Abstract
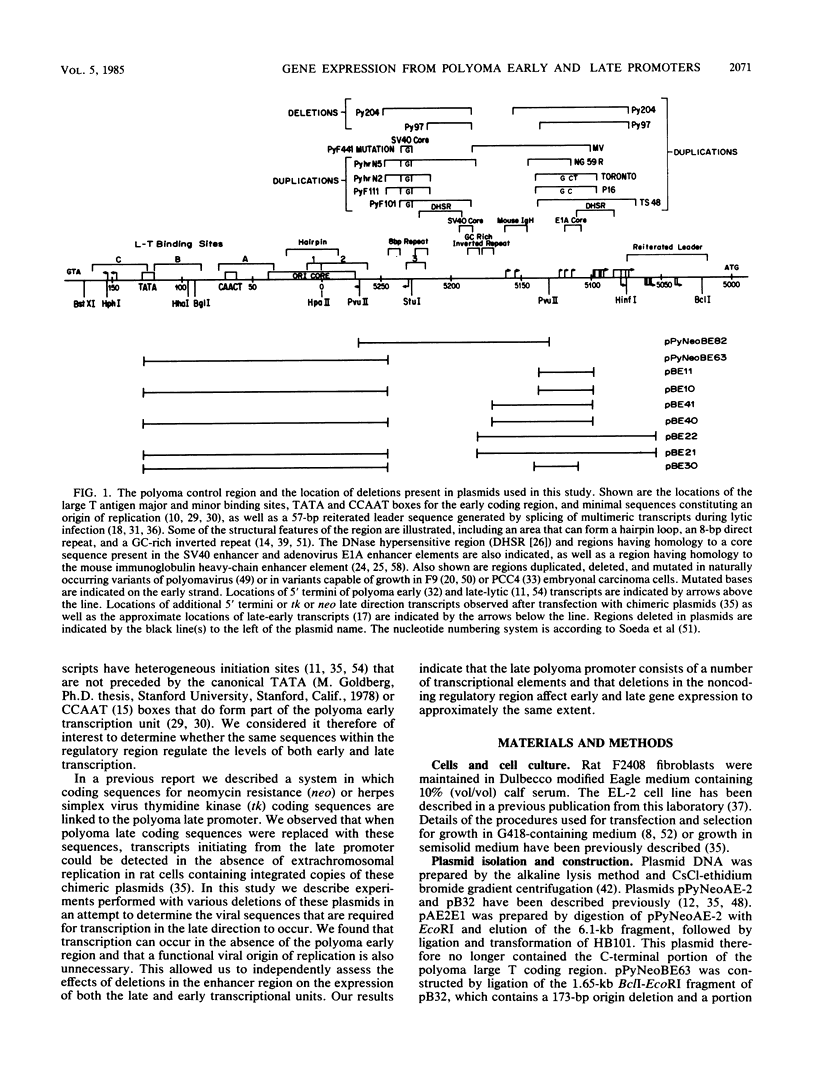
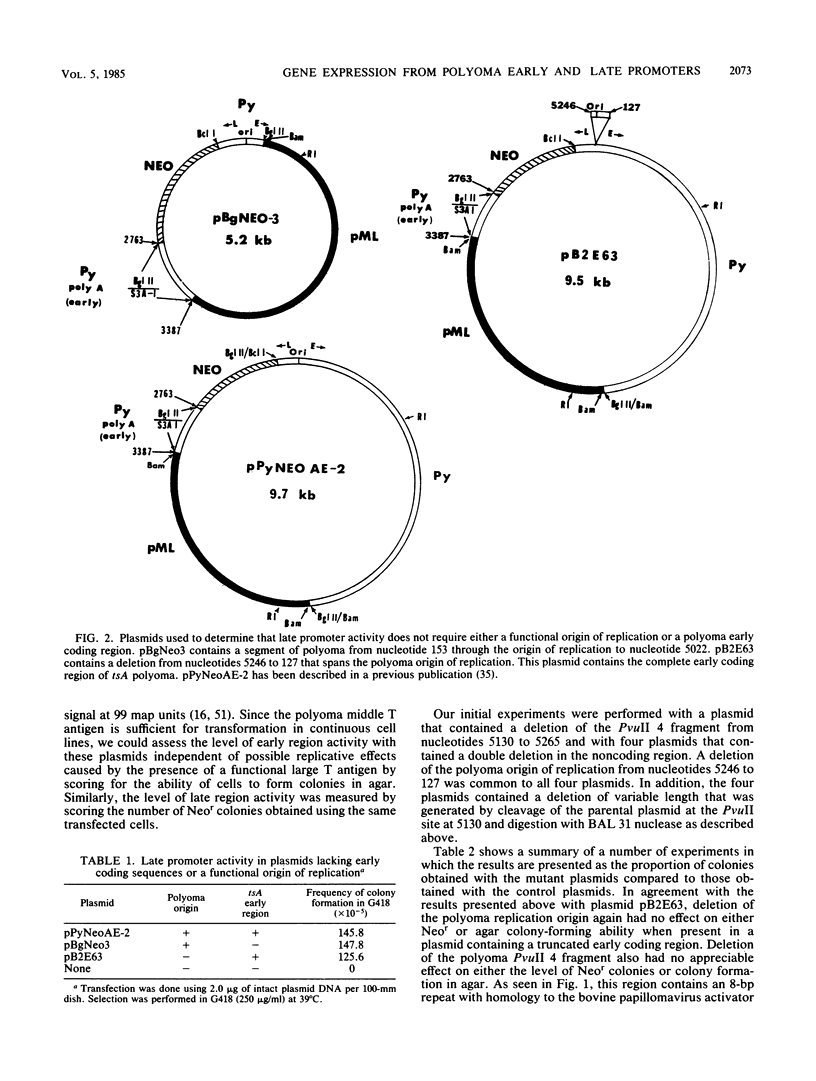
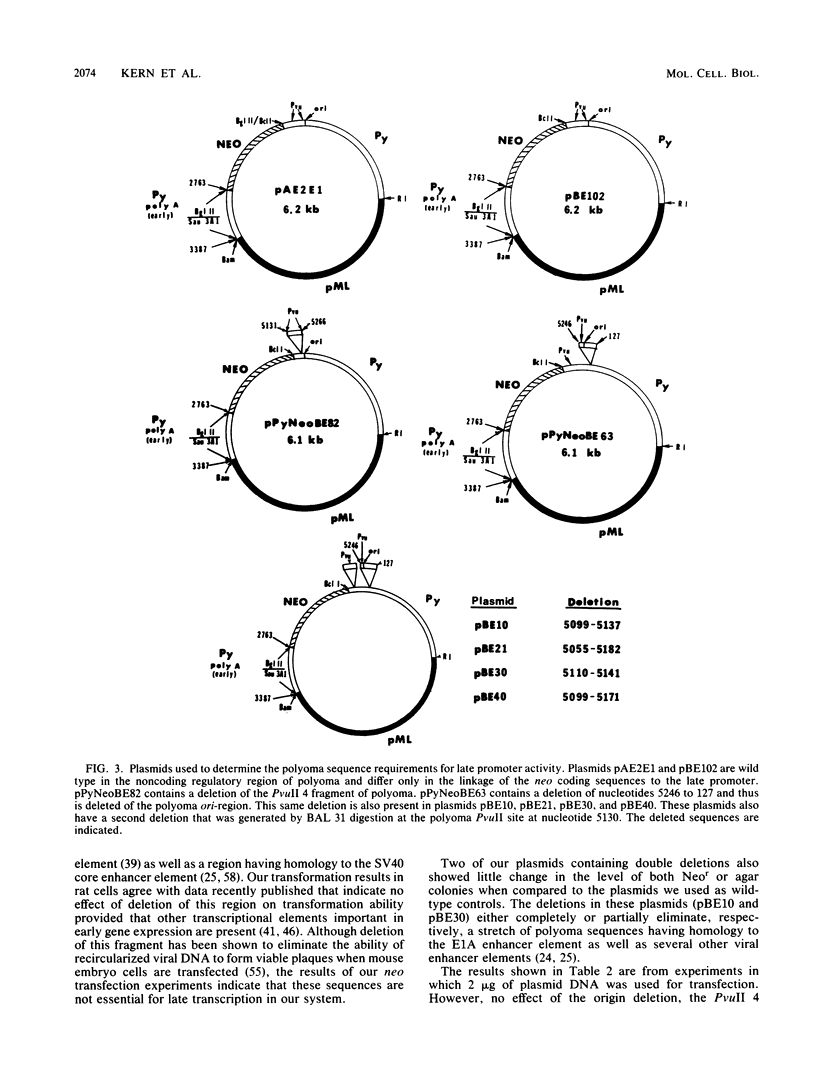
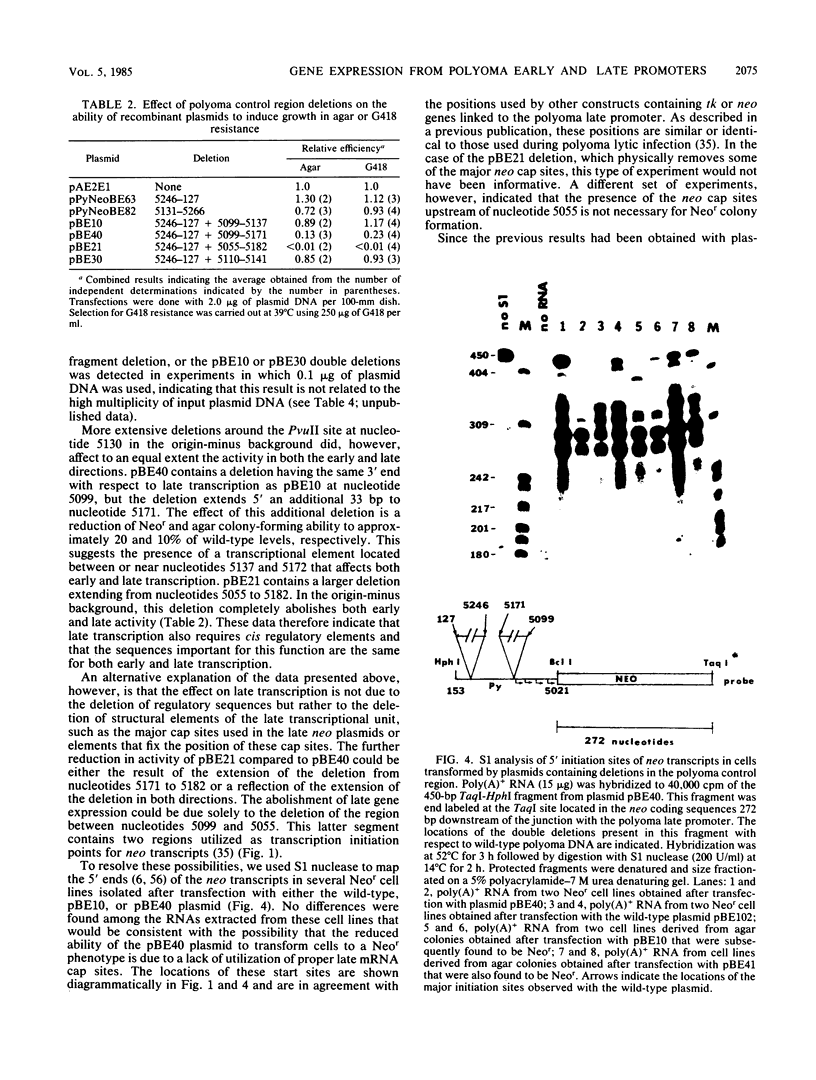
In a previous report we showed that transcripts initiating from the late promoter of integrated polyoma plasmids could be detected at significant levels when neomycin resistance (neo) coding sequences were linked to this promoter. In this report we used chimeric plasmids that contain either a limited portion of the polyoma genome or deletions within the polyoma noncoding regulatory region to determine the sequence requirements for late promoter activity in this system. We observed no absolute requirement for either the polyoma early coding region or the origin of DNA replication for Neo-r colony formation. We were therefore able to independently assess the effects of deletions in the polyoma enhancer region on gene activity in both the early and late directions. We measured the ability of cells transfected with plasmids containing deletions in this region to form colonies in either semisolid or G418-containing medium under nonreplicative conditions. Our results indicate that either the PvuII 4 fragment, which contains the simian virus 40 core enhancer sequence, or a region from nucleotides 5099 to 5142, which contains the adenovirus type 5 E1A core enhancer sequence, can be deleted without significantly affecting gene expression in either direction. However, a deletion of nucleotides 5099 to 5172 reduced activities to similar extents in both directions, and a plasmid containing a larger deletion of nucleotides 5055 to 5182 showed a further reduction in activity. Although having no effect by itself, a second origin region deletion of nucleotides 5246 to 127 when present in these mutant backgrounds caused either a further reduction or elimination, respectively, of both G418 and agar colony-forming ability, suggesting the presence of an additional common regulatory element within this region. A comparison of 5' ends of neo transcripts present in cells transformed by these plasmids suggested that the reduction in activity was due to deletion of regulatory rather than structural elements of the late promoter. Our results indicate that the noncoding region of polyoma contains multiple complementing regulatory elements that control the level of both early and late gene expression.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Beck E., Ludwig G., Auerswald E. A., Reiss B., Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982 Oct;19(3):327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Colbère-Garapin F., Horodniceanu F., Kourilsky P., Garapin A. C. A new dominant hybrid selective marker for higher eukaryotic cells. J Mol Biol. 1981 Jul 25;150(1):1–14. doi: 10.1016/0022-2836(81)90321-1. [DOI] [PubMed] [Google Scholar]
- Contreras R., Gheysen D., Knowland J., van de Voorde A., Fiers W. Evidence for the direct involvement of DNA replication origin in synthesis of late SV40 RNA. Nature. 1982 Dec 9;300(5892):500–505. doi: 10.1038/300500a0. [DOI] [PubMed] [Google Scholar]
- Cowie A., Kamen R. Multiple binding sites for polyomavirus large T antigen within regulatory sequences of polyomavirus DNA. J Virol. 1984 Dec;52(3):750–760. doi: 10.1128/jvi.52.3.750-760.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowie A., Tyndall C., Kamen R. Sequences at the capped 5'-ends of polyoma virus late region mRNAs: an example of extreme terminal heterogeneity. Nucleic Acids Res. 1981 Dec 11;9(23):6305–6322. doi: 10.1093/nar/9.23.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey L., Pellegrini S., Basilico C. Deletion of the origin of replication impairs the ability of polyomavirus DNA to transform cells and to form tandem insertions. J Virol. 1984 Mar;49(3):984–987. doi: 10.1128/jvi.49.3.984-987.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- FRIED M. CELL-TRANSFORMING ABILITY OF A TEMPERATURE-SENSITIVE MUTANT OF POLYOMA VIRUS. Proc Natl Acad Sci U S A. 1965 Mar;53:486–491. doi: 10.1073/pnas.53.3.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton R. G., Basilico C. Changes in the topography of early region transcription during polyoma virus lytic infection. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7142–7146. doi: 10.1073/pnas.79.23.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton R. G., Basilico C. Viral gene expression in polyoma virus-transformed rat cells and their cured revertants. J Virol. 1981 Oct;40(1):150–163. doi: 10.1128/jvi.40.1.150-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell A. J., Cowie A., Arrand J. R., Kamen R. Localization of three major cappe 5' ends of polyoma virus late mRNA's within a single tetranucleotide sequence in the viral genome. J Virol. 1980 Feb;33(2):902–908. doi: 10.1128/jvi.33.2.902-908.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura F. K., Deininger P. L., Friedmann T., Linney E. Mutation near the polyoma DNA replication origin permits productive infection of F9 embryonal carcinoma cells. Cell. 1981 Mar;23(3):809–814. doi: 10.1016/0092-8674(81)90445-1. [DOI] [PubMed] [Google Scholar]
- Gattoni S., Colantuoni V., Basilico C. Relationship between integrated and nonintegrated viral DNA in rat cells transformed by polyoma virus. J Virol. 1980 Jun;34(3):615–626. doi: 10.1128/jvi.34.3.615-626.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell S. W., Byrne B. J., Subramanian K. N. Mapping of the late promoter of simian virus 40. Proc Natl Acad Sci U S A. 1984 Jan;81(1):23–27. doi: 10.1073/pnas.81.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell S. W., Byrne B. J., Subramanian K. N. The simian virus 40 minimal origin and the 72-base-pair repeat are required simultaneously for efficient induction of late gene expression with large tumor antigen. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6335–6339. doi: 10.1073/pnas.81.20.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Saragosti S., Blangy D., Yaniv M. Fine structure of the origin-proximal DNAase I-hypersensitive region in wild-type and EC mutant polyoma. Cell. 1981 Sep;25(3):651–658. doi: 10.1016/0092-8674(81)90172-0. [DOI] [PubMed] [Google Scholar]
- Hutchinson M. A., Hunter T., Eckhart W. Characterization of T antigens in polyoma-infected and transformed cells. Cell. 1978 Sep;15(1):65–77. doi: 10.1016/0092-8674(78)90083-1. [DOI] [PubMed] [Google Scholar]
- Ito Y., Spurr N., Dulbecco R. Characterization of polyoma virus T antigen. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1259–1263. doi: 10.1073/pnas.74.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jat P., Novak U., Cowie A., Tyndall C., Kamen R. DNA sequences required for specific and efficient initiation of transcription at the polyoma virus early promoter. Mol Cell Biol. 1982 Jul;2(7):737–751. doi: 10.1128/mcb.2.7.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jat P., Roberts J. W., Cowie A., Kamen R. Comparison of the polyoma virus early and late promoters by transcription in vitro. Nucleic Acids Res. 1982 Feb 11;10(3):871–887. doi: 10.1093/nar/10.3.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Favaloro J., Parker J. Topography of the three late mRNA's of polyoma virus which encode the virion proteins. J Virol. 1980 Feb;33(2):637–651. doi: 10.1128/jvi.33.2.637-651.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamen R., Jat P., Treisman R., Favaloro J., Folk W. R. 5' termini of polyoma virus early region transcripts synthesized in vivo by wild-type virus and viable deletion mutants. J Mol Biol. 1982 Aug 5;159(2):189–224. doi: 10.1016/0022-2836(82)90493-4. [DOI] [PubMed] [Google Scholar]
- Katinka M., Yaniv M., Vasseur M., Blangy D. Expression of polyoma early functions in mouse embryonal carcinoma cells depends on sequence rearrangements in the beginning of the late region. Cell. 1980 Jun;20(2):393–399. doi: 10.1016/0092-8674(80)90625-x. [DOI] [PubMed] [Google Scholar]
- Keller J. M., Alwine J. C. Activation of the SV40 late promoter: direct effects of T antigen in the absence of viral DNA replication. Cell. 1984 Feb;36(2):381–389. doi: 10.1016/0092-8674(84)90231-9. [DOI] [PubMed] [Google Scholar]
- Kern F. G., Basilico C. Transcription from the polyoma late promoter in cells stably transformed by chimeric plasmids. Mol Cell Biol. 1985 Apr;5(4):797–807. doi: 10.1128/mcb.5.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legon S., Flavell A. J., Cowie A., Kamen R. Amplification in the leader sequence of late polyoma virus mRNAs. Cell. 1979 Feb;16(2):373–388. doi: 10.1016/0092-8674(79)90013-8. [DOI] [PubMed] [Google Scholar]
- Liboi E., Caruso M., Basilico C. New rat cell line that is highly susceptible to transformation by several oncogenes. Mol Cell Biol. 1984 Dec;4(12):2925–2928. doi: 10.1128/mcb.4.12.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linney E., Donerly S. DNA fragments from F9 PyEC mutants increase expression of heterologous genes in transfected F9 cells. Cell. 1983 Dec;35(3 Pt 2):693–699. doi: 10.1016/0092-8674(83)90102-2. [DOI] [PubMed] [Google Scholar]
- Lusky M., Berg L., Weiher H., Botchan M. Bovine papilloma virus contains an activator of gene expression at the distal end of the early transcription unit. Mol Cell Biol. 1983 Jun;3(6):1108–1122. doi: 10.1128/mcb.3.6.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusky M., Botchan M. Inhibition of SV40 replication in simian cells by specific pBR322 DNA sequences. Nature. 1981 Sep 3;293(5827):79–81. doi: 10.1038/293079a0. [DOI] [PubMed] [Google Scholar]
- Luthman H., Nilsson M. G., Magnusson G. Non-contiguous segments of the polyoma genome required in cis for DNA replication. J Mol Biol. 1982 Nov 15;161(4):533–550. doi: 10.1016/0022-2836(82)90406-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller C. R., Mes-Masson A. M., Bouvier M., Hassell J. A. Location of sequences in polyomavirus DNA that are required for early gene expression in vivo and in vitro. Mol Cell Biol. 1984 Dec;4(12):2594–2609. doi: 10.1128/mcb.4.12.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W. J., Mueller C. R., Mes A. M., Hassell J. A. Polyomavirus origin for DNA replication comprises multiple genetic elements. J Virol. 1983 Sep;47(3):586–599. doi: 10.1128/jvi.47.3.586-599.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini S., Dailey L., Basilico C. Amplification and excision of integrated polyoma DNA sequences require a functional origin of replication. Cell. 1984 Apr;36(4):943–949. doi: 10.1016/0092-8674(84)90044-8. [DOI] [PubMed] [Google Scholar]
- Ruley H. E., Fried M. Sequence repeats in a polyoma virus DNA region important for gene expression. J Virol. 1983 Jul;47(1):233–237. doi: 10.1128/jvi.47.1.233-237.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekikawa K., Levine A. J. Isolation and characterization of polyoma host range mutants that replicate in nullipotential embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1981 Feb;78(2):1100–1104. doi: 10.1073/pnas.78.2.1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeda E., Arrand J. R., Smolar N., Walsh J. E., Griffin B. E. Coding potential and regulatory signals of the polyoma virus genome. Nature. 1980 Jan 31;283(5746):445–453. doi: 10.1038/283445a0. [DOI] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Treisman R. Characterisation of polyoma late mRNA leader sequences by molecular cloning and DNA sequence analysis. Nucleic Acids Res. 1980 Nov 11;8(21):4867–4888. doi: 10.1093/nar/8.21.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyndall C., La Mantia G., Thacker C. M., Favaloro J., Kamen R. A region of the polyoma virus genome between the replication origin and late protein coding sequences is required in cis for both early gene expression and viral DNA replication. Nucleic Acids Res. 1981 Dec 11;9(23):6231–6250. doi: 10.1093/nar/9.23.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., de Villiers J., Schaffner W. An SV40 "enhancer trap" incorporates exogenous enhancers or generates enhancers from its own sequences. Cell. 1984 Apr;36(4):983–992. doi: 10.1016/0092-8674(84)90048-5. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W. A small segment of polyoma virus DNA enhances the expression of a cloned beta-globin gene over a distance of 1400 base pairs. Nucleic Acids Res. 1981 Dec 11;9(23):6251–6264. doi: 10.1093/nar/9.23.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W., Tyndall C., Lupton S., Kamen R. Polyoma virus DNA replication requires an enhancer. Nature. 1984 Nov 15;312(5991):242–246. doi: 10.1038/312242a0. [DOI] [PubMed] [Google Scholar]




