidarian (Cnidaria, Anthozoa, Hexacorallia, Actiniaria) common to tidally restricted pools in high marsh environments (Hand & Uhlinger 1994). In recent years, N. vectensis has emerged as a model cnidarian in molecular biology and comparative genomics due to ease of laboratory culture and the publication of its genome (Putnam et al. 2007), the first for a cnidarian. Sexual reproduction and developmental stages have been well characterized in a laboratory cultures (Reitzel et al. 2007). Eggs of N. vectensis are released in a gelatinous mass by female anemones and then externally fertilized by males. Development progresses from a fertilized egg to an early embryo within the egg mass. Subsequently, early larvae swim from the degraded egg jelly, develop into an elongated late larval stage, and then settle as a four-tentacle juvenile stage within seven days. This species holds great promise as a useful model for understanding the ecological genomics of coastal species (Darling et al. 2005) given that it is found in high marsh estuaries that are impacted by human encroachment, has been repeatedly introduced to non-native locations, and has a broad geographic range likely resulting in local adaptation. N. vectensis has been collected in salt marshes along the Pacific and Atlantic coast of North America, a portion of England (Hand & Uhlinger 1994; Reitzel et al. 2008), and Brazil (Silva et al. 2010). Previous research on the population genetic structure of N. vectensis, using RAPDs, AFLPs, and microsatellites, has identified significant genetic differences among major coastline regions, estuaries within each region, and even among subpopulations within a single estuary (Pearson et al. 2002; Darling et al. 2004; Reitzel et al. 2008; Darling et al. 2009). These studies have also shown high variation in the relative contribution of clonal reproduction to resident populations throughout its range (Darling et al. 2009). In addition, available data suggest that N. vectensis has been introduced from the west Atlantic to the west coast of North America and England, where it receives protective status under the Wildlife and Countryside Act. Despite these insights, we lack an understanding of the phylogeography of populations in the native range due to the low resolution provided by these traditional markers. High resolution data are critical for testing hypotheses about the historical distribution of this species, the connectivity of current populations, and the source locations for introduced populations in non-native habitats. Moreover, there are currently few data to test for potential genetic adaptation in natural populations that span its large geographic range. Two previous studies (Sullivan et al. 2009; Reitzel et al. 2010) utilized expressed sequence tags generated during the sequencing of the N. vectensis genome to document polymorphisms in coding regions, particularly nonsynonymous substitutions in conserved protein domains. Their findings suggest that SNPs are present in genes that could exert a large influence on protein function. More recent work has identified substantial phenotypic variation in natural populations (Reitzel et al. in revision) highlighting the need for high-density genomic markers to provide the tools for linking genetic and phenotypic diversity in populations occupying environmental gradients that may result in phenotypic clines.
Our understanding of the genetic diversity and coarse population-level relationships for N. vectensis is representative of the general population genetic data for other cnidarians. Within the marine environment, cnidarians represent a critical taxonomic group of benthic and pelagic species for both ecological function and conservation management. The phylum Cnidaria contains corals that are ecosystem engineers and support a rich biodiversity in shallow and deep marine habitats (Jones et al. 1994; Roberts et al. 2006), but are frequently threatened by anthropogenic activities. Moreover, jellyfish have emerged as common nuisance species where population blooms dramatically impact fisheries and pelagic biodiversity (Purchell et al. 2007). For species of conservation concern, resolving genetic diversity and its structure is critical to understand the impact of human activities as well as the opportunity for recovery after disturbances (Palumbi 2003; Baums 2008). In addition, understanding genetic diversity will assist in assessing the opportunity for adaptation of populations to changing environments (Hughes et al. 2003) and, with the availability of a genome, identification of genomic regions under selection. For groups, like jellyfish, that are exerting negative impacts on marine communities and human economies, high-resolution characterization of genetic diversity would markedly improve our understanding of the impacts derived from the introduction of these species to non-native areas and the composition of blooms that develop in particular locations. Despite the clear need for data to understand phylogeography and the particular regions of the genome undergoing selection, population genetic studies of cnidarians often are unable to resolve many of these questions due to the availability of only few allele-based markers (e.g., microsatellites), with the exception of a small number of species, as well as the near absence of variable sequence-based markers (Shearer et al. 2002; Bilewitch & Degnan 2011). Thus, the development and application of next-generation sequencing to the population genetics of cnidarians will bridge these critical gaps. In this respect, N. vectensis is an ideal cnidarian model in which to assess how RAD-seq, or similar genomic methods, can be utilized to characterize phylogeographic relationships among populations as well as regions of the genome under selection.
In this study we utilized RAD-seq to characterize the genetic diversity and population genetic structure of N. vectensis individuals collected along the Atlantic coast of North America. We compared our results with and without the use of the available reference genome to assess the potential impacts of utilizing RAD-seq in non-model species with limited genomic data. Finally, we mapped the SNPs inferred to be under selection to the reference genome in order to identify genes that are likely under selection, and then we grouped them based on potential biological function. Together, our study provides one of the first applications of RAD-seq to a marine invertebrate (see De Wit & Palumbi 2012) and highlights the utility of a reference genome in generating hypotheses for linking population and functional genomics.
METHODS
Collection
Adults of Nematostella vectensis were collected from three estuaries along the Atlantic coast of North America (Peggy’s Cove, Nova Scotia; Sippewissett, Massachusetts; Baruch, South Carolina; see Reitzel et al. (2008) for details). Briefly, individuals were sieved from loose sediments, transferred to 13%thou (parts per thousand) artificial seawater, and transported to the laboratory. Individuals were maintained under a standard culturing protocol for N. vectensis (13%thou artificial seawater, fed 2-3 times per week with freshly hatched Artemia sp.). Individuals from a common laboratory stock maintained in the Martindale lab (Kewalo Marine Laboratory, University of Hawaii) were originally collected from Rhode River, Maryland. In addition, this laboratory culture served as the source population from which the N. vectensis genome was sequenced. When necessary, individual clonal lines were generated by transverse bisection to yield adequate genomic DNA.
Molecular laboratory methods
Individual anemones or pooled individuals developed through bisected clonal lines were starved for at least three days prior to genomic DNA extraction to minimize potential contamination from food sources. Genomic DNA for nine individuals from each of the Nova Scotia and Massachusetts populations, and six from the Maryland and South Carolina populations, was extracted with the Qiagen DNAeasy kit (Qiagen). Genomic DNA quality was checked by visual inspection on an agarose gel and with a ND-1000 Nanodrop spectrophotometer (Nanodrop Technologies). DNA concentration was also determined with a Nanodrop spectrophotometer. Ten micrograms of high quality (260/280 > 1.8) genomic DNA per individual was submitted to Floragenex Inc. for library preparation and sequencing. Individual libraries were produced from DNA digested with a high-fidelity SbfI restriction enzyme and barcoded with 5-base pair sequence tags. Libraries were sequenced on a single-lane of an Illumina GAIIX sequencer.
Data QC & QA and SNP calling
Sequencing data were filtered using the program PRINSEQ v0.18 (Schmieder & Edwards 2011). All sequence reads (i.e., individual fragments of contiguous nucleotide bases) were trimmed to a length of 31bp; shorter reads were discarded. Reads with ambiguous characters or with mean Phred quality score (Ewing & Green 1998; Ewing et al. 1998) lower than 20 (base call accuracy lower than 99%) were also discarded (Huse et al. 2007).
Reads were aligned to the reference genome of N. vectensis (v1.0, http://genome.jgi-psf.org/Nemve1/Nemve1.home.html) using BOWTIE v0.12.7 (Langmead et al. 2009). Only reads that produced a unique best alignment to the genome (in terms of the smallest number of mismatches and the highest Phred score of the mismatch positions), with at most 3 mismatches, were retained. Aligned reads were processed in the program STACKS v0.998 (Catchen et al. 2011) – a tool used to form stacks of identical unique sequences from each individual, identify loci by aligning homologous stacks, generate genotypes, and match loci among individuals. High-confidence SNP calls in STACKS are performed using a maximum-likelihood framework that accounts for sources of error inherent to RAD markers (i.e., sequencing error, variable depth of coverage) (Hohenlohe et al. 2010; Catchen et al. 2011). A minimum depth of 4 reads per stack (i.e., 8 per locus) was enforced. Significantly high-repetitive stacks were discarded by implementing the deleveraging algorithm, as these likely represent sequencing errors, duplications, or repetitive regions. The deleveraging algorithm assumes similar depths for stacks originating from a common locus (Catchen et al. 2011). No mismatches among loci were allowed when creating the catalog of all the loci identified among the sampled individuals. In a similar manner, reads were processed without the use of a reference genome in order to evaluate the effects of the lack of this resource in downstream analysis. The maximum number of mismatches allowed among loci within each individual was 2. Loci with more than 2 alleles per SNP per individual were discarded as these are considered methodological artifacts in diploid organisms or products from multiple-copy elements in the genome. Hereafter we refer to the loci identified in this analysis as RAD markers.
The reference genome of N. vectensis (Putnam et al. 2007) was sequenced from the offspring of two parent strains originally collected from Rhode River, Maryland, USA, which is one of the populations sampled for this study. The use of this reference genome to process sequence reads by retaining only those that produce unique alignments to it could introduce a form of ascertainment bias (i.e., markers present in individuals from the Maryland population being more likely to be included in the analyses than others). To assess the effect of this potential source of bias we tested for significant differences in average number of reads with one reported alignment to the genome and the number of RAD markers, per individual, among populations. To account for the variability in number of reads among individuals we randomly resampled the sets of reads in each individual in order to normalize them to the set with the smallest number of reads (56 851), using the PERL-script DAISYCHOPPER v0.6 (available from www.genomics.ceh.ac.uk/GeneSwytch).
Clone detection
Due to the capability of N. vectensis to reproduce asexually we tested for the presence of clones in our dataset by comparing the percentage of genotypic distances among individuals within each population. To account for the possibility that the observed differences were caused by variability in sequencing coverage of particular markers among individuals and/or SNP calling errors, we established an arbitrary cutoff value of 95% for the percentage of genotypic pairwise distances (i.e., individuals with genotypic distances smaller than 5% are considered potential clones). This is a conservative threshold considering that the probability of a given genotype for any individual in our study was calculated to be less than 1×10−9 (Arnaud-Haond & Belkhir 2007; Arnaud-Haond et al. 2007). To evaluate the effect of the presence of potential clones in the dataset, all subsequent analyses were performed comparatively using genome aligned or unaligned reads and with or without potential clone individuals.
Detection of markers under selection
To identify potential markers in genomic regions subject to selection we used the FST outlier method (Beaumont & Nichols 1996) implemented in the program LOSITAN (Antao et al. 2008). This method utilizes the observed allele frequencies of SNPs to estimate expected heterozygosities and global unbiased FST values (Weir & Cockerham 1984; Cockerham & Weir 1993) to simulate an expected neutral distribution for FST, assuming an island model of migration (Wright 1931). One million simulations were performed assuming an infinite alleles mutation model. 95% confidence intervals were built around the simulated mean neutral FST. SNPs with FST values significantly greater than expected under neutrality were considered candidates for positive selection. Conversely, SNPs with FST values significantly smaller than expected under neutrality were considered candidates for balancing selection (Beaumont & Nichols 1996). RAD makers containing SNPs with conflicting selection classifications (e.g., one SNP candidate neutral and another candidate balancing, in the same marker) were excluded from the analyses to avoid ambiguities.
Candidate markers under selection
Candidate markers under balancing selection that were common among all four analyses (genome aligned or unaligned reads, with or without potential clones) were mapped to the reference genome of N. vectensis (Putnam et al. 2007). Position of each marker was annotated whether it was located in an annotated gene body (intron or exon) or close to the nearest annotated gene in the current version of the genome. When the marker was located in an intergenic region, we identified the closest gene and quantified the distance to this gene. Selected genes were then tentatively assigned a name based on U.S. Department of Energy Joint Genome Institute (JGI) annotations, or on sequence similarity to available protein sequences assessed through BLASTp searches at the U.S. National Center for Biotechnology Information (NCBI) REFSEQ. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway assignments for each selected protein were identified using the program Blast2GO v2.5.1 (Conesa et al. 2005). Results from GO analysis were grouped based on ‘biological process’ to cluster potential shared functions for these proteins.
Demographic inferences
Inferences of demographic parameters were carried out using candidate neutral markers only. Only one SNP per RAD marker was taken into account to avoid violating the assumption of independence among markers. Only biallelic SNP were included in order to simplify the calculations and fit the assumptions of the software utilized for the analyses. As indicated above, all inferences were performed comparatively using genome aligned or unaligned reads and with or without potential clone individuals.
To evaluate the validity of putative populations defined by their sampling location, we inferred population structuring through a principal component analysis (PCA) using the software EIGENSOFT v4.2 (Patterson et al. 2006; Price et al. 2006). We evaluated the significance of the identified principal components through Tracy-Widom statistics (Tracy & Widom 1994; Johnstone 2001). The statistical significance of the differences between identified populations was evaluated via a chi-square test. The summing of ANOVA statistics of genetic differentiation between pairs of populations along each eigenvector approximates a chi-square distribution with degrees of freedom equal to the number of eigenvectors (Patterson et al. 2006; see EIGENSOFT documentation). We also inferred population structuring (historical lineages) by maximizing the posterior probability of the genotypic data, given a set number of clusters (K). This method is known as Bayesian population clustering and is implemented in the program STRUCTURE v2.3.2 (Pritchard et al. 2000; Falush et al. 2003) available in the Bioportal (Kumar et al. 2009). The admixture model was used with uncorrelated allele frequencies. The MCMC was run for 1 100 000repetitions (burnin period 1,000 000). Values for K were evaluated from 1 to 5 (10 replicates each). The optimal value of K was selected using the program STRUCTURE HARVESTER v0.6.92 (Earl & Vonholdt 2012) according to the ad hoc ΔK statistic (Evanno et al. 2005), which is the second order rate of change of the likelihood function. STRUCTURE results were visualized using the program DISTRUCT v1.1 (Rosenberg 2004).
The overall RAD marker variability was compared among individuals within each population. A quantitative measure of this variation was obtained by estimating four commonly used genetic diversity indexes: the proportion of polymorphic SNPs, the mean observed heterozygosity, the mean expected heterozygosity, and the mean number of alleles. These indexes were calculated with the R-package POPGENKIT v1.0 (Rioux Paquette 2011).
Genetic differentiation among populations was measured using the unbiased FST estimator θ□ (Weir & Cockerham 1984) (here referred to as FSTW&C) and the asymptotically consistent estimator F□ (Reich et al. 2009) (here referred to as FSTR) using custom scripts in R. F□ has been shown to consistently yield accurate estimates of population differentiation at small sample sizes (n < 6) when large numbers of loci (> 100) are available (Willing et al. 2012). A correction that accounts for potential inbreeding effects on F□ (Reich et al. 2009) (here referred to as FSTRcor ), which could be prevalent in N. vectensis due to possible small effective population sizes, was also applied. Confidence intervals were calculated for each estimator based on 1 000 bootstrap replicates.
In order to generate useful sequence matrices for phylogeographic analyses, the nucleotide identity data from individual homozygous SNP loci (variable among individuals) were sorted and concatenated following procedures suggested by Emerson et al. (2010). Phylogenetic inferences of evolutionary relationships were performed through the implementation of statistical methods following the maximum likelihood criterion as implemented in PHYML v3.0 (Guindon et al. 2010). The general time-reversible model (GTR) of nucleotide substitution (Tavare 1986) was assumed. Topological robustness was assessed through 1,000 non-parametric bootstrap replicates. Trees were visualized and edited in the program FigTree v1.3.1 (Rambaut 2009).
RESULTS
SNP discovery and clone detection
The mean number of sequence reads obtained per individual was 160 409 (95% CI ± 31 084; SD = 85 127; n = 30), and individual values ranged between 56 851 and 353 084 reads. On average, 1,721 reads (1%; 95% CI ± 31 084; SD = 85 127; n = 30) were discarded as low quality (Figure S1). An average of 114 071 reads (95% CI ± 22 240; SD = 60 907; n = 30) had a unique alignment to the reference genome, representing ca. 71% of all reads (Figure S1). Approximately 4% of the reads failed to produce an alignment, and 24% were discarded due to having more than one reportable alignment. To avoid any possible downstream analytical biases on the accuracy of population parameter estimates and their uncertainty, e.g., Hinrichs & Suarez (2005), only markers present in all individuals were retained. Reads processed without alignment to the reference genome yielded 20% more RAD markers than the genome aligned reads (see Table 1 and Figure S2 for additional details). The percentage increase in the number of polymorphic RAD markers and the number of SNPs per individual was 88% and 107%, respectively. However, there was an overall slight reduction in the number of polymorphic RAD markers (14% less) and the number of SNPs (18% less) that were shared among all individuals in this unaligned analysis.
Table 1.
RAD marker statistics per analyses with or without clones, and using genome aligned or unaligned reads.
| Potential clones included (n=30) | Potential clones removed (n=22) | |||||||
|---|---|---|---|---|---|---|---|---|
| Genome Aligned | No Genome Aligned | Genome Aligned | No Genome Aligned | |||||
| Mean number of RAD markers per individual | 2 305 | (95% CI ± 71; SD = 193) | 2 759 | (95% CI ± 89; SD = 243) | 2 330 | (95% CI ± 75; SD = 206) | 2 737 | (95% CI ± 90; SD = 246) |
| Mean depth of coverage per RAD marker per individual | 48X | (95% CI ± 8; SD = 22) | 49X | (95% CI ± 9; SD = 23) | 51X | (95% CI ± 8; SD = 23) | 54X | (95% CI ± 9; SD = 24) |
| Mean number of polymorphic RAD markers per individual | 139 | (95% CI ± 18; SD = 50) | 261 | (95% CI ± 24; SD = 65) | 142 | (95% CI ± 21; SD = 57) | 252 | (95% CI ± 26; SD = 72) |
| Mean number of SNPs per individual | 174 | (95% CI ± 24; SD = 66) | 360 | (95% CI ± 24; SD = 66) | 179 | (95% CI ± 27; SD = 75) | 356 | (95% CI ± 33; SD = 90) |
| Total Number of RAD markers in the catalog | 2 | 987 | 4 | 065 | 2 | 978 | 3 | 925 |
| Total Number of RAD markers present in all individuals | 1 | 297 | 1 | 251 | 1 | 351 | 1426 | |
| Total number of polymorphic markers present in all individuals | 287 | 248 | 304 | 310 | ||||
| Number of polymorphic RAD markers with 1 SNP | 220 | 204 | 232 | 250 | ||||
| RAD markers candidate neutral | 164 | 145 | 167 | 169 | ||||
| RAD markers candidate balancing selection | 56 | 59 | 65 | 79 | ||||
| RAD markers candidate positive selection | 0 | 0 | 0 | 2 | ||||
| Number of polymorphic RAD markers with >1 SNP | 67 | 44 | 72 | 60 | ||||
| RAD markers candidate neutral* | 47 | 27 | 46 | 31 | ||||
| RAD markers candidate balancing selection* | 6 | 7 | 8 | 13 | ||||
| RAD markers candidate positive selection* | 0 | 0 | 0 | 1 | ||||
| Number of markers candidate neutral | 211 | 172 | 213 | 200 | ||||
| Number of markers candidate balancing selection | 62 | 66 | 73 | 92 | ||||
| Number of markers candidate positive selection | 0 | 0 | 0 | 3 | ||||
| Total number of SNPs present in all individuals | 365 | 298 | 388 | 374 | ||||
| SNPs candidate neutral | 266 | 199 | 269 | 231 | ||||
| SNPs candidate balancing selection* | 68 | 74 | 81 | 107 | ||||
| SNPs candidate positive selection* | 0 | 0 | 0 | 4 | ||||
| Biallelic SNPs candidate neutral + | 209 | 172 | 212 | 200 | ||||
Markers containing SNPs with conflicting classifications (e.g. one SNP candidate neutral and another candidate balancing, in the same locus) were excluded from the analyses
To avoid violations of the assumption of independence, only one SNP per RAD marker was used for the demographic analyses
The ascertainment bias analysis performed to address the possible effect of using the reference genome to process the sequence reads showed that, when comparing among populations, individuals from Maryland (same population as the source of the reference genome) had the largest number of retained reads. However, there were no significant differences in the average number of identified RAD markers between the Maryland and Massachusetts populations, and only marginal differences between these and the populations from Nova Scotia or South Carolina (α = 0.05, see 95% confidence intervals in Figure S3).
There were eight individuals identified as potential clones in three populations: one in Massachusetts, five in Nova Scotia, and one in Maryland. Not a single potential clone pair shared identical genotypes. The percentage of pairwise genotypic similarities among potential clones ranged between 99.0 and 99.9% (mean = 99.5%, 95% CI ± 0.1; SD = 0.3; n = 13). In contrast, the genotypic distances among non-potential clones ranged between 61.2 and 86.5% (mean = 73.3%, 95% CI ± 1.5; SD = 7.3; n = 89). As mentioned in the Methods section, all analyses were also performed after excluding these potential clone individuals from the dataset. The results of these analyses were very similar to the ones obtained when all sampled individuals were included (including potential clones). When the potential clones were excluded the sequence reads processed without alignment to the reference genome yielded 17% more RAD markers than the genome aligned reads (see Table 1). This produced a percentage increase in the number of polymorphic RAD markers and the number of SNPs, per individual, of 77% and 99%, respectively. However, the number of polymorphic RAD markers and the number of SNPs shared among all individuals remained virtually unaltered (changes were less than 4%).
Detection of markers under selection
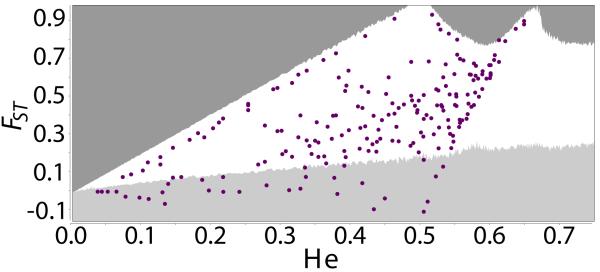
Overall there were approximately 200 candidate neutral markers and 70 candidate balancing selection markers identified in each analysis using genome aligned or unaligned reads and with or without potential clones (Table 1, Figure 1, and Figure S4). Most RAD markers (ca. 80%) contained exactly one SNP position. The majority of SNPs were biallelic (> 99% overall) and none contained more than 3 alleles.
Figure 1.
Scatter plot of FST vs. expected heterozygosity (He) for the biallelic SNP loci in the analysis of genome aligned reads without potential clones. Shaded boundaries indicate the 95% confidence intervals obtained through simulations in LOSITAN. Dark gray region indicates candidates for positive selection, and light gray regions candidates for balancing selection.
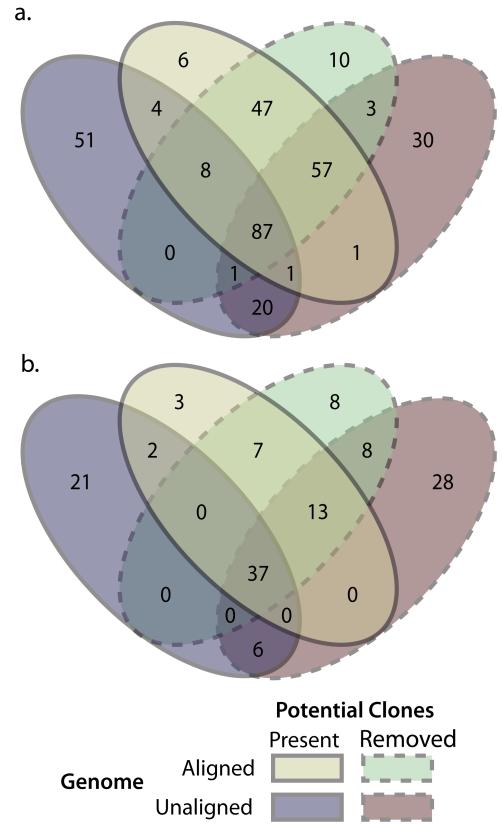
Approximately 40% of the candidate neutral and balancing selection markers identified in the presence of potential clones were shared between analyses with and without genome aligned reads (Figure 2). When the potential clones were removed this percentage of shared markers between analyses increased slightly to 56%. Comparisons of analyses using genome aligned reads showed that most identified markers (88% of neutral and 73% of balancing) are shared among the analyses with and without potential clones. These percentages drop to 42% for neutral markers and 38% for balancing selection markers when using unaligned reads. Eighty-seven candidate neutral markers and 37 candidate balancing markers were common among all analyses.
Figure 2.
Venn diagrams showing the number of markers that were unique to, and common among, the four analyses using genome aligned or unaligned reads and with or without potential clone individuals. (a) Candidate neutral markers. (b) Candidate balancing selection markers.
Candidate loci under selection
Thirty percent (n = 37 of 124) of markers common among all four analyses, using genome aligned or unaligned reads and with or without potential clone individuals, were inferred to be under balancing selection based on statistical comparisons. All 37 markers represent unique loci and mapped closely to single proteins in the current version of the genome of N. vectensis. Thirty-three of these were located within a gene body (19 in exons, 14 in introns, Table S1). The four remaining SNPs were located within a few kilobases to an annotated coding sequence. The SNP located furthest from a coding sequence was a polymorphism in locus number 584, which was 9 067 bp from an open reading frame for a forkhead transcript factor (JGI: 239634).
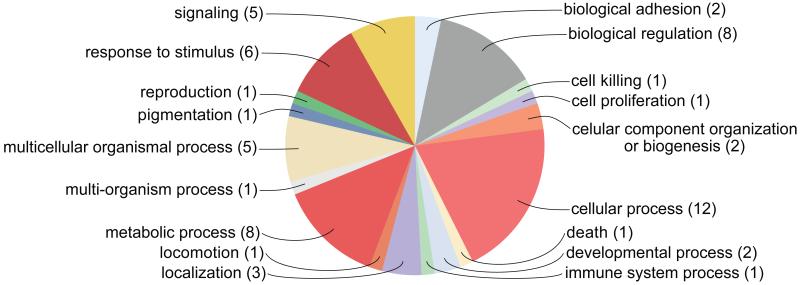
All but three of the coding sequences containing or nearest to these 37 markers were annotated based on either JGI identification or through BLAST similarity. Categorization of these proteins by GO annotation suggested a diverse set of biological processes, including cellular processes, metabolic processes, and response to stimulus (Figure 3). No particular process or function (data not shown) were particularly enriched in the GO annotation, instead these data suggest that the proteins are involved in a broad set of molecular, cellular, and organismal processes. Numerically, the GO categories with largest representation were cellular processes (n = 12, e.g., centrosomal protein), metabolic processes (n = 8, e.g., GTP binding protein), and biological regulation (n = 8, e.g., phosphatidic acid phosphatase). KEGG annotation identified two pathways, each with one N. vectensis protein: DNA methyltransferase 1 (locus 813) involved in cysteine and methionine metabolism and natriuretic peptide receptor (locus 551) with a function in purine metabolism.
Figure 3.
Distribution of GO categories (‘biological process’, level 2) for proteins coded by genes containing or most closely positioned in the genome to SNPs inferred to be under balancing selection. Analysis utilized only the 37 markers that were common among all the four analyses using genome aligned or unaligned reads and with or without potential clone individuals. The numbers in parentheses after the GO category refer to the number of proteins annotated for each category.
For proteins near to or containing these markers under balancing selection, we identified a few proteins of particular interest due to their role in gene regulation. Two markers were located near or in a gene body for two transcription factors: locus 301 was located in an exon of the nuclear receptor co-repressor (N-CoR1) and locus 383 was 2 kb from heat shock factor 1 (HSF1). One other notable protein, a TGFβ receptor (locus 729), contained one SNP located in an exon.
Demographic inferences
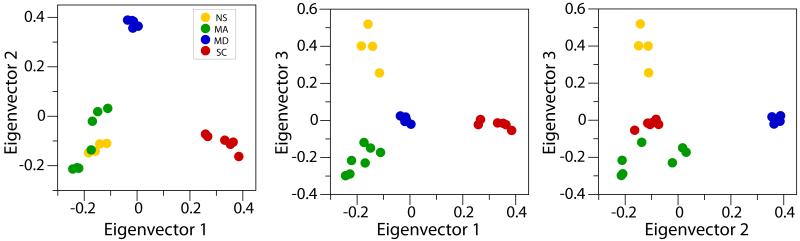
Principal component analyses identified three large eigenvectors (axes of variation) revealing the presence of four distinct clusters (Figures 4, S5-S7). This same result was found in all analyses using genome aligned or unaligned reads and with or without potential clones. The eigenvector 1, with the largest eigenvalue, was not significant (p = 0.097), and the eigenvector 2, with the second largest eigenvalue, was marginally significant (p = 0.045, Table S2). The eigenvector 3, with the third largest eigenvalue, was highly significant (p < 0.001). All differences among identified clusters were also highly significant (Table S3). The results from the STRUCTURE analyses are congruent with the inferences made from the PCA (data not shown). The four identified clusters unambiguously matched the a priori population assignments based on the geographic origin of the samples (Figures 4, S5-S7).
Figure 4.
Estimated population structure of N. vectensis according to the principal component analysis (PCA). Each dot represents an individual. Colors indicate the geographic site locations: Nova Scotia (NS), Massachusetts (MA), Maryland (MD), and South Carolina (SC). The three principal axes of variation are shown.
Genetic diversity in all four analyses was highest in the Massachusetts population and lowest in the Maryland population, as measured by the proportion of polymorphic markers, the expected and observed heterozygosities, and the average number of alleles (Table 2). Genetic diversities for the Nova Scotia and South Carolina populations were similar, although higher in Nova Scotia. The genetic diversity estimates between analyses with or without potential clones were very similar, but slightly higher overall when potential clones were removed.
Table 2.
Estimates of genetic diversity per population for four analyses (with or without clones, genome aligned or unaligned reads).
| Nova Scotia (NS) |
Massachusetts (MA) |
Maryland (MD) | South Carolina (SC) |
|
|---|---|---|---|---|
| Potential Clones Included | ||||
| Number of samples | 9 | 9 | 6 | 6 |
| Genome-aligned | ||||
| RAD markers with biallelic SNPs (neutral) | 209 | |||
| Proportion of polymorphic SNP | 0.517 | 0.612 | 0.239 | 0.445 |
| Mean observed heterozygosity | 0.213 | 0.290 | 0.112 | 0.161 |
| Mean expected heterozygosity | 0.169 | 0.205 | 0.082 | 0.143 |
| Mean number of alleles per SNP | 1.517 | 1.612 | 1.239 | 1.445 |
| Number of private alleles | 23 | 22 | 4 | 53 |
| Unaligned | ||||
| RAD markers with biallelic SNPs (neutral) | 172 | |||
| Proportion of polymorphic SNP | 0.500 | 0.703 | 0.320 | 0.407 |
| Mean observed heterozygosity | 0.201 | 0.378 | 0.153 | 0.162 |
| Mean expected heterozygosity | 0.163 | 0.258 | 0.110 | 0.148 |
| Mean number of alleles per SNP | 1.500 | 1.703 | 1.320 | 1.407 |
| Number of private alleles | 14 | 12 | 8 | 25 |
| Potential Clones Exclued | ||||
| Number of samples | 4 | 7 | 5 | 6 |
| Genome-aligned | ||||
| RAD markers with biallelic SNPs (neutral) | 212 | |||
| Proportion of polymorphic SNP | 0.505 | 0.632 | 0.250 | 0.434 |
| Mean observed heterozygosity | 0.217 | 0.321 | 0.121 | 0.176 |
| Mean expected heterozygosity | 0.183 | 0.230 | 0.092 | 0.154 |
| Mean number of alleles per SNP | 1.505 | 1.632 | 1.250 | 1.434 |
| Number of private alleles | 22 | 28 | 4 | 40 |
| Unaligned | ||||
| RAD markers with biallelic SNPs (neutral) | 200 | |||
| Proportion of polymorphic SNP | 0.535 | 0.705 | 0.345 | 0.420 |
| Mean observed heterozygosity | 0.261 | 0.402 | 0.176 | 0.178 |
| Mean expected heterozygosity | 0.206 | 0.276 | 0.128 | 0.158 |
| Mean number of alleles per SNP | 1.535 | 1.705 | 1.345 | 1.420 |
| Number of private alleles | 11 | 16 | 8 | 29 |
Overall, the pairwise FST values suggest that differentiation was significant (i.e., 95% confidence intervals did not contain the value of the null hypothesis = 0) among all populations (Table 3). Genetic differentiation was greatest between the Maryland and South Carolina populations. Large genetic differentiation was also found between the Nova Scotia population and the southern populations (Maryland and South Carolina). The most similar populations were Massachusetts and Nova Scotia. None of the FST estimators yielded significantly different values between the analyses using genome aligned or unaligned reads. When using genome unaligned reads we observed significantly greater FST values when potential clones were included in the analyses than when they were not. Similarly, when using genome aligned reads we also observed greater FST values when potential clones were included in the analyses than when they were not; however, these differences were not statistically significant. No significant differences were found among different FST estimators.
Table 3.
Pairwise FST estimates for four analyses (with or without clones, genome aligned or unaligned reads).
| FST W&C | 95% CI | FST R | 95% CI | FST Rcor | 95% CI | |
|---|---|---|---|---|---|---|
| Potential Clones Included | ||||||
| Genome-aligned | ||||||
| NS vs. MA | 0.298 | (0.254, 0.351) | 0.286 | (0.239, 0.340) | 0.298 | (0.256, 0.350) |
| NS vs. MD | 0.544 | (0.477, 0.601) | 0.556 | (0.488, 0.617) | 0.563 | (0.498, 0.629) |
| NS vs. SC | 0.518 | (0.459, 0.572) | 0.517 | (0.461, 0.575) | 0.521 | (0.461, 0.575) |
| MA vs. MD | 0.474 | (0.426, 0.518) | 0.485 | (0.431, 0.536) | 0.497 | (0.450, 0.548) |
| MA vs. SC | 0.480 | (0.425, 0.529) | 0.480 | (0.429, 0.533) | 0.487 | (0.435, 0.538) |
| MD vs. SC | 0.622 | (0.562, 0.681) | 0.617 | (0.560, 0.670) | 0.622 | (0.564, 0.680) |
| Unaligned | ||||||
| NS vs. MA | 0.316 | (0.270, 0.371) | 0.303 | (0.252, 0.361) | 0.316 | (0.268, 0.371) |
| NS vs. MD | 0.592 | (0.534, 0.648) | 0.595 | (0.534, 0.653) | 0.602 | (0.547, 0.659) |
| NS vs. SC | 0.560 | (0.498, 0.627) | 0.559 | (0.499, 0.618) | 0.561 | (0.502, 0.624) |
| MA vs. MD | 0.460 | (0.413, 0.507) | 0.467 | (0.417, 0.519) | 0.481 | (0.437, 0.533) |
| MA vs. SC | 0.434 | (0.387, 0.483) | 0.438 | (0.386, 0.489) | 0.446 | (0.397, 0.502) |
| MD vs. SC | 0.637 | (0.580, 0.697) | 0.632 | (0.567, 0.688) | 0.637 | (0.579, 0.695) |
| Potential Clones Exclued | ||||||
| Genome-aligned | ||||||
| NS vs. MA | 0.230 | (0.185, 0.275) | 0.218 | (0.174, 0.270) | 0.231 | (0.183, 0.279) |
| NS vs. MD | 0.522 | (0.456, 0.590) | 0.502 | (0.438, 0.571) | 0.508 | (0.438, 0.578) |
| NS vs. SC | 0.479 | (0.422, 0.536) | 0.468 | (0.411, 0.527) | 0.471 | (0.415, 0.535) |
| MA vs. MD | 0.431 | (0.380, 0.477) | 0.437 | (0.389, 0.490) | 0.451 | (0.398, 0.503) |
| MA vs. SC | 0.446 | (0.402, 0.492) | 0.440 | (0.398, 0.485) | 0.449 | (0.408, 0.496) |
| MD vs. SC | 0.569 | (0.506, 0.636) | 0.572 | (0.504, 0.637) | 0.576 | (0.516, 0.640) |
| Unaligned | ||||||
| NS vs. MA | 0.218 | (0.179, 0.258) | 0.201 | (0.161, 0.247) | 0.221 | (0.181, 0.267) |
| NS vs. MD | 0.489 | (0.430, 0.545) | 0.467 | (0.410, 0.525) | 0.479 | (0.419, 0.538) |
| NS vs. SC | 0.494 | (0.438, 0.550) | 0.479 | (0.431, 0.541) | 0.485 | (0.428, 0.538) |
| MA vs. MD | 0.397 | (0.354, 0.442) | 0.395 | (0.350, 0.440) | 0.413 | (0.368, 0.454) |
| MA vs. SC | 0.402 | (0.361, 0.446) | 0.394 | (0.349, 0.436) | 0.406 | (0.365, 0.450) |
| MD vs. SC | 0.576 | (0.520, 0.630) | 0.573 | (0.522, 0.627) | 0.579 | (0.530, 0.640) |
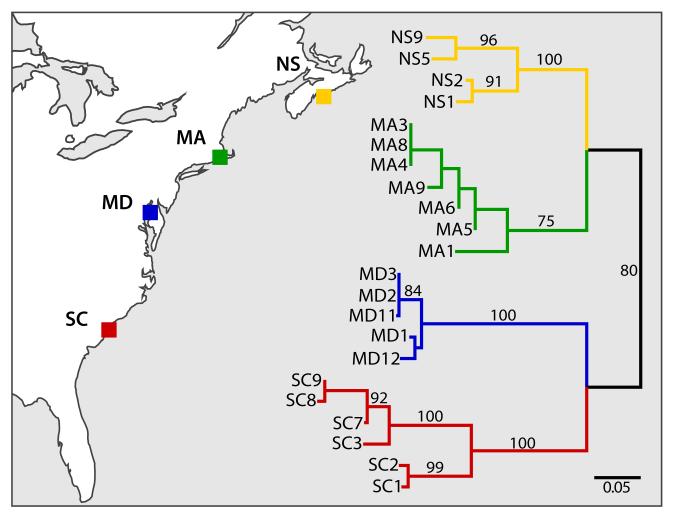
The inferred phylogeographic hypotheses clustered individuals according to their sampling location, indicating that individuals in each population share a most recent common ancestor not shared with individuals from other populations (Figure 5). The Nova Scotia and Massachusetts populations form a monophyletic group with respect to the other two southern populations, which is consistent with the sorting of historical lineages inferred by the PCA and STRUCTURE clustering. Tree topologies of the phylogenies inferred in all analyses using genome aligned or unaligned reads and with or without potential clones are virtually identical (Figures 5 and S8), with minor differences in the bootstrap support values of the most poorly supported branches. The number of characters used to perform the phylogenetic analyses was roughly 430 (Table 4). Approximately 36% of the characters were invariable in the analyses of genome-aligned reads and 47% in the analyses without genome alignment. As expected, the proportion of autapomorphic characters was greater in the analyses where potential clones were excluded.
Figure 5.
Phylogeography of N. vectensis. (left) Map showing the location of the sampled populations: Nova Scotia (NS), Massachusetts (MA), Maryland (MD), and South Carolina (SC). (right) Maximum likelihood tree showing the most-likely phylogeographic hypothesis inferred in the analysis of genome aligned reads without potential clones. Branches are labeled and colored to indicate site of collection. Numbers indicate bootstrap support values. Scale bar indicates substitutions per site.
Table 4.
Statistics of the matrices used for four phylogeographic analyses (with or without clones, genome aligned or unaligned reads).
| Potential Clones Included | |
| Genome-aligned | |
| Total number of characters | 472 |
| Proportion of invariable characters | 0.364 |
| Proportion of parsimony-informative characters | 0.553 |
| Proportion of autapomorphic characters | 0.083 |
| Proportion of missing data | 0.327 |
| Unaligned | |
| Total number of characters | 344 |
| Proportion of invariable characters | 0.471 |
| Proportion of parsimony-informative characters | 0.439 |
| Proportion of autapomorphic characters | 0.090 |
| Proportion of missing data | 0.335 |
| Potential Clones Exclued | |
| Genome-aligned | |
| Total number of characters | 484 |
| Proportion of invariable characters | 0.362 |
| Proportion of parsimony-informative characters | 0.519 |
| Proportion of autapomorphic characters | 0.120 |
| Proportion of missing data | 0.334 |
| Unaligned | |
| Total number of characters | 432 |
| Proportion of invariable characters | 0.477 |
| Proportion of parsimony-informative characters | 0.398 |
| Proportion of autapomorphic characters | 0.125 |
| Proportion of missing data | 0.408 |
DISCUSSION
In this study we have performed one of the first applications of RAD-seq to a marine invertebrate and examined genome-wide distribution of polymorphisms in natural populations of a coastal cnidarian. Together, our data reveal strong population genetic structure and clear phylogeographic relationships. Additionally, through statistical analyses of FST outliers, we have identified candidate regions of the genome of N. vectensis likely undergoing balancing selection in these populations. Our findings were largely insensitive to the availability of a reference genome and to the possible presence of clone individuals. These results further highlight the application of RAD-seq, and other population genomic approaches, towards understanding the genetic relationships of marine invertebrate populations and generating hypotheses about functional portions of the genome being shaped by natural selection.
RAD sequencing
Remarkably, 99% of the reads produced in this study passed as high-quality reads after our conservative filtering criteria (Figure S1), indicating that RAD-seq data from cnidarians can be of extremely high quality. Given the fact that ca. 96% of the reads had a positive alignment to the reference genome it can be inferred that the amount of foreign DNA contributing to the pool of RAD tags was extremely low; demonstrating that cnidarian DNA purified from non-sterile tissues (e.g., whole individuals) is suitable for RAD sequencing. As with other population genetic studies, additional factors that could have contributed to the 4% of reads unable to produce an alignment to the reference genome include: PCR errors, sequencing errors, genetic divergence, and completeness of the reference genome. However, the greatest loss of data arose from reads with multiple alignments to the reference genome (24% of all reads, see Figure S1). This phenomenon can be attributed to the presence of repetitive elements in the genome, which could comprise significantly large fractions of eukaryote genomes (de Koning et al. 2011), and recently duplicated genomic regions, which lack sufficient divergence for unique identification. For comparison, in the threespine stickleback study from Hohenlohe et al. (2010) 61% of the reads generated by RAD-seq (SbfI, 28-44bp read length) produced unique alignments to the genome. Nelson et al. (2011) found that only 86% of RAD-seq reads sampled in silico from the sorghum genome (PstI and BsrFI, 36-76bp read length) could be uniquely aligned. Thus, it is clear that there is a limit to the maximum fraction of RAD tag reads that can be uniquely aligned to a given reference genome. Increasing the read lengths should theoretically increase this fraction.
Based on the number of SbfI cut sites counted in the genome of N. vectensis (ca. 2 000) it was expected that roughly 4 000 RAD markers would be obtained after sequencing. In our dataset, 58% of the expected RAD markers were covered when these were identified from reads with unique alignments to the reference genome. This coverage would increase given a larger sequencing effort; plots of number of reads vs. number of RAD markers suggest that the cumulative number of covered RAD markers is close- to, but has not yet reached an asymptotic value (see Figure S2). The maximum number of RAD markers that can be recovered via the analytical methods employed in this study is significantly smaller than the actual number of RAD markers present in a given genome of an N. vectensis individual. Specifically, RAD markers from repetitive regions cannot be appropriately accounted for with current methodologies. It is thus possible, if not likely, that the ratio of expected-to-observed number of RAD markers varies across different taxa with different genome architectures and with the kind of restriction enzyme employed. As an example, Nelson et al. (2011) achieved 57% and 73% coverage of the expected number of RAD markers in the sorghum reference genome for the BsrFI and PstI enzymes, respectively. In contrast, Hohenlohe et al. (2010) achieved approximately 94% coverage of RAD markers generated with SbfI in the threespine stickleback genome.
Population differentiation
The demographic inferences based on the neutral biallelic SNP markers derived from RAD loci indicated that there is strong structuring among the examined populations of N. vectensis, which span over 2 000 km of coastline. Strong population differentiation and complete monophyly of populations are consistent with limited dispersal and low connectivity, as previously inferred for N. vectensis (Reitzel et al. 2008). The pairwise FST values calculated from genome-aligned markers ranged between 0.218 and 0.61. Hohenlohe et al. (2010) reported a genome-wide average FST value of 0.01 between oceanic highly-dispersing threespine stickleback populations collected 1 000 km apart, whereas the values for this statistic ranged from 0.05 to 0.15 between pairs of oceanic vs. freshwater populations that have been separated for less than 10 000 years. Similarly Roesti et al. (2012) reported FST values of 0.00 to 0.15 between pairs of stream vs. freshwater stickleback populations. The relatively high FST values calculated for the examined populations of N. vectensis could be inflated if the sampled individuals were close relatives (Allendorf-Phelps effect, see Allendorf & Phelps 1981; Waples 1998). In the extreme and unlikely case that the effective number of breeders responsible for the sampled individuals in a given population (Nb) was only 2, then the maximum magnitude of the contribution of this Allendorf-Phelps effect to the observed FST values would be 0.25, calculated as 1/2(Nb) (Waples 1998 and citations therein). This value is smaller than most of the estimated pairwise FST values among populations of N. vectensis in this study. Another potentially important source of bias on the estimation of FST values can arise from the variable and relatively small sample sizes. The contribution of this sampling error to raw FST estimates has been shown to be approximately 1/(2S) (Waples 1998 and citations therein), where S is the number of individuals sampled from a population. However, the FST estimators used in this study (Weir & Cockerham 1984; Reich et al. 2009) explicitly account for this source of bias. Furthermore, a recent simulation study showed that the FSTR estimator is extremely accurate even when sample sizes are very small (n < 6), and that its precision is great provided that a large number of independent markers are employed (> 100) (Willing et al. 2012). Therefore, the significant, strong differentiation among populations of N. vectensis found in this study does not seem to be a methodological or analytical artifact, but in fact a real pattern.
Possible clone individuals
The potential presence of clones among the sampled individuals had no dramatic effects on the overall population demographic inferences in this study. The main parameters that showed consistent, yet small, changes in the analyses excluding potential clones vs. analyses including potential clones were genetic diversity (Table 2) and genetic differentiation (FST values, see Table 3). Overall, the smaller genetic diversity and larger genetic differentiation observed when potential clones were included could be caused by the overrepresentation of particular genotypes and biases in the allelic differences among populations. The decrease in population sample sizes after the exclusion of potential clones could have also magnified the effect of sampling error and thus contributed to the observed small parameter changes.
Phylogeography
We found a clear genetic break between northern (NS, MA) and southern (MD, SC) populations, but a colonization scenario that could explain this pattern is unclear. Principally, we were unable to root the tree due to the uncertainty in the history of these populations and the lack of a clear outgroup species. Because the northern portion of the range of N. vectensis was covered during the last glacial maximum, a reasonable hypothesis would be that populations recolonized estuaries north of Cape Cod after the glaciers receded, similar to other coastal invertebrates (Jennings et al. 2009). Thus, we would expect reduced genetic diversity in these higher latitude populations. However, genetic diversity was overall higher in these more northern populations. Similarly, genetic diversity assayed with AFLPs (Reitzel et al. 2008) and by sequence-based markers (Reitzel et al. 2008; Sullivan et al. 2009; Reitzel et al. 2010) also suggested that genetic diversity is similar or even higher in populations north of Cape Cod. Previous research with the estuarine fishes Fundulus heteroclitus (Adams et al. 2006; Williams & Oleksiak 2008) and Menidia menidia (Mach et al. 2011), both of which have overlapping ranges with N. vectensis, has also observed similar genetic diversity among populations along the Atlantic coast of North America. In these fish species, the absence of reduced diversity in higher latitude populations is in part a result of local adaptation along environmental clines and in response to anthropogenic stressors. These environmental variables have shaped the regional genetic diversity despite the movement of populations during glacial periods. Future research with N. vectensis incorporating additional locations along the Atlantic coast of North America may help resolve the directionality of population colonization and the importance of genetic adaptation to regional environmental conditions.
Utility and promise for non-model organisms
Local physical oceanographic processes and human-mediated introductions can greatly influence the population connectivity dynamics among estuarine communities. The life history of N. vectensis, containing an egg mass that retains embryos, a demersal larva with a short swimming period (< 7 days), and an infaunal adult, would likely promote limited dispersal of adults and developmental stages. Consistent with this expectation, surveys of genetic structure within estuaries and between adjacent locations have identified significant structure (Reitzel et al. 2008). Previous genetic research has indicated that anthropogenic dispersal has played an important role shaping the broad geographic scale distribution and resulting population genetic relationships in N. vectensis (Darling et al. 2004; Reitzel et al. 2008; Darling et al. 2009). Similar to a number of other coastal invertebrates in North America, N. vectensis appears to have been introduced from the Atlantic coast to the Pacific coast, potentially through the transport of commercial shellfish. The addition of the high density SNP data generated in this study to previous data will provide a high degree of analytical power to understand both genetic partitioning in the small spatial scales of natural dispersal and large scales of long-distance anthropogenic dispersal. Even more so, these methods hold great opportunity for understanding similar processes in other coastal species. Despite the differences in the number of loci and SNP recovered when reads were filtered with the genome and when they were not, the results from the demographic inferences were overall identical. Furthermore, the use of the reference genome did not substantially affect the number of retrieved RAD loci across populations, thus avoiding the introduction of an ascertainment bias. Our results highlight the usefulness of RAD sequencing for population genetics and evolutionary studies with or without the availability of a reference genome (for more examples see Baird et al. 2008; Emerson et al. 2010; Amores et al. 2011; Baxter et al. 2011; Dasmahapatra et al. 2012; Peterson et al. 2012). Because most coastal and oceanic species from shallow and deep environments lack genomic resources, RAD-seq offers a valuable tool for the identification of native source locations for introduced species, and a tremendous opportunity for the characterization of genetic diversity in other species of ecological or conservation interest, especially those for which basic taxonomic and population structure knowledge has been particularly challenging to obtain (e.g., octocorals, see Herrera et al. 2010; McFadden et al. 2010 and references therein; Herrera et al. 2012).
Selection
High density SNP maps generated from field-sampled populations can be used to identify genomic regions potentially under selection. When correlated with known phenotypic diversity, linkage studies provide a powerful tool in functional genomics to bridge genetic and phenotypic variation (Feder & Mitchell-Olds 2003; Mitchell-Olds et al. 2008; Stinchcombe & Hoekstra 2008; Nadeau & Jiggins 2010). RAD-seq and similar methods, e.g., restriction-site tiling analysis (Pespeni et al. 2010), that generate large number of SNPs provide the technological approaches to produce these data for non-model species. For example, studies in stickleback (Hohenlohe et al. 2010) and the purple sea urchin (Pespeni et al. 2012) have each identified novel genomic regions under selection, which correlate with differential phenotypes in natural populations. Given the extensive latitudinal range and high degree of genetic structure of N. vectensis, it is reasonable to expect local adaptation in its populations.
Two previous studies with N. vectensis mined SNPs from Sanger-sequenced expressed sequence tags and identified geographically segregated polymorphisms in highly conserved regions of genes (Reitzel et al. 2010), one of which has dramatic functional impacts on protein function (NF-κB, Sullivan et al. 2009). This previous approach has clear limitations because SNPs could only be identified in coding regions, which are certainly important in adaptive evolution (Hoekstra & Coyne 2007), but would not identify SNPs in non-coding regions that are also of functional importance (Wray 2007). Furthermore, this approach introduces biases, such as ascertainment bias, because all source sequences for SNP identification are generated from individuals collected at one geographic location. In this study, we have identified SNPs throughout the genomes of individuals collected from four geographic locations. We utilized the restriction enzyme SbfI to generate the RAD tags, which would at most produce ca. 2 000 cuts based on counts from the reference genome of N. vectensis. This number is considerably smaller than the number of cut sites in the genomes of teleost fishes (ca. 25 000 to 30 000), such as the threespine stickleback (Hohenlohe et al. 2010; Amores et al. 2011), which makes it impossible to generate equivalent high-density mapping for N. vectensis from data generated with this same restriction enzyme. To achieve a higher mapping density, additional, more frequently cutting restriction enzymes would be required (e.g., EcoRI). However, even under this restriction, we identified 37 polymorphic sites common among all analyses that were inferred to be under balancing selection. Perhaps surprisingly, a large majority of these SNPs were in gene bodies, many of which have clear orthology to proteins of known function in other animals. For example, one SNP was located in an intron of a single ortholog to DNA methyltransferase 1, an enzyme that establishes and regulates tissue-specific patterns of cytosine methylation, and an intergenic SNP located nearest to heat shock factor I, the principle transcription factor that regulates downstream expression of genes involved in temperature stress. Future research utilizing a more frequently cutting enzyme will generate a higher density SNP map, which will facilitate a more thorough analysis of genomic regions under selection in these populations.
Future directions for Nematostella
In addition to resolving population relationships and identification of genomic regions undergoing selection, RAD-seq identification of SNPs can be used as a tool to push functional molecular studies in N. vectensis. Identification of SNPs linked to a particular genomic region will allow researchers to identify and test the relationship of candidate genomic loci to phenotypes of interest. N. vectensis has emerged as a premier model in cnidarian developmental biology and is a prime candidate as an experimental system in functional molecular genetics. Experimentally induced mutations combined with SNP profiling are a powerful tool that can be used to identify mutations underlying novel phenotypes in N. vectensis with high resolution. Researchers would be able to exploit the asexual reproductive biology of N. vectensis to perpetually maintain deleterious alleles in heterozygous individuals, which would facilitate conducting forward genetic screens to investigate molecular mechanisms governing development of particular morphological characters or differences in physiology. This unbiased forward approach would be an influential technological leap for evolutionary developmental biology and evolutionary ecology of cnidarians, which, until now, rely heavily on candidate gene approaches. Such unbiased approaches would inherently investigate novel mechanisms governing biological processes.
CONCLUSION
We have presented the broad utility of RAD-seq to characterize the genome-wide distribution of polymorphisms in a coastal invertebrate. Our data reveal strong population genetic structure, clear phylogeographic relationships, and candidate regions of the genome undergoing selection in natural populations. This approach holds tremendous promise towards understanding the genetic relationships and phylogeography of other marine invertebrates, including those of conservation concern that have traditionally be difficult to study due to lack of genetic variation (e.g., corals). Population genomic approaches will also facilitate collection of necessary data for empirically measuring the role of the environment selecting for local adaptation via ecologically important regions of the genome to generate hypotheses about functional portions of the genome being shaped by natural and anthropogenic selection.
Supplementary Material
Figure S1. Percentages of the number of reads retained after each major filtering step. Circled areas are proportional to the percentages.
Figure S2. Scatter plots of (top) the coverage per locus and (bottom) the number of identified RAD markers a functions of the number of reads generated per individual. Each point represents a single individual. Solid squares represent the average coverage per marker achieved from reads not aligned to the reference genome, and open squares represent their respective standard deviations. Open circles indicate the number of markers identified without alignments to the reference genome. Conversely, closed circles indicate the number of markers identified with alignments to the reference genome. Colors indicate the geographic site locations of each individual: Nova Scotia (yellow), Massachusetts (green), Maryland (blue), and South Carolina (red).
Figure S3. Box and whisker plots showing the effect of utilizing the N. vectensis reference genome to filter sequence reads. Data are from resampled datasets and are grouped by population: Nova Scotia (NS), Massachusetts (MA), Maryland (MD), and South Carolina (SC). (a) Number of reads with a unique alignment to the reference genome. (b) Number of RAD markers built from the genome-aligned reads. Boxes indicate the 25th and 75th percentiles. Black horizontal lines indicate the median. Open circles indicate the mean and red bars indicate the 95% confidence intervals. Black whiskers indicate maximum and minimum values.
Figure S4. Scatter plots of FST vs. expected heterozygosity (He) for the biallelic SNP in the four analyses. (top left) Analysis of genome aligned reads with potential clones. (top right) Analysis of genome aligned reads without potential clones. (bottom left) Analysis of genome unaligned reads with potential clones (same as Figure 1). (bottom right) Analysis of genome unaligned reads without potential clones. Colored boundaries indicate the 95% confidence intervals obtained through simulations in LOSITAN. Red region indicates candidates for positive selection, and yellow regions candidates for balancing selection.
Figure S5. PCA eigenvector 1 vs. eigenvector 2, results of the four analyses. (top left) Analysis of genome aligned reads with potential clones. (top right) Analysis of genome aligned reads without potential clones. (bottom left) Analysis of genome unaligned reads with potential clones (same as Figure 4 left). (bottom right) Analysis of genome unaligned reads without potential clones. Each dot represents an individual. Colors indicate the geographic site locations: Nova Scotia (NS; yellow), Massachusetts (MA; green), Maryland (MD; blue), and South Carolina (SC; red).
Figure S6. PCA eigenvector 1 vs. eigenvector 3, results of the four analyses. (top left) Analysis of genome aligned reads with potential clones. (top right) Analysis of genome aligned reads without potential clones. (bottom left) Analysis of genome unaligned reads with potential clones (same as Figure 4 middle). (bottom right) Analysis of genome unaligned reads without potential clones. Each dot represents an individual. Colors indicate the geographic site locations: Nova Scotia (NS; yellow), Massachusetts (MA; green), Maryland (MD; blue), and South Carolina (SC; red).
Figure S7. PCA eigenvector 2 vs. eigenvector 3, results of the four analyses. (top left) Analysis of genome aligned reads with potential clones. (top right) Analysis of genome aligned reads without potential clones. (bottom left) Analysis of genome unaligned reads with potential clones (same as Figure 4 right). (bottom right) Analysis of genome unaligned reads without potential clones. Each dot represents an individual. Colors indicate the geographic site locations: Nova Scotia (NS; yellow), Massachusetts (MA; green), Maryland (MD; blue), and South Carolina (SC; red).
Figure S8. Phylogeography of N. vectensis, results of the four analyses. Presented trees show the most-likely phylogeographic hypotheses resulting from maximum likelihood analyses. (top left) Analysis of genome aligned reads with potential clones. (top right) Analysis of genome aligned reads without potential clones. (bottom left) Analysis of genome unaligned reads with potential clones (same as Figure 4 left). (bottom right) Analysis of genome unaligned reads without potential clones. Branch colors are equivalent and match the ones in Figures 4, 5, S2, S5, S6 and S7. Numbers indicate percentage of 1000 bootstrap. Scale bars indicates substitutions per site.
ACKNOWLEDGEMENTS
AMR was supported by Ruth L. Kirschstein National Research Service Award F32HD062178, National Institutes of Health, NICHD. We are grateful for the support provided by the Office of Ocean Exploration, National Oceanic and Atmospheric Administration (NA05OAR4601054) the National Science Foundation (OCE-0624627; OCE-1131620), and the Academic Programs Office (Ocean Ventures Fund award to SH), the Deep Ocean Exploration Institute (Fellowship support to TMS) and the Ocean Life Institute of the Woods Hole Oceanographic Institution. MJL was supported by Ruth L. Kirschstein National Research Service Award FHD0550002, National Institutes of Health, NICHD. Partial funding for data generation was provided by the Woods Hole Oceanographic Institution to Dr. Ann Tarrant (WHOI). We thank members of the Shank lab for proofreading earlier versions of this manuscript. The comments from three anonymous reviewers substantially improved this manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
AMR, SH, and MJL designed the experiment. AMR and SH analyzed the data and drafted the manuscript. MQM and TMS participated in design of the study and interpretation of results. All authors approved the final manuscript.
DATA ACCESSIBILITY
-Raw DNA sequence reads are available at the U.S. National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) accession number SRA055050.
-Alignments of RAD sequence reads to the reference genome, as produced by BOWTIE, and the RAD markers used in the population genomic analyses are available in DRYAD doi:10.5061/dryad.gk2vc.
-Genomic positions, protein IDs, population statistics and GO results for each one of the candidate loci under selection are available as supplementary material.
REFERENCES
- Adams SM, Lindmeier JB, Duvernell DD. Microsatellite analysis of the phylogeography, Pleistocene history and secondary contact hypotheses for the killifish, Fundulus heteroclitus. Molecular Ecology. 2006;15:1109–1123. doi: 10.1111/j.1365-294X.2006.02859.x. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Phelps SR. Use of allelic frequencies to describe population-structure. Canadian Journal of Fisheries and Aquatic Sciences. 1981;38:1507–1514. [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the Teleost genome duplication. Genetics. 2011;188:799–U779. doi: 10.1534/genetics.111.127324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao T, Lopes A, Lopes R, Beja-Pereira A, Luikart G. LOSITAN: A workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud-Haond S, Belkhir K. GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Molecular Ecology Notes. 2007;7:15–17. [Google Scholar]
- Arnaud-Haond S, Duarte CM, Alberto F, Serrao EA. Standardizing methods to address clonality in population studies. Molecular Ecology. 2007;16:5115–5139. doi: 10.1111/j.1365-294X.2007.03535.x. [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, et al. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS One. 2008;3:3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baums IB. A restoration genetics guide for coral reef conservation. Molecular Ecology. 2008;17:2796–2811. doi: 10.1111/j.1365-294X.2008.03787.x. [DOI] [PubMed] [Google Scholar]
- Baxter SW, Davey JW, Johnston JS, et al. Linkage mapping and comparative genomics using next-generation RAD sequencing of a non-model organism. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bay L, Crozier R, Caley M. The relationship between population genetic structure and pelagic larval duration in coral reef fishes on the Great Barrier Reef. Marine Biology. 2006;149:1247–1256. [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society of London Series B-Biological Sciences. 1996;263:1619–1626. [Google Scholar]
- Bilewitch JP, Degnan SM. A unique horizontal gene transfer event has provided the octocoral mitochondrial genome with an active mismatch repair gene that has potential for an unusual self-contained function. BMC Evolutionary Biology. 2011;11:228. doi: 10.1186/1471-2148-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botsford LW, White JW, Coffroth MA, et al. Connectivity and resilience of coral reef metapopulations in marine protected areas: matching empirical efforts to predictive needs. Coral Reefs. 2009;28:327–337. doi: 10.1007/s00338-009-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury IR, Laurel B, Snelgrove PVR, Bentzen P, Campana SE. Global patterns in marine dispersal estimates: the influence of geography, taxonomic category and life history. Proceedings of the Royal Society B-Biological Sciences. 2008;275:1803–1809. doi: 10.1098/rspb.2008.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito PH, Edwards SV. Multilocus phylogeography and phylogenetics using sequence-based markers. Genetica. 2008;135:439–455. doi: 10.1007/s10709-008-9293-3. [DOI] [PubMed] [Google Scholar]
- Brumfield R. The utility of single nucleotide polymorphisms in inferences of population history. Trends in Ecology & Evolution. 2003;18:249–256. [Google Scholar]
- Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH. Stacks: building and genotyping Loci de novo from short-read sequences. G3. 2011;1:171–182. doi: 10.1534/g3.111.000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciannelli L, Knutsen H, Olsen EM, et al. Small-scale genetic structure in a marine population in relation to water circulation and egg characteristics. Ecology. 2010;91:2918–2930. doi: 10.1890/09-1548.1. [DOI] [PubMed] [Google Scholar]
- Cockerham CC, Weir BS. Estimation of gene flow from F-statistics. Evolution. 1993;47:855–863. doi: 10.1111/j.1558-5646.1993.tb01239.x. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cowen RK, Lwiza KMM, Sponaugle S, Paris CB, Olson DB. Connectivity of marine populations: open or closed? Science. 2000;287:857–859. doi: 10.1126/science.287.5454.857. [DOI] [PubMed] [Google Scholar]
- Cowen RK, Sponaugle S. Larval dispersal and marine population connectivity. Annual Review of Marine Science. 2009;1:443–466. doi: 10.1146/annurev.marine.010908.163757. [DOI] [PubMed] [Google Scholar]
- Darling JA, Kuenzi A, Reitzel AM. Human-mediated transport determines the non-native distribution of the anemone Nematostella vectensis, a dispersal-limited estuarine invertebrate. Marine Ecology Progress Series. 2009;380:137–146. [Google Scholar]
- Darling JA, Reitzel AM, Finnerty JR. Regional population structure of a widely introduced estuarine invertebrate: Nematostella vectensis Stephenson in New England. Molecular Ecology. 2004;13:2969–2981. doi: 10.1111/j.1365-294X.2004.02313.x. [DOI] [PubMed] [Google Scholar]
- Darling JA, Reitzel AR, Burton PM, et al. Rising starlet: the starlet sea anemone, Nematostella vectensis. Bioessays. 2005;27:211–221. doi: 10.1002/bies.20181. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra KK, Walters JR, Briscoe AD, et al. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012:1–5. doi: 10.1038/nature11041. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. Plos Genetics. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit P, Palumbi SR. Transcriptome-wide polymorphisms of red abalone (Haliotis rufescens) reveal patterns of gene flow and local adaptation. Molecular Ecology. 2012 doi: 10.1111/mec.12081. [DOI] [PubMed] [Google Scholar]
- Diniz-Filho JAF, de Campos Telles MP, Bonatto SL, Eizirik E, De Freitas TRO, De Marco P, Santos FR, Sole-Cava A, Soares TN. Mapping the evolutionary twilight zone: molecular markers, populations and geography. Journal of Biogeography. 2008;35:753–763. [Google Scholar]
- Earl DA, Vonholdt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources. 2012;4:359–361. [Google Scholar]
- Emerson KJ, Merz CR, Catchen JM, et al. Resolving postglacial phylogeography using high-throughput sequencing. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2010;107:16196–16200. doi: 10.1073/pnas.1006538107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Research. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurby S, Barber PH. Theoretical limits to the correlation between pelagic larval duration and population genetic structure. Molecular Ecology. 2012;21:3419–3432. doi: 10.1111/j.1365-294X.2012.05609.x. [DOI] [PubMed] [Google Scholar]
- Feder ME, Mitchell-Olds T. Evolutionary and ecological functional genomics. Nature Reviews Genetics. 2003;4:651–657. doi: 10.1038/nrg1128. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hand C, Uhlinger K. The unique, widely distributed sea anemone, Nematostella vectensis Stephenson: A review, new facts, and questions. Estuaries. 1994;17:501–508. [Google Scholar]
- Hauser L, Carvalho GR. Paradigm shifts in marine fisheries genetics: ugly hypotheses slain by beautiful facts. FishandFisheries. 2008;9:333–362. [Google Scholar]
- Herrera S, Baco A, Sánchez JA. Molecular systematics of the bubblegum coral genera (Paragorgiidae, Octocorallia) and description of a new deep-sea species. Molecular Phylogenetics and Evolution. 2010;55:123–135. doi: 10.1016/j.ympev.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Herrera S, Shank TM, Sanchez JA. Spatial and temporal patterns of genetic variation in the widespread antitropical deep-sea coral Paragorgia arborea. Molecular Ecology. 2012 doi: 10.1111/mec.12074. doi: 10.1111/mec.12074. [DOI] [PubMed] [Google Scholar]
- Hinrichs AL, Suarez BK. Genotyping errors, pedigree errors, and missing data. Genetic Epidemiology. 2005;29:S120–S124. doi: 10.1002/gepi.20120. [DOI] [PubMed] [Google Scholar]
- Hoekstra HE, Coyne JA. The locus of evolution: Evo devo and the genetics of adaptation. Evolution. 2007;61:995–1016. doi: 10.1111/j.1558-5646.2007.00105.x. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, et al. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. Plos Genetics. 2010:6. doi: 10.1371/journal.pgen.1000862. - [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TP, Baird AH, Bellwood DR, et al. Climate change, human impacts, and the resilience of coral reefs. Science. 2003;301:929–933. doi: 10.1126/science.1085046. [DOI] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Mark Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biology. 2007:8. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings RM, Shank TM, Mullineaux LS, Halanych KM. Assessment of the Cape Cod phylogeographic break using the bamboo worm Clymenella torquata reveals the role of regional water masses in dispersal. Journal of Heredity. 2009;100:86–96. doi: 10.1093/jhered/esn067. [DOI] [PubMed] [Google Scholar]
- Johnstone IM. On the distribution of the largest eigenvalue in principal components analysis. Annals of Statistics. 2001;29:295–327. [Google Scholar]
- Jones CG, Lawton JH, Shachak M. Organisms as ecosystem engineers. Oikos. 1994;69:373–386. [Google Scholar]
- Kumar S, Skjaeveland A, Orr RJS, et al. AIR: A batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics. 2009:10. doi: 10.1186/1471-2105-10-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009:10. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart G, England PR, Tallmon D, Jordan S, Taberlet P. The power and promise of population genomics: from genotyping to genome typing. Nature Reviews Genetics. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- Mach M, Sbrocco E, Hice L, et al. Regional differentiation and post-glacial expansion of the Atlantic silverside, Menidia menidia, an annual fish with high dispersal potential. Marine Biology. 2011;158:515–530. doi: 10.1007/s00227-010-1577-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariette S, Le Corre V, Austerlitz F, Kremer A. Sampling within the genome for measuring within-population diversity: trade-offs between markers. Molecular Ecology. 2002:11. doi: 10.1046/j.1365-294x.2002.01519.x. [DOI] [PubMed] [Google Scholar]
- McFadden CS, Sanchez JA, France SC. Molecular phylogenetic insights into the evolution of Octocorallia: a review. Integrative and Comparative Biology. 2010;50:389–410. doi: 10.1093/icb/icq056. [DOI] [PubMed] [Google Scholar]
- Mitchell-Olds T, Feder M, Wray G. Evolutionary and ecological functional genomics. Heredity. 2008;100:101–102. doi: 10.1038/sj.hdy.6801015. [DOI] [PubMed] [Google Scholar]
- Morin PA, Luikart G, Wayne RK, Grp SW. SNPs in ecology, evolution and conservation. Trends in Ecology & Evolution. 2004;19:208–216. [Google Scholar]
- Nadeau NJ, Jiggins CD. A golden age for evolutionary genetics? Genomics studies of adaptation in natural populations. Trends in Genetics. 2010;26:484–492. doi: 10.1016/j.tig.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Nelson JC, Wang SC, Wu YY, et al. Single-nucleotide polymorphism discovery by high-throughput sequencing in sorghum. Bmc Genomics. 2011:12. doi: 10.1186/1471-2164-12-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbi SR. Population genetics, demographic connectivity, and the design of marine reserves. Ecological Applications. 2003;13:S146–S158. [Google Scholar]
- Palumbi SR. Marine reserves and ocean neighborhoods: The spatial scale of marine populations and their management. Annual Review of Environment and Resources. 2004;29:31–68. [Google Scholar]
- Parker PG, Snow AA, Schug MD, Booton GC, Fuerst PA. What molecules call tell us about populations: choosing and using a molecular marker. Ecology. 1998;79:361–382. [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. Plos Genetics. 2006;2:2074–2093. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CVM, Rogers AD, Sheader M. The genetic structure of the rare lagoonal sea anemone, Nematostella vectensis Stephenson (Cnidaria; Anthozoa) in the United Kingdom based on RAPD analysis. Molecular Ecology. 2002;11:2285–2293. doi: 10.1046/j.1365-294x.2002.01621.x. [DOI] [PubMed] [Google Scholar]
- Pespeni MH, Garfield DA, Manier MK, Palumbi SR. Genome-wide polymorphisms show unexpected targets of natural selection. Proceedings of the Royal Society B: Biological Sciences. 2012;279:1412–1420. doi: 10.1098/rspb.2011.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pespeni MH, Oliver TA, Manier MK, Palumbi SR. Method Restriction Site Tiling Analysis: accurate discovery and quantitative genotyping of genome-wide polymorphisms using nucleotide arrays. Genome Biology 11. 2010;11:R44. doi: 10.1186/gb-2010-11-4-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One. 2012;7:e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit RJ, Duminil J, Fineschi S, et al. Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Molecular Ecology. 2004;14:689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purchell JE, Uye S, Lo W-T. Anthropogenic causes of jellyfish blooms and their direct consequences for humans: a review. Marine Ecology Progress Series. 2007;350:153–174. [Google Scholar]
- Putnam NH, Srivastava M, Hellsten U, et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- Rambaut A. FigTree: Tree figure drawing tool, version 1.3.1. Institute of Evolutionary Biology, University of Edinburgh; 2009. [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–U450. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Burton PM, Krone C, Finnerty JR. Comparison of developmental trajectories in the starlet sea anemone Nematostella vectensis: embryogenesis, regeneration, and two forms of asexual fission. Invertebrate Biology. 2007;126:99–112. [Google Scholar]
- Reitzel AM, Darling JA, Sullivan JC, Finnerty JR. Global population genetic structure of the starlet anemone Nematostella vectensis: multiple introductions and implications for conservation policy. Biological Invasions. 2008;10:1197–1213. [Google Scholar]
- Reitzel AM, Sullivan JC, Finnerty JR. Discovering SNPs in protein coding regions with StellaSNP: Illustrating the characterization and geographic distribution of polymorphisms in the estuarine anemone Nematostella vectensis. Estuaries and coasts. 2010;33:930–943. [Google Scholar]
- Rioux Paquette S. PopGenKit: Useful Functions for File Conversion and Data Resampling in Microsatellite Datasets. R package, version 1.0. 2011 Available at http://cran.r-project.org/web/packages/PopGenKit/index.html.
- Roberts JM, Wheeler AJ, Freiwald A. Reefs of the deep: The biology and geology of cold-water coral ecosystems. Science. 2006;312:543–547. doi: 10.1126/science.1119861. [DOI] [PubMed] [Google Scholar]
- Roesti M, Hendry AP, Salzburger W, Berner D. Genome divergence during evolutionary diversification as revealed in replicate lake-stream stickleback population pairs. Molecular Ecology. 2012;21:2852–2862. doi: 10.1111/j.1365-294X.2012.05509.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Molecular Ecology Notes. 2004;4:137–138. [Google Scholar]
- Rowe HC, Renaut S, Guggisberg A. RAD in the realm of next-generation sequencing technologies. Molecular Ecology. 2011;20:3499–3502. doi: 10.1111/j.1365-294x.2011.05197.x. [DOI] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer T, Van Oppen M, Romano S, Worheide G. Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria) Molecular Ecology. 2002;11:2475–2487. doi: 10.1046/j.1365-294x.2002.01652.x. [DOI] [PubMed] [Google Scholar]
- Silva JF, Lima CA, Perez CD, Gomes PB. First record of the sea anemone Nematostella vectensis (Actiniaria: Edwardsiidae) in Southern Hemisphere waters. Zootaxa. 2010;2343:66–68. [Google Scholar]
- Stinchcombe JR, Hoekstra HE. Combining population genomics and quantitative genetics: finding the genes underlying ecologically important traits. Heredity. 2008;100:158–170. doi: 10.1038/sj.hdy.6800937. [DOI] [PubMed] [Google Scholar]
- Sullivan JC, Wolenski FS, Reitzel AM, et al. Two alleles of NF-κB in the sea anemone Nematostella vectensis are widely dispersed in nature and encode proteins with distinct activities. PLoS One. 2009;4:e7311. doi: 10.1371/journal.pone.0007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnucks P. Efficient genetic markers for population biology. Trends in Ecology & Evolution. 2000;15:199–203. doi: 10.1016/s0169-5347(00)01825-5. [DOI] [PubMed] [Google Scholar]
- Tavare S. Lectures on Mathematics in the Life Sciences (American Mathematical Society) 1986. Some Probabilistic and Statistical Problems in the Analysis of DNA Sequences; pp. 57–86. [Google Scholar]
- Tracy CA, Widom H. Level-spacing distributions and the airy kernel. Communications in Mathematical Physics. 1994;159:151–174. [Google Scholar]
- Waples RS. Separating the wheat from the chaff: Patterns of genetic differentiation in high gene flow species. Journal of Heredity. 1998;89:438–450. [Google Scholar]
- Weersing K, Toonen RJ. Population genetics, larval dispersal, and connectivity in marine systems. Marine Ecology Progress Series. 2009;393:1–12. [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-Statistics for the analysis of population-structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Williams LM, Oleksiak MF. Signatures of selection in natural populations adapted to chronic pollution. BMC Evolutionary Biology. 2008;8:282. doi: 10.1186/1471-2148-8-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing EM, Dreyer C, van Oosterhout C. Estimates of genetic differentiation measured by F(ST) do not necessarily require large sample sizes when using many SNP markers. PLoS One. 2012;7:e42649. doi: 10.1371/journal.pone.0042649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nature Reviews Genetics. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Percentages of the number of reads retained after each major filtering step. Circled areas are proportional to the percentages.
Figure S2. Scatter plots of (top) the coverage per locus and (bottom) the number of identified RAD markers a functions of the number of reads generated per individual. Each point represents a single individual. Solid squares represent the average coverage per marker achieved from reads not aligned to the reference genome, and open squares represent their respective standard deviations. Open circles indicate the number of markers identified without alignments to the reference genome. Conversely, closed circles indicate the number of markers identified with alignments to the reference genome. Colors indicate the geographic site locations of each individual: Nova Scotia (yellow), Massachusetts (green), Maryland (blue), and South Carolina (red).
Figure S3. Box and whisker plots showing the effect of utilizing the N. vectensis reference genome to filter sequence reads. Data are from resampled datasets and are grouped by population: Nova Scotia (NS), Massachusetts (MA), Maryland (MD), and South Carolina (SC). (a) Number of reads with a unique alignment to the reference genome. (b) Number of RAD markers built from the genome-aligned reads. Boxes indicate the 25th and 75th percentiles. Black horizontal lines indicate the median. Open circles indicate the mean and red bars indicate the 95% confidence intervals. Black whiskers indicate maximum and minimum values.
Figure S4. Scatter plots of FST vs. expected heterozygosity (He) for the biallelic SNP in the four analyses. (top left) Analysis of genome aligned reads with potential clones. (top right) Analysis of genome aligned reads without potential clones. (bottom left) Analysis of genome unaligned reads with potential clones (same as Figure 1). (bottom right) Analysis of genome unaligned reads without potential clones. Colored boundaries indicate the 95% confidence intervals obtained through simulations in LOSITAN. Red region indicates candidates for positive selection, and yellow regions candidates for balancing selection.
Figure S5. PCA eigenvector 1 vs. eigenvector 2, results of the four analyses. (top left) Analysis of genome aligned reads with potential clones. (top right) Analysis of genome aligned reads without potential clones. (bottom left) Analysis of genome unaligned reads with potential clones (same as Figure 4 left). (bottom right) Analysis of genome unaligned reads without potential clones. Each dot represents an individual. Colors indicate the geographic site locations: Nova Scotia (NS; yellow), Massachusetts (MA; green), Maryland (MD; blue), and South Carolina (SC; red).
Figure S6. PCA eigenvector 1 vs. eigenvector 3, results of the four analyses. (top left) Analysis of genome aligned reads with potential clones. (top right) Analysis of genome aligned reads without potential clones. (bottom left) Analysis of genome unaligned reads with potential clones (same as Figure 4 middle). (bottom right) Analysis of genome unaligned reads without potential clones. Each dot represents an individual. Colors indicate the geographic site locations: Nova Scotia (NS; yellow), Massachusetts (MA; green), Maryland (MD; blue), and South Carolina (SC; red).
Figure S7. PCA eigenvector 2 vs. eigenvector 3, results of the four analyses. (top left) Analysis of genome aligned reads with potential clones. (top right) Analysis of genome aligned reads without potential clones. (bottom left) Analysis of genome unaligned reads with potential clones (same as Figure 4 right). (bottom right) Analysis of genome unaligned reads without potential clones. Each dot represents an individual. Colors indicate the geographic site locations: Nova Scotia (NS; yellow), Massachusetts (MA; green), Maryland (MD; blue), and South Carolina (SC; red).
Figure S8. Phylogeography of N. vectensis, results of the four analyses. Presented trees show the most-likely phylogeographic hypotheses resulting from maximum likelihood analyses. (top left) Analysis of genome aligned reads with potential clones. (top right) Analysis of genome aligned reads without potential clones. (bottom left) Analysis of genome unaligned reads with potential clones (same as Figure 4 left). (bottom right) Analysis of genome unaligned reads without potential clones. Branch colors are equivalent and match the ones in Figures 4, 5, S2, S5, S6 and S7. Numbers indicate percentage of 1000 bootstrap. Scale bars indicates substitutions per site.