Abstract
Nowadays, Enterococcus faecium is one of the leading nosocomial pathogens worldwide. Strains causing clinical infections or hospital outbreaks are enriched in the enterococcal surface protein (Esp) encoding ICEEfm1 mobile genetic element. Previous studies showed that Esp is involved in biofilm formation, endocarditis and urinary tract infections. In this study, we characterized the role of the putative AraC type of regulator (locus tag EfmE1162_2351), which we renamed ebrB and which is, based on the currently available whole genome sequences, always located upstream of the esp gene, and studied its role in Esp surface exposure during growth. A markerless deletion mutant of ebrB resulted in reduced esp expression and complete abolishment of Esp surface exposure, while Esp cell-surface exposure was restored when this mutant was complemented with an intact copy of ebrB. This demonstrates a role for EbrB in esp expression. However, during growth, ebrB expression levels did not change over time, while an increase in esp expression at both RNA and protein level was observed during mid-log and late-log phase. These results indicate the existence of a secondary regulation system for esp, which might be an unknown quorum sensing system as the enhanced esp expression seems to be cell density dependent. Furthermore, we determined that esp is part of an operon of at least 3 genes putatively involved in biofilm formation. A semi-static biofilm model revealed reduced biofilm formation for the EbrB deficient mutant, while dynamics of biofilm formation using a flow cell system revealed delayed biofilm formation in the ebrB mutant. In a mouse intestinal colonization model the ebrB mutant was less able to colonize the gut compared to wild-type strain, especially in the small intestine. These data indicate that EbrB positively regulates the esp operon and is implicated in biofilm formation and intestinal colonization.
Introduction
For long, the Gram-positive species Enterococcus faecium was considered a harmless commensal of the mammalian intestinal tract. However, in the last two decades E. faecium emerged as one of the leading nosocomial pathogens [1]. Molecular epidemiological studies using multilocus sequence typing (MLST) identified host-specific genogroups including three hospital-associated E. faecium (HA-Efm) lineages designated lineage-17, -18 and -78 [1]–[4]. HA-Efm are characterized by multidrug-resistance including ampicillin and quinolone resistance and enrichment of several (putative) virulence genes [5]–[8], including a gene encoding the approximately 200 kDa surface protein Esp, which is located on a pathogenicity island (PAI) [9]. Recently, we showed that this PAI is self-transmissible and contained the characteristics of an integrative conjugative element [10], and was therefore renamed in ICEEfm1. By constructing an esp insertion-deletion mutant (E1162Δesp:cat) and by using different animal models, we and others demonstrated that Esp is involved in biofilm formation [12] and contributes to the pathogenesis of urinary tract infection [13] and endocarditis [14]. Furthermore, van Wamel et al. [15] demonstrated variable cell surface expression of Esp among different strains, growth condition (temperature and presence of air oxygen) dependent expression of Esp and a correlation between expression levels and initial adherence and biofilm formation in a polystyrene binding assay. Further analysis of ICEEfm1 encoded genes identified upstream of esp an ORF with locus tag EfmE1162_2351 with similarity to the family of AraC type of transcriptional regulators. Here we report functional analysis of EfmE1162_2351, which we renamed ebrB for enterococcal biofilm regulator B and performed additional analysis to determine dynamics of Esp expression.
Materials and Methods
Ethics Statement
The Animal Care and Use Committee of the University of Amsterdam approved the mouse intestinal colonization experiment.
Bacterial strains, plasmids and growth conditions
E. faecium and Escherichia coli strains and plasmids used in this study are listed in Table 1. The E. coli strains DH5α (Invitrogen) and EC1000 [16] were grown in Luria-Bertani (LB) medium. Enterococci were grown in brain heart infusion (BHI) medium, Tryptic soy broth supplemented with 1% glucose (TSBg) or Tryptic soy agar supplemented with 5% sheep red blood cells (BA) (BD, Alphen aan den Rijn, The Netherlands) at 37°C. For enterococci, the antibiotics gentamicin and spectinomycin were used in concentrations of 300 µg/ml. For E. coli, gentamicin and spectinomycin were used in concentrations of 30 µg/ml and 100 µg/ml, respectively. All antibiotics were obtained from Sigma-Aldrich (Saint Louis, MO). The genome sequence of E. faecium strain E1162 is available (GenBank: ABQJ00000000), including the esp containing ICEEfm1 encoded on contig156 (ABQJ01000139).
Table 1. Bacterial strains and plasmids.
| Strain or plasmid | Relevant characteristics a | Reference or source |
| Strains | ||
| E. faecium | ||
| E1162 | Clinical blood isolate; AmpR, VanS, ChlS, GenS, SpcS; ICEEfm1+ | [11] |
| E1162ΔebrB | Markerless deletion mutant of ebrB of E1162; GenS; ICEEfm1+ | This study |
| E1162ΔebrB+pEF25 | Complementation strain of E1162ΔebrB, harboring "empty" vector pEF25; SpcR, GenS; ICEEfm1+ | This study |
| E1162ΔebrB+ebrB | Complementation strain of E1162ΔebrB, harboring pEF25-ebrB; SpcR, GenS; ICEEfm1+ | This study |
| E1162Δesp | Markerless deletion mutant of esp of E1162; GenS; ICEEfm1+ | This study |
| E1162Δesp+pEF25 | Complementation strain of E1162Δesp, harboring "empty" vector pEF25; SpcR, GenS; ICEEfm1+ | This study |
| E1162Δesp+esp | Complementation strain of E1162Δesp, harboring pEF25-esp; SpcR, GenS; ICEEfm1+ | This study |
| E0305 | Hospital outbreak isolate, ebrB natural mutant by insertion of IS256 | [24] |
| E0305+pEF25 | Complementation strain of E0305, harboring "empty" vector pEF25 | This study |
| E0305+ebrB | Complementation strain of E0305, harboring pEF25-ebrB | This study |
| E. coli | ||
| DH5α | E. coli host strain for routine cloning | Invitrogen |
| EC1000 | MC1000 glgB:repA | [16] |
| Plasmids | ||
| pWS3 | Shuttle plasmid; ts in gram-positive hosts; SpcR | [21] |
| pEF39 | pWS3: ebrB fusion with gentamicin resistance casssette cloned in the EcoRI site of the ebrBgene fusion fragment | This study |
| plasmid for generating an ebrB marked mutation; SpcR, GenR | ||
| pEF40 | pWS3: esp fusion with gentamicin resistance casssette cloned in the EcoRI site of the espgene fusion fragment | This study |
| plasmid for generating an esp marked mutation; SpcR, GenR | ||
| pWS3-Cre | pWS3 derivative expressing Cre in E. faecium | [20] |
| pAT18 | shuttle plasmid; EryR | [22] |
| pEF25 | shuttle plasmid pAT18 with spectinomycin resistance cassette cloned in the EcoRI site; SpcR, EryR | This study |
| pEF25-esp | Complementation plasmid for esp; pEF25 carrying gene esp; SpcR, EryR | This study |
| pEF25-ebrB | Complementation plasmid for ebrB; pEF25 carrying gene ebrB; SpcR, EryR | This study |
Amp, ampicillin; Van, vancomycin; Chl, chloramphenicol; Gen, gentamycin; Spc, spectinomycin; ICEEfm1+, E. faecium ebrB containing pathogenicity island
Standard molecular techniques
Plasmid DNA purification (Qiagen, Venlo, The Netherlands), digestion with restriction endonuclease (New England Biolabs, Leusden, The Netherlands), amplification of DNA by PCR performed in 25 µl volumes with HotStarTaq Master Mix (Qiagen, Venlo, The Netherlands), AccuPrime Taq DNA polymerase High Fidelity with buffer 1 (Invitrogen, Breda, The Netherlands) or Expand Long Template PCR system with buffer 3 (Roche Applied Sciences, Almere, The Netherlands) and ligation of DNA fragments with T4 DNA ligase (Invitrogen) were performed according to the manufacturers' instructions. E. faecium chromosomal DNA was purified using the Wizard Genomic DNA purification kit (Promega, Leiden, The Netherlands) according to the protocol with minor modifications. In brief, 1 ml of overnight culture was harvested by centrifugation and resuspended in 580 µl 50mM EDTA. Lysozyme (20 µl, 50 mg/ml) was added and the suspension was incubated at 37°C for 1 h. The sequential steps were according to the manufacturers' protocol. Primers were purchased from Invitrogen and are listed in Table 2. PCRs were performed with a 9800 Fast Thermal Cycler (Applied Biosystems, Life technologies, The Netherlands). PCR amplification conditions using HotStarTaq and performed in a volume of 25 µl were as follows: initial denaturation at 95°C for 15 min, followed by 30 cycles of 30 s at 94°C, 30 s at 53°C and 72°C (the time depending on the size of the PCR product). For AccuPrime™ Taq polymerase the following PCR conditions were used, initial denaturation at 94°C for 2 min, followed by 30 cycles of 30 s at 94°C, 30 s at 53°C and 68°C for 1 min per kb of PCR product. The PCR conditions for the Expand Long Template PCR systems included an initial denaturation at 95°C for 2 min, followed by 10 cycles of 10 s at 94°C, 30 s at 53°C and 68°C (the time depending on the size of the PCR product), followed by 25 cycles with the same denaturing and annealing and elongation conditions, with an increased elongation time of 20 s per cycle. Plasmids were introduced into E. faecium by electroporation using a Gene Pulser unit (Bio-Rad Laboratories, Richmond, CA) as described elsewhere [17].
Table 2. Primers used in this study.
| Primer name | Primer sequencea,b | Startpositionc |
| pAW068-spcF | 5′-GGAATTCTTTTGTTTCGAAGCAGCAGAT | |
| pAW068-spcR | 5′-GGAATTCGGACGCTTTATTCTTCCCAAA | |
| ebrB-1F | 5′-TCTGTCGTTCAATTCATCG | 69605 |
| ebrB-2F | 5′-CGGATCATAATAATTATTGTCTTTG | 68488 |
| ebrB-1R | 5′-GTCATATTCATTTAACACACTATTATTACC | 70088 |
| esp-1R | 5′-AATACTCTCTTATTATTCTTGCTAACC | 71021 |
| hyp-1R | 5′-ATTGGAGTTATCAACATTTTTTC | 70515 |
| Deletion/complementation | ||
| ebrBUp-F-XhoI | 5′-CCGCTCGAGCATATATCTTCTTAAATATCAAACATG | 68219 |
| ebrBUp-R-EcoRI | 5′-AGTATGGTTTGAATTCTAATAAGACTTCTTTATCTGAAAACAC | 68747 |
| ebrBDn-F-EcoRI | 5′-CTTATTAGAATTCAAACCATACTATCAGTGAAGTTTC | 69784 |
| ebrBDn-R-SmaI | 5′-CCCCCCGGGCGTAATAATCTTCCCAGCTTTC | 70302 |
| ebrB-check up | 5′-GTATTAGCGGTGTTCAAAATG | 67950 |
| ebrB-check down | 5′-ATTGGAGTTATCAACATTTTTTC | 70515 |
| ebrBcompF | 5′-GCGGAGCTCGTTAGCTTATTTTGACAGAGGAATAG | 68634 |
| ebrBcompR | 5′-GGTACGCCCGGGTCAGCTAATGTTGTTGAAATTG | 69925 |
| espUp-F-XhoI | 5′-CCGCTCGAGGTTGATAACTCCAATCATTCG | 70501 |
| espUp-R-EcoRI | 5′-GATTGTCAGGAATTCTCTCTTATTATTCTTGCTAACCAT | 71017 |
| espDn-F-EcoRI | 5′-ATAATAAGAGAGAATTCCTGACAATCAAGGTAGCAAC | 76742 |
| espDn-R-SmaI | 5′-CCCCCCGGGCTCAGAATTTAGTGTCATTCTATTTG | 77264 |
| esp-check up | 5′-TCTGTCGTTCAATTCATCG | 69605 |
| esp-check down | 5′-ATGTATTCCATTTTTTGATAGTATTTC | 77674 |
| espcompF | 5′-CCGGAATTCGCTTGCATCAAAATAAACTACATGGGTATAAT | 70993 |
| AGCAATGAAATGCATTTCAAAAATATTTTGAGGAGAATTT | ||
| AGTATGGTTAGCAAGAATAATAAGAG | ||
| espcompR | 5′-CCGGAATTCCCTCTTTTCAGAGAAGATT | 76954 |
| qRT-PCR | ||
| ebrB-RT-F | 5′-TGAGGGATTCTGGGATTGTTT | 69528 |
| ebrB-RT-R | 5′-GCCGATGAATTGAACGACAGA | 69625 |
| esp-RT-F | 5′-CCACGAGTTAGCGGGAACAG | 72498 |
| esp-RT-R | 5′-TTGGAGCCCCATCTTTTTCA | 72599 |
| nox-RT-F | 5′-AGCCGCAGCTCGATTTCTAA | 77095 |
| nox-RT-R | 5′-AACGATGTCCCACATTCCAA | 77193 |
| mur-RT-F | 5′-GGTGAGCCGATTCATGCAGT | 79670 |
| mur-RT-R | 5′-AACGCGGTTGATCCATCTTC | 79776 |
| 1542-RT-F | 5′-TGGTCACCTTACTGTTGTTGAGGA | 80323 |
| 1542-RT-R | 5′-CGTTTCATTCCCACAGTCACA | 80404 |
| efflux-RT-F | 5′-ACGGGTGGTACAAGCCATTG | 81857 |
| efflux-RT-R | 5′-GCCCGACCACGTTCATGTAT | 81949 |
| tufA-RT-F | 5′-TACACGCCACTACGCTCAC | |
| tufA-RT-R | 5′-AGCTCCGTCCATTTGAGCAG | |
| 5′-RACE | ||
| ebrB-GSP1 | 5′-CAGACCGAATCGTATCTCCA | 69383 |
| ebrB-GSP2 | 5′-ATCAGCCATTGCAACATTCA | 69043 |
| esp-GSP1 | 5′-GGTTTGCGTATCGGTTGTTT | 71577 |
| esp-GSP2 | 5′-TTCTGCCCCAGCAAATAATC | 71559 |
Restriction sites are boldface
Regions -35, -10, the ribosome binding site from the bacA promoter and the ATG startcodon of esp are underlined [12]
Nucleotide reference positions relative to ABQJ01000139
Bioinformatic analysis
The putative helix-turn-helix (HTH) motif of EbrB encompassing amino acids 345-386 was aligned with a sequence logo generated using a Prosite database containing 310 HTH motifs (accession number PS00041) in Weblogo, Version 2.8.2 [18]. Presence of putative transcription terminators were predicted using RNAfold (http://rna.tbi.univie.ac.at/) [19].
Generation of targeted markerless deletion mutants
Recently, in our group the first targeted markerless deletion mutants were constructed based on the Cre-lox system [20], [21]. Here, we used this method to generate markerless deletion mutants of the esp and ebrB gene. Previously, Heikens et al. constructed an esp insertion-deletion mutant E1162Δesp:cat [12]. In order to compare Esp expression levels with the newly generated E1162ΔebrB independently of a putative effect of the inserted chloramphenicol cassette, we also constructed a markerless deletion mutant in esp. For the amplification of the 5′-flanking regions of ebrB and esp, we used primers ebrBUp-F-XhoI, ebrBUp-R-EcoRI, espUp-F-XhoI and espUp-R-EcoRI, respectively, while for the 3′-flanking regions primers ebrBDn-F-EcoRI, ebrBDn-R-SmaI, espDn-F-EcoRI and espDn-R-SmaI were used (Table 2). Generation of a marked deletion mutant was performed as described [20] and was confirmed by PCR using the ebrB and esp check up and check down primers (Table 2). Removal of the gentamicin resistance marker was obtained by electroporation of pWS3-Cre into the marked deletion mutants as described [20]. Loss of the marker was confirmed by PCR using the ebrB and esp check up and check down primers.
In trans complementation of mutants
A modified pAT18 vector [22] designated pEF25 was used for complementation studies (Table 1). In this vector a spectinomycin resistance cassette amplified from vector pAW068 [23] using primers pAW068-spcF and pAW068-spcR both containing EcoRI restriction sites, was cloned into the EcoRI site of pAT18.
To complement E1162▵ebrB, ebrB was amplified from E1162 genomic DNA using AccuPrime™ Taq Polymerase with primers ebrBcompF and ebrBcompR (Table 2). The forward primer, which contained a SacI restriction site to facilitate cloning of the fragment, was located 86 bp upstream the startcodon of ebrB, to ensure that expression of ebrB is driven by its native promoter, which was mapped by 5′ RACE as described below. The reverse primer included a SmaI restriction site. The resulting ebrB containing PCR product was digested with SacI and SmaI and ligated to a similar digested pEF25 resulting in pEF25-ebrB. The recombinant plasmid pEF25-ebrB and the negative control pEF25 were introduced into the E1162▵ebrB by electroporation resulting in E1162?ebrB+ebrB and E1162?ebrB+pEF25, respectively (Table 1).
A screen for the presence of ebrB using primers ebrB-2F and ebrB-1R (Table 2) on a selection of isolates revealed that strain E0305 [24] was a natural mutant for ebrB as it contained an insertion of IS256 in ebrB as determined by sequencing of the obtained PCR fragment using the same primers ebrB-2F and ebrB-1R. To complement E0305 with an intact copy of ebrB, we also introduced pEF25 and pEF25-ebrB by electroporation resulting in E305+pEF25 and E0305+ebrB, respectively (Table 1).
Complementation of the E1162▵esp was performed as described for the ebrB mutant using primers espcompF, including the promoter regions of the bacA gene of Enterococcus faecalis and espcompR as described previously [12]. Several attempts to transform the ligation mixture in E. coli failed, which was also reported for other large repeat containing genes [25] though direct electroporation of the ligation mixture in the E1162▵esp resulted in stable complementation. The complemented strain of the E1162▵esp mutant was designated E1162▵esp+esp (Table 1). For comparison E1162▵esp was complemented with the empty vector pEF25 resulting in E1162▵esp+pEF25 (Table 1).
Determination of growth curves
A BioScreen C instrument (Oy Growth Curves AB, Helsinki, Finland) was used to monitor effects of esp or ebrB deletion on bacterial growth. Wild-type E. faecium E1162 and mutants were grown overnight in BHI and TSBg, while the complemented strains were grown in BHI and TSBg with the addition of spectinomycin. Cells were inoculated at an initial OD660 of 0.0025 into 300 µl BHI and TSBg and incubated in the Bioscreen C system at 37°C with continuous shaking and absorbance of 600 nm (A600) was recorded every 15 min for 9 hours. Each experiment was performed in triplicate.
Determination of (cell-surface) expression of Esp
Dynamics of Esp (cell surface) expression was determined for wild-type E1162, E1162▵ebrB, E1162▵esp, E0305 and the complemented strains grown on blood agar plate and in broth, including BHI and TSBg using flow cytometry, electron microscopy and Western blotting. Flow cytometry, electron microscopy and Western blotting were performed as previously described [12], [15]. Flow cytometry experiments were performed in triplicate.
Transcriptome profiling
In order to compare the transcriptomes of E. faecium E1162 and E1162▵ebrB, microarray analysis was performed on four independent biological replicates using a custom made 8×15K Agilent E. faecium E1162 microarray as previously described [20]. E1162 and E1162▵ebrB were grown in TSBg for 18 hours. Cultures were then diluted to OD660 0.025 in 20 ml of prewarmed TSBg and grown to OD660 0.3. RNA isolation, cDNA synthesis, labeling and hybridization were performed as previously described [20]. After removal of the data for the different controls printed on the microarray slides, the normalized data for each spot from the microarrays were analyzed for statistical significance using the Web-based VAMPIRE microarray suite [26], [27]. A spot was found to be differentially expressed between two samples using the threshold of a false discovery rate smaller than 0.05. Changes of ≥ 2-fold for up- and down regulated genes in the parental strain were introduced as additional significance limits. A gene with two identical probes or all four probes meeting this criterion were classified as differentially expressed. The microarray data generated in this study have been deposited in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-3801.
Reverse transcription and (quantitative) real-time RT-PCR
In all cases, RNA was isolated as described for the microarray. In order to investigate whether esp and four downstream of esp located genes with locus-tags EfmE1162_1544, EfmE1162_1543, EfmE1162_1542 and EfmE1162_1541 are part of an operon, we isolated RNA from wild-type strain E1162 (four biological replicates), performed first strand synthesis using Maxima reverse transcriptase (Fermentas, Thermo Scientific, St. Leon-Rot, Germany) in combination with 5′-end located gene specific primers 1544-1R, 1543-1R, 1542-1R and 1541-1R, respectively (Table 2, Fig. 1B) on each individual gene. The presence of intergenic cDNA was subsequently determined by PCR using the same gene specific primer in combination with a 3′-end located primer of its upstream located gene, i.e. esp-1F/1544-1R, 1544-1F/1543-1R, 1543-1F/1542-1R and 1542-1F/1541-1R (Fig.1B). As negative control the same procedure for cDNA synthesis was followed but without adding reverse transcriptase. As positive control for the PCR we included purified genomic E1162 DNA.
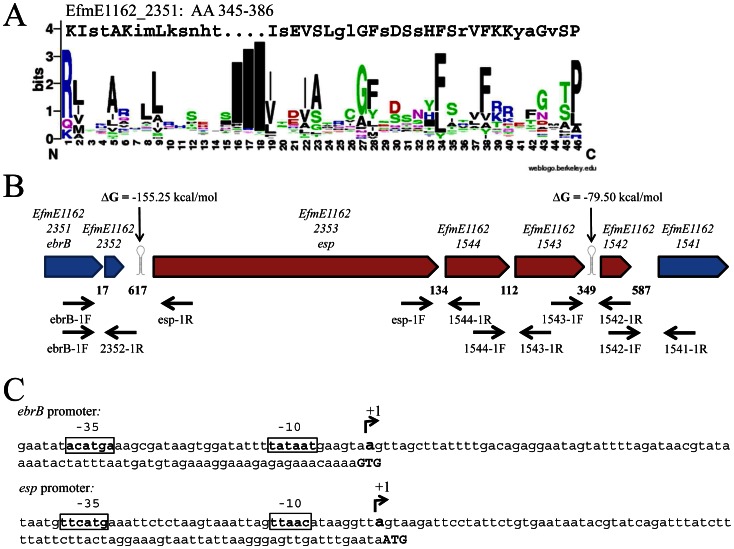
Figure 1. Structural organization of the ebrB region.
(A) Alignment of the putative helix-turn-helix motif, amino acids 345-386, of EbrB to a sequence logo generated using a Prosite database containing 310 HTH motifs (accession number PS00041) in Weblogo. Conserved amino acids are depicted in capital letter. (B) Overview ebrB region, including the esp operon in red, primer sites and predicted transcription terminators (RNAfold). Numbers indicate the intergenic distances in bp (C) ebrB and esp promoter mapping, in bold/box putative -35 and -10 sequence and transcription start (+1).
Quantitative real time PCR (qRT-PCR) was used to confirm the microarray data, growth condition dependent expression of esp and ebrB in wild-type strain E1162 and to determine whether esp is part of an operon by comparison of differential expression of esp and its previously mentioned four downstream of esp located genes, between strains E1162▵ebrB+ebrB and E1162▵ebrB+pEF25 and E1162▵esp+esp and E1162▵esp+pEF25. cDNA was synthesized from RNA using Maxima First strand cDNA synthesis kit for RT-qPCR (Fermentas, Thermo Scientific, St. Leon-Rot, Germany) and 1 µg of total RNA. Quantitative PCRs using primers indicated with RT (Table 2) on the synthesized cDNAs were performed using Maxima® SYBR Green/ROX qPCR Master Mix (Fermentas) using a StepOne™ Realtime PCR system (Applied Biosystems, Nieuwekerk a/d IJssel, The Netherlands) with the following program: 95°C for 10 min, and subsequently 40 cycles of 95°C for 15 sec, 55°C for 1 min. The expression of the tufA gene was used as a reference for the determination of relative expression levels (Table 2) [28]. For the confirmation of the microarray data and the analysis of the esp operon, relative transcript levels were calculated by using the relative expression software tool (REST (Qiagen)) [29]. For the growth condition dependent expression of esp and ebrB, we normalized the Ct values of each sample by the amplification efficiency. The relative transcript level (fold difference relative to tufA) of the esp and ebrB genes were calculated by using the normalized Ct value of the tufA housekeeping control minus the normalized Ct value of the esp and ebrB genes. The resulting value represents a log2 transformed fold difference in gene expression. Statistical significance between wild-type and mutant was assessed by the unpaired two-tailed Student′s t-test. This experiment was performed with two biological replicates and qRT-PCR performed in duplo.
Promoter mapping of ebrB and esp using 5′RACE
Total RNA was isolated as previously described [20]. We used the 5′ RACE kit (Rapid amplification of cDNA ends, Invitrogen, The Netherlands) to map the promoter of ebrB and esp according to the manufacturers′ protocol. After first strand synthesis using gene specific primers 1 (GSP1) (Table 2), a nested PCR with GSP2 primers was performed to amplify the product and cloned in pGEM-T Easy TA cloning vector (Promega, Madison, WI). Inserts were sequenced to determine the cDNA end.
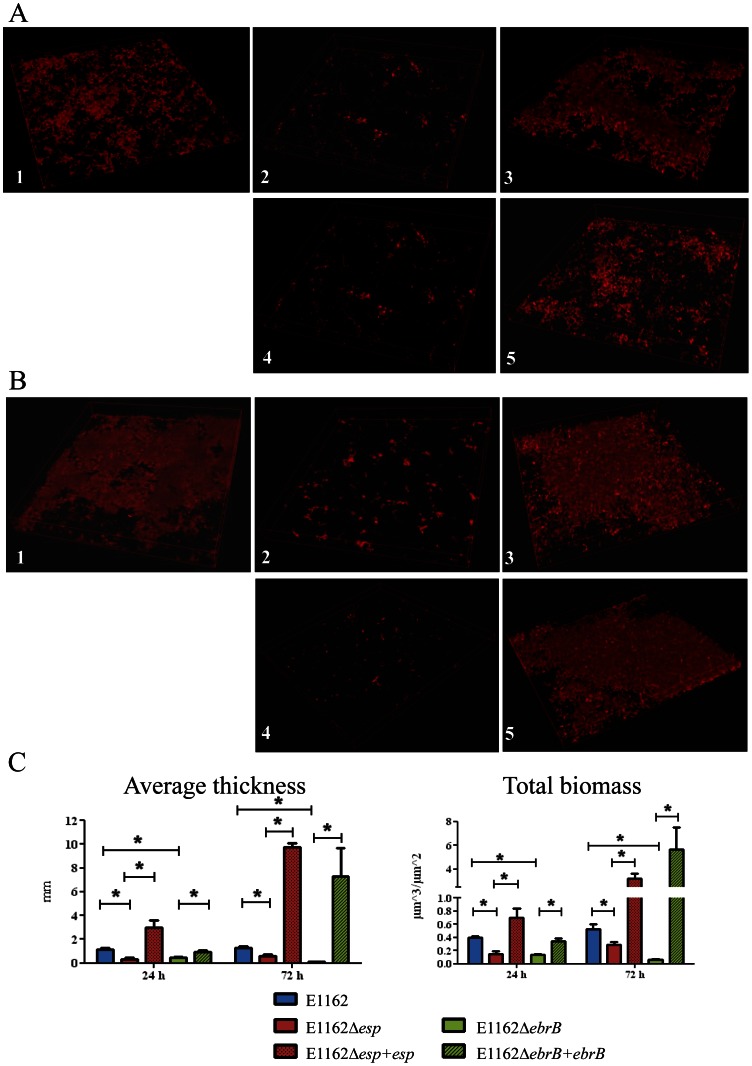
Biofilm semi-static model and confocal laser scanning microscopy (CLSM)
For the semi-static biofilm model, overnight in TSBg grown wild-type E1162, E1162Δesp and E1162ΔebrB and complemented strains E1162▵esp+esp, E1162▵esp+pEF25, E1162▵ebrB+ebrB and E1162▵ebrB+pEF25 were diluted to OD660 0.01 in a 6-well polystyrene plate (Corning Inc.) containing a poly-L-lysin coated coverslip (0.45 µm; diameter, 12 mm; Becton Dickinson) and 6 ml TSBg to facilitate attachment and biofilm formation. The 6-well plates were incubated at 37°C under gently shaking at 120 rpm, for 24 h or 72 h. The coverslips were washed two times with 0.85% NaCl, and the biofilms were chemically fixed using 8% glutaraldehyde (Merck) in 0.85% NaCl for 20 min and washed again two times with 0.85% NaCl. The biofilms were stained using 15 µg/ml propidium iodide (PI) in 0.85% NaCl for 15 min. After incubation, the stain was removed and coverslips were transferred to glass microscope slides. Biofilms were analyzed by a confocal laser scanning microscope (CLSM) (Leica SP5) equipped with an oil plan-neofluor 63x/1.4 objective. PI was excited at 633 nm. Z-stacks were taken with an interval of 0.42 µm. Pictures were analyzed with LAS AF software (Leica) and biofilm formation was quantified using Comstat (Heydorn et al., 2000)/Matlab R2010b software (The MathWorks). The average thickness and total biomass of the biofilms was measured at five randomly chosen positions.
Biofilm flow cell model
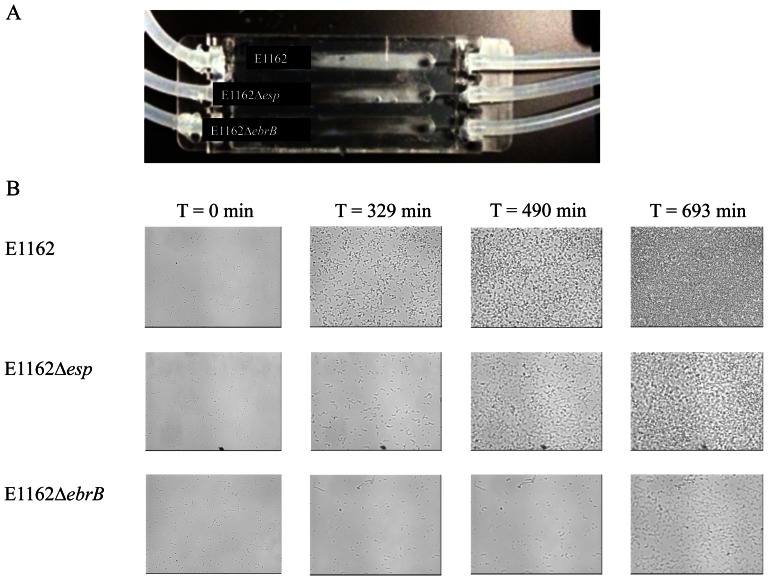
Dynamics of biofilm formation were studied in a Stovall flow cell system (Life Science, Inc., Greensboro, N.C.) for E1162 (wild-type), E1162Δesp and E1162ΔebrB in TSB diluted in PBS (1:10, v:v) with 1% glucose. After inoculation of the flow chambers bacterial cells were allowed to adhere for 1h in the absence of flow. Biofilms were grown under a flow of 0.13 ml/min for 17 h. Biofilm development was scanned at regular intervals of 7 min (40×objective), with a DFC360 FX Digital Camera Kit SP5 (Leica) using CLSM (Leica SP5).
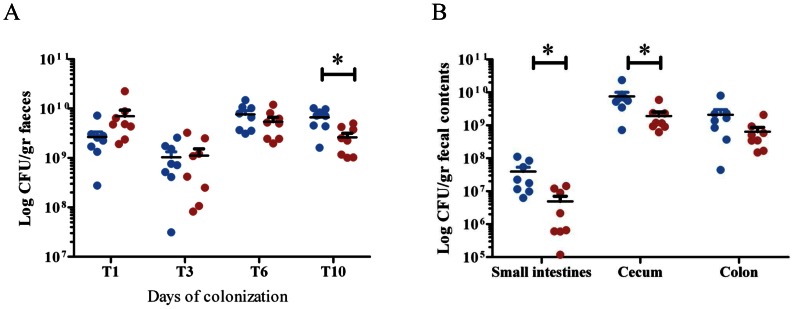
In vivo colonization model
Intestinal colonization by wild-type E1162 and E1162ΔebrB was tested as previously described [30], but with a modification of the decolonization regimen as recently described by Zhang et al. [31]. In brief, specific pathogen free 10-week-old male Balb/c mice (16 mice in total) were purchased from Charles River Laboratories Inc. (Maastricht, the Netherlands) and housed as described previously. Two days before inoculation of bacteria, mice were administered subcutaneous injections of ceftriaxone (Roche, Woerden, The Netherlands; 100 µl per injection, 12 mg/ml) two times daily and one time at the day of inoculation. For the remaining duration of the experiment, cefoxitin (0.125 g/l) was added to sterile drinking water. The inoculum of 2×109 CFU/300 µl Todd Hewitt Broth E1162 or E1162ΔebrB was prepared as described previously [30]. Collection of samples and determination of bacterial outgrowth was performed as previously described [30].
Statistical analysis
For analysis of cell surface expression of Esp, biofilm formation and intestinal colonization an unpaired two-tailed Student's t-test was applied.
Results
Bioinformatic analysis of EbrB
Upstream of esp we identified a gene, efmE1162_2351, renamed ebrB for reasons described later, which was annotated to belong to the AraC family of transcriptional regulators. BLAST analysis identified an AraC type helix-turn-helix (HTH) motif in the C-terminal region of the protein. These HTH domains are known to be involved in binding to promoter regions of genes [32], [33]. Comparison of the putative HTH motif encompassing amino acids 345-386 with a sequence logo generated using a Prosite database containing 310 HTH motifs identified 24 conserved amino acids known to be important for the structure of the helices (Fig. 1A) [34]. Furthermore, EbrB was also annotated to belong to the TIGR04094 protein family of transcriptional regulators that are located adjacent to proteins with the YSIRK variant form of signal peptide. Indeed, the downstream located Esp encoding gene has a variant YSIRK motif, YSIKK, in its signal peptide. Based on these genetic features, we hypothesized that EbrB is involved in the transcriptional regulation of esp expression.
Transcriptional organization of the ebrB-esp region
To investigate the transcriptional organization of the ebrB-esp region, we first determined whether ebrB and esp are transcribed as a single RNA molecule. A PCR using primers ebrB-1F and esp-1R (Fig. 1B, Table 2) on synthesized cDNA obtained from E1162 RNA yielded no product (data not shown). A positive PCR result was obtained on cDNA using primers in ebrB (ebrB-1F ) and in a downstream located small coding sequence (locus tag EfmE1162_2352) of unknown function (2352-1R) (Fig. 1B, Table 2, data not shown). These PCR results in combination with the presence of a predicted transcription terminator downstream the hypothetical CDS with a ▵G of -155.25 kcal/mol (Fig. 1B), indicated that erbB and esp are not part of the same operon. The transcription start sites of ebrB and esp were identified at 86-bp and 89-bp upstream their respective startcodon using 5′-RACE analysis (Fig. 1C). A putative promoter region, including -10 and -35 boxes were identified 99-bp and 124-bp upstream of the ebrB startcodon and 102-bp and 128-bp upstream of the esp startcodon (Fig. 1C).
EbrB is involved in cell-surface expression of Esp
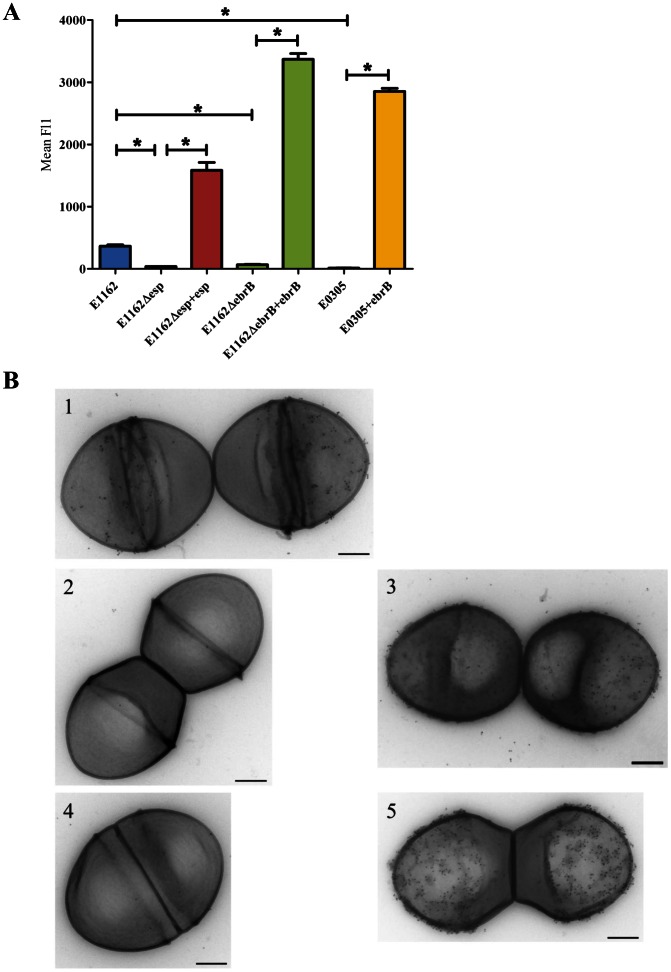
To investigate the role of EbrB in surface expression of Esp we constructed markerless EbrB deficient and Esp deficient mutants and determined first growth rates of these mutants. Growth curves of wild-type and the mutant strains grown in either BHI or TSBg using the Bioscreen were similar (Fig. S1). Next, wild-type E1162, E1162Δesp and E1162ΔebrB were grown on BA plates to determine cell-surface expression of Esp by flow cytometry using rabbit α-Esp immune serum (Fig. 2A). Esp expression was significantly reduced in both E1162Δesp and E1162ΔebrB compared to wild-type (p<0.01). Transmission electron microscopy confirmed the absence of Esp on the cell surface in both mutants (Fig. 2B). Esp expression was restored and highly expressed in the esp and ebrB complemented strains E1162Δesp+esp and E1162▵ebrB+ebrB (Fig. 2), but not in the complemented strains with the empty vector pEF25 (data not shown), demonstrating that EbrB is involved in regulation of Esp expression.
Figure 2. Cell surface expression of Esp.
(A) Cell surface expression of Esp on plate grown cells was analyzed by flow cytometry using rabbit α-Esp immune serum for wild-type strain E1162, esp mutant strain (E1162▵esp) and the esp complemented strain (E1162▵esp+esp), ebrB mutant strain (E1162▵ebrB) and the ebrB complemented strain (E1162▵ebrB+ebrB) and a natural ebrB mutant strain (E0305) and complemented strain (E0305+ebrB). The means of mean Fl1 from three independent experiments are shown. Asterisks represent significant differences (*p<0.01) as determined by an unpaired two-tailed Student's t-test) between the indicated samples. (B) Shown are transmission electron micrographs at a magnification of 60,000x. The wild-type strain (E1162) (1), the esp mutant strain (E1162▵esp) (2), the esp complemented strain (E1162▵esp +esp) (3), the ebrB mutant strain (E1162▵ebrB) (4) and the ebrB complemented strain (E1162▵ebrB+ebrB) (5) were incubated with rabbit α-Esp immune serum, followed by protein-A-Gold. Bars, 200nm.
In a previous screen for the presence of ebrB using ebrB specific primers, we identified a strain in which the ebrB PCR yielded a product that was 1.1 kbp larger than expected. Sequencing of this PCR product revealed that this strain, coded E0305 and originating from a hospital outbreak from Detroit area, USA [24] contained an insertion of IS256 in ebrB, thus represents a natural ebrB mutant (data not shown). In line with observations for E1162ΔebrB we also observed abolished cell surface expression of Esp in E0305, while Esp expression was restored in the ebrB complemented strain E0305+ebrB (Fig. 2A), but not in E0305+pEF25 (data not shown). All these findings support our hypothesis that EbrB regulates esp expression.
Growth phase dependent cell-surface Esp expression
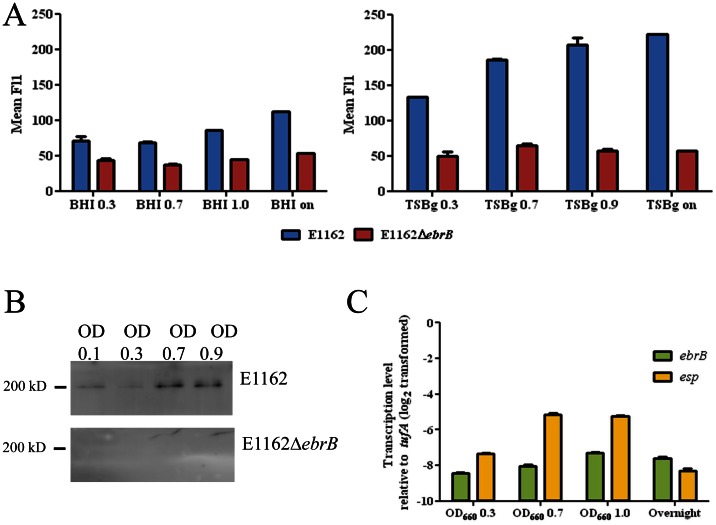
Previously, van Wamel et al. demonstrated temperature and air-oxygen dependent cell-surface expression of Esp using BA plate grown cells [15]. In order to determine dynamics of Esp expression in different growth phases, E1162 and E1162ΔebrB were grown in BHI and TSBg broth and Esp expression were measured using flow cytometry at OD660 0.3, 0.7, 1.0 (BHI) or 0.9 (TSBg) and from overnight culture (Fig. 3A). Increased Esp expression was observed in both BHI and TSBg during growth, although growth in BHI resulted in only a small increase in Esp expression. In TSBg, the highest increase of Esp expression was observed between OD660 0.3 and 0.7 resulting in higher levels of Esp expression in late log and stationary phase. As expected, only low background levels of fluorescence were measured in E1162ΔebrB. These levels were comparable to the background levels observed in the Esp deficient mutant (data not shown), which indicates that Esp is not expressed on the cell surface of the EbrB deficient mutant. In order to determine whether the addition of glucose in TSBg explained the difference with BHI grown cells, 1% glucose was also added to BHI, but did not result in higher Esp expression levels (data not shown).
Figure 3. Dynamics of Esp expression.
(A) Cell surface Esp expression for wild-type strain (E1162) in blue and the ebrB mutant strain (E1162▵ebrB) in red, during growth in BHI and TSBg at OD660 0.3, 0.7 and 1.0 and from an overnight culture analyzed by flow cytometry using rabbit α-Esp immune serum. (B) Esp expression in E1162 and E1162▵ebrB cell extracts obtained during growth in TSBg at OD660 0.1, 0.3, 0.6 and 0.9 analyzed by Western blot using rabbit α-Esp immune serum. (C) qRT-PCR analysis of ebrB and esp expression ratios in E1162 at OD660 0.3, 0.7, 1.0 and overnight culture. The data from the qRT-PCR were normalized using tufA as an internal standard. The differences in gene expression (log2-transformed data) relative to tufA are shown.
Western blot analysis using α-Esp antibodies on cell lysates from E1162 and E1162ΔebrB grown in TSBg at OD660 0.1, 0.3, 0.7 and 0.9 was performed to determine whether Esp might be expressed in early and mid-log phase but not transported to the cell surface. This revealed a clear increase of Esp at higher OD660, while Esp was absent in E1162ΔebrB (Fig. 3B), which means that increased cell surface expression of Esp at higher cell densities is correlated with total Esp protein expression levels.
Dynamics of ebrB and esp expression was further investigated on RNA level using qRT-PCR on synthesized cDNA obtained from RNA extracted in the different growth phases in TSBg. Relative to tufA, transcription levels of both ebrB and esp were lower in all growth phases, i.e. for ebrB between 7.3- and 8.4-fold and for esp between 5.1- and 8.3-fold lower expression (Fig. 3C). However, while only small fluctuation in transcription levels were observed for ebrB, increased transcription levels of esp were observed at OD660 0.7 and OD660 1.0 compared to OD660 0.3 (Fig. 3C). After overnight growth expression levels of ebrB and esp were comparable. The finding of increased esp transcription correlate with the increase in cell surface expression of Esp during late-log and stationary phase. Interestingly, as the higher Esp expression was found in growth phases with increased cell densities including late log and stationary phase, this finding suggests that Esp expression is cell density dependent which seems to be independent of EbrB, while ebrB expression did not change during increased cell density.
Comparative analysis of the transcriptome of E. faecium E1162 and E1162ΔebrB
In order to determine whether deletion of ebrB also influenced expression levels of other genes and might act as a global regulator, we used microarray-based transcriptome analysis on exponentially grown (OD660 = 0.3) E. faecium E1162 and E1162ΔebrB cultures in TSBg medium. Surprisingly, compared to E1162 wild-type, only esp, in addition to ebrB, was significantly down-regulated (2.3 fold) in E1162ΔebrB. qRT-PCR confirmed a 2-fold lower expression of esp (p<0.001) in E1162ΔebrB using the same cDNA. These results indicate that EbrB is not a global regulator in E. faecium.
Esp is part of an operon that is controlled by ebrB
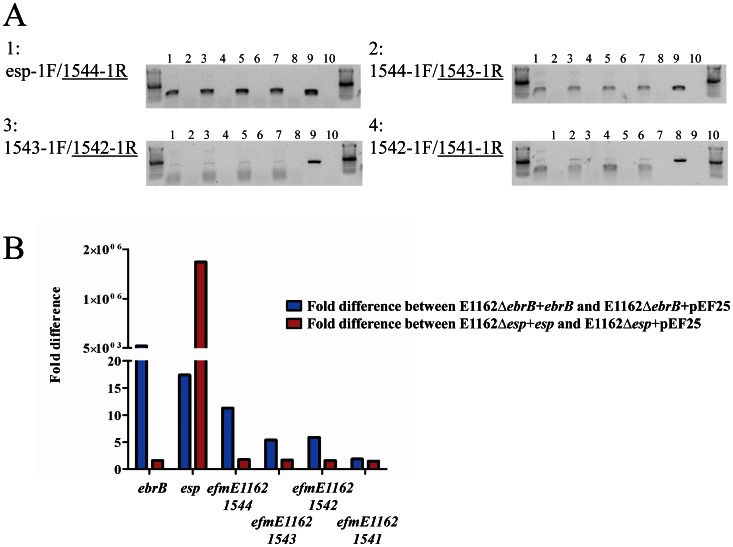
Analysis of the genetic organization of the esp region suggests that esp is part of an operon of in total five genes including efmE1162_1544, previously annotated as nox encoding a putative NADH oxidase, efmE1162_1543, previously annotated as mur encoding a putative muramidase, efmE1162_1542, previously annotated as phage encoding a hypothetical protein and efmE1162_1541, previously annotated as permease encoding a putative drug resistance transporter (Fig. 1B) [9]. To determine whether these genes are transcribed as a single RNA molecule, we performed intergenic PCRs on synthesized cDNA using gene specific primers on RNA obtained from E1162. In all cases PCR on genomic DNA was positive and as expected positive PCR results were obtained for the intergenic region between esp and efmE1162_1544 and efmE1162_1544 and efmE1162_1543, although the latter signal was a little bit lower (Fig. 4A). A very weak PCR product was obtained for the intergenic region of genes efmE1162_1543 and efmE1162_1542, which makes is dubious that efmE1162_1542 is indeed part of this operon. The fact that the intergenic region between efmE1162_1543 and efmE1162_1542 is larger than that between the more upstream located genes (349 bp compared to 134 bp and 112 bp, respectively, Fig.1B) and that RNAfold predicted a relative weak transcriptional terminator with a ▵G of -79.50 kcal/mol in the efmE1162_1543 and efmE1162_1542 intergenic region (Fig. 1B) also points towards the fact that efmE1162_1542 may not part of the esp operon. On the other hand, the weak PCR product we observed for the efmE1162_1543 and efmE1162_1542 intergenic region of genes could be the result of the fact that the RNA polymerase may read through this weak terminator or because of decline in expression due to degradation of a very large polycistronic mRNA (almost 10 Kb) that is transcribed from this operon, which may suggests that efmE1162_1542 is part of the esp operon. A positive though weak PCR product was also obtained between efmE1162_1542 and efmE1162_1541 (Fig. 4A), but this expression might be independent of EbrB as it is not likely that also this gene would be part of the operon based on the relative large intergenic region of 587 bp (Fig. 1B). In order to determine whether EbrB regulates this entire operon, we compared expression levels of ebrB, esp, efmE1162_1544, efmE1162_1543, efmE1162_1542 and efmE1162_1541 in the ebrB mutant strain complemented with wild-type ebrB (E1162ΔebrB+ebrB), in which Esp expression was highly restored, with the ebrB mutant strain complemented with empty vector (E1162ΔebrB+pEF25) using qRT-PCR on synthesized cDNA. As a control, the same qRT-PCRs were performed on synthesized cDNA isolated from the esp complemented strains E1162Δesp+esp and E1162Δesp+pEF25. As expected, ebrB and esp, encoded from multicopy plasmids in the complemented strains E1162ΔebrB+ebrB and E1162Δesp+esp were highly expressed compared to the strains complemented with empty vector pEF25, i.e. 4.7×104-fold and 1.7×106-fold, respectively (Fig. 4B). Interestingly, in the ebrB complemented strain overexpression of EbrB resulted not only in much higher expression levels of esp (16-fold), but also of the downstream encoded genes, i.e. efmE1162_1544 (11-fold)and efmE1162_15443 (5-fold) (Fig. 4B). As we also observed a 5-fold increased expression of efmE1162_1542, it is not unlikely that also this gene is part of the esp operon. In contrast, only a<2-fold difference in expression was observed for ORF efmE1162_1541, which is comparable with the fold difference identified in the esp complemented strain (Fig. 4B). If true, this decline in expression could be the result of degradation of the large polycistronic mRNA that is transcribed from this operon. In the esp complemented strains, there was, except for esp, no differential expression observed for ebrB and the downstream of esp encoded genes (all<2-fold). All together these data indicate that EbrB regulates not only esp but at least two and possibly three downstream located genes. The fact that we did not find differential expression of these three genes in our microarray experiments might be due to relative low transcription levels of ebrB in broth as also observed in our experiments where we assessed growth phase dependent expression of ebrB (Fig. 3C).
Figure 4. Determination of the esp operon.
(A) RT-PCR analysis on synthesized cDNA on RNA obtained from wild-type E1162 using gene specific primers (underlined). Panel 1: intergenic region esp and efmE1162_1544. Panel 2: intergenic region efmE1162_1544 and efmE1162_1543. Panel 3: intergenic region efmE1162_1543 and efmE1162_1542. Panel 4: intergenic region efmE1162_1542 and efmE1162_1541. RT-PCRs were performed on 4 biological replicates represented by lanes 1, 3, 5 and 7. Lanes 2, 4, 6, 8 represent negative controls in which incubation with reverse transcriptase was omitted. Lane 9 is the positive genomic DNA control and lane 10 negative water control. (B) Expression levels of ebrB and the esp operon in ebrB and esp complemented strains. Expression of ebrB, esp, efmE1162_1544, efmE1162_1543, efmE1162_1542 and efmE1162_1541 was analyzed using qRT-PCR on synthesized cDNA from the ebrB complemented strains (E1162▵ebrB+ebrB and E1162▵ebrB+pEF25) in blue and esp complemented strains (E1162▵esp+esp and E1162▵esp+pEF25) in red.
Dynamics of biofilm formation
Previously, Heikens et al. showed that Esp is involved in initial adherence and biofilm formation to polystyrene using E1162Δesp:cat. Here we confirm abolished biofilm formation in the newly constructed esp markerless mutant, but also in the ebrB mutant using CLSM in a 24 h and 72 h semi-static biofilm model (Fig. 5A and B). Comstat/Matlab analysis revealed significant decreased average thickness (p<0.01) and total biomass (p<0.01) for both mutants at 24 h and 72 h, although at 72 h the decrease in biofilm formation in the ebrB mutant seemed to be more pronounced than in the esp mutant (Fig. 5C). Biofilm formation was restored even to higher levels than wild-type in the esp and ebrB complemented strains (Fig.5), while biofilm formation of the controls (esp and ebrB mutant complemented with the empty vector) was comparable to the respective esp and ebrB mutant strains (data not shown). Furthermore, a difference in biofilm composition can be observed between wild-type and complemented strains (Fig. 5A and B). In the biofilms of both complemented strains spots with higher intensities can be observed suggesting cell aggregation. Interestingly, a similar phenomenon was also observed when growing the strains in TSBg broth. After overnight growth both complemented strains appear to aggregate, while this was not observed in mutants complemented with the empty vector or with all strain grown in BHI broth (Fig. S2). This phenomenon is likely due to overexpression of Esp in the complemented strains. Because of the clear biofilm deficient phenotype of E1162▵ebrB we refer to this AraC-type regulator here as EbrB, for Enterococcal biofilm regulator B.
Figure 5. Biofilm semi-static model using confocal laser scanning microscopy (CLSM).
Biofilms were grown in TSBg on coverslips coated with poly-L-lysin for (A) 24h and (B) 72h, including E1162 wild-type strain (1), esp mutant E1162Δesp (2), the esp complemented strain E1162Δesp+esp (3), ebrB mutant E1162ΔebrB (4) and ebrB complemented strain E1162ΔebrB+ebrb (5). (C) Quantification of the average thickness and total biomass of biofilms using Comstat/Matlab analysis. Asterisks represent significant differences (*p<0.01) as determined by an unpaired two-tailed Student's t-test) between the indicated samples. Pictures were taken at 63×magnification with 2.5 optical zoom.
Dynamics of biofilm formation of strains wild-type E1162, E1162Δesp and E1162▵ebrB were further investigated using flow cells. Although some biofilm formation was observed for all strains, visual inspection of the flow chambers after 17 h of growth clearly revealed the highest amount of biofilm formation for wild-type strain E1162 (Fig. 6A). Furthermore, for wild-type strain E1162 development of biofilm formation was observed after ∼329 min, while biofilm formation of the esp and ebrB mutants was less and clearly delayed at ∼490 min and ∼693 min, respectively (Fig. 6B). Also here, biofilm formation in the ebrB mutant seemed to be more attenuated than in the esp mutant. This may be due to the fact that EbrB also affects expression of the four genes located downstream of esp. The observed difference is likely not due to differences in growth rates as the growth curves were highly comparable when grown in TSBg (Fig. S1).
Figure 6. Biofilm formation using a flow cell system.
Wild-type strain (E1162), esp mutant (E1162Δesp) and ebrB mutant (E1162ΔebrB) were grown in TSB diluted in PBS (1:10, v:v) with 1% glucose in a flow cell system. (A) Flow chambers after 17 h of biofilm formation using Stovall flow cell system. (B) Pictures at predefined position of time point zero and time points when start of biofilm formation for individual strains was observed, i.e. for E1162 at 329 min, for E1162▵esp at 490 min and for E1162▵ebrB at 693 min.
Intestinal colonization
Next we investigated whether the ebrB mutant was attenuated in a mouse model of intestinal colonization. Both the wild type and the ebrB mutant were able to colonize the intestinal tract in comparable high numbers until day 6 after inoculation (Fig. 7A). Ten days after inoculation, a significant difference was observed between E1162 and E1162ΔebrB with slightly but significantly higher CFU counts for the wild-type strain (7.2 (4.4 – 10.0)×109 CFU/gram feces) than for the ebrB mutant (2.4 (1.0 – 5.0)×109 CFU/gram feces) (p = 0.004) (Fig. 7A). Furthermore, significant lower amounts of E1162ΔebrB compared to E1162 were present in the small intestine (1.4 (0.1 – 14)×106 CFU/gram and 2.0 (0.6 – 11)×107 CFU/gram, p = 0.02, respectively) and cecum (1.7 (0.6 – 5.9)×109 CFU/gram and 6.1 (0.7 – 24)×109 CFU/gram, p = 0.04) (Fig. 7B). Lower amounts of E1162ΔebrB were also observed in de colon (4.3 (1.5 – 21)×108 CFU/gram) compared to wild-type (15 (0.4 – 80)×108 CFU/gram) although this difference is not significant (p = 0.13) (Fig. 7B). These results suggest that EbrB is also implicated in intestinal colonization.
Figure 7. Intestinal colonization.
Mice were orally inoculated with E1162 (blue dots) and ebrB mutant (E1162ΔebrB, red dots). (A) Numbers of E1162 and E1162ΔebrB were determined in stool of mice at different time point after E. faecium inoculation. (B) After 10 days of colonization numbers of E1162 and E1162ΔebrB were determined in the small intestines, cecum and colon. Data are expressed as CFU per gram of stool/fecal contents and means are shown for 8 mice per group. Asterisks represent significant differences (*p<0.01) as determined by an unpaired two-tailed Student's t-test) between the indicated samples.
Discussion
Esp is a surface protein of E. faecium that is contained on the integrative conjugative element ICEEfm1, which is specifically found in hospital-associated E. faecium lineages. Previously, it has been demonstrated that Esp was involved in biofilm formation and virulence [12]–[14], [30]. In the current study, we identified a regulator of esp, designated EbrB for enterococcal biofilm regulator B, which is located just upstream of esp and characterized its role in Esp expression, biofilm formation, and intestinal colonization. Furthermore, we showed that esp is part of an operon that includes at least two and possibly three additional downstream located genes.
Esp of both E. faecium and E. faecalis belongs to the Alp family of proteins, where Alp stands for α-like proteins, which are characterized by the presence of long, completely identical repeats [35]. Alp proteins were first identified in Streptococcus agalactiae, including the α, Rib, R28 and Alp2 proteins, but were later also identified in other gram positive bacteria like R28 in Streptococcus pyogenes and Bap in Staphylococcus aureus (see Lindahl et al. [36] for a review on these proteins). So far nothing is known about transcriptional regulation of the genes encoding this group of proteins. Remarkably, the availability of whole genome sequences revealed that several of these Alp proteins (e.g. Rib and Alp2 in S. agalactiae) are located either up- or downstream of an AraC-type of regulator. In 47/53 esp containing E. faecium whole genome sequences (date December 2012), an AraC-type of regulator is located upstream of esp. In the remaining 6 esp containing whole genome sequences, this regulator was also present, but on the border of a contig. It is very likely that also in these strains this regulator is located upstream of esp. Therefore, it has been suggested that these proteins have a role in the transcriptional regulation of expression of these genes [36]. We, now, demonstrate for the first time that EbrB, a protein that contains the characteristic C-terminal HTH of an AraC-type of regulator, regulates expression of the Alp family protein Esp in E. faecium.
A BLAST search revealed that EbrB had the highest identity (60% AA) with the E. faecalis pathogenicity island encoded regulator, PerA, which is also located adjacent to esp, though downstream located [37]. Recently the regulon of the E. faecalis PerA was studied by comparing the transcriptome of an E. faecalis wild-type strain (E99) with its isogenic perA insertion mutant (DBS01) using microarray and qRT-PCR [37]. Despite the fact that PerA and EbrB display 60% similarity and are both araC-type regulator genes located adjacent to esp our transcriptome results for ebrB are different to those found for perA. Microarray analysis revealed that EbrB regulates only one gene, esp, while additional qRT-PCR experiments revealed that EbrB also controls expression of three additional genes located just downstream of esp on the esp containing ICEEfm1 of E. faecium and that are part of a single operon. In contrast, microarray analysis of PerA identified in total 151 differentially expressed genes in mid- and late exponential and stationary phase, which are mainly encoded outside the esp containing pathogenicity island of E. faecalis. Consequently, PerA was suggested to act as global regulator and importantly PerA did not seem to control esp expression [37]. We cannot exclude that also other genes might be regulated by EbrB, but were not detected using microarray analysis due to low level of ebrB expression at OD660 0.3.
To investigate involvement of EbrB in regulation of esp expression, we constructed an EbrB deficient mutant and found that esp expression, based on microarray and qRT-PCR, as well as Esp protein expression and surface exposure of Esp was reduced in the mutant and that this phenotype could be restored by complementing the mutant with full length EbrB. The reduction in Esp surface exposure in the EbrB deficient mutant was comparable with that observed in a newly constructed markerless E1162▵esp mutant. Also in the natural ebrB mutant E. faecium strain E0305, complementation with ebrB resulted in increased Esp expression levels. These experiments demonstrate involvement of EbrB in esp expression, but not direct binding of EbrB to the promoter of esp or its own promoter. This is currently under investigation. Sequence analysis of the promoter region did not identify an obvious EbrB binding site.
Previously, temperature and air oxygen dependent cell surface Esp expression was studied using plate grown cells [15]. Here, we investigated growth phase dependent cell surface expression of Esp using different growth media and showed a clear growth-phase dependent Esp expression in TSBg broth and not in BHI broth, though expression levels were much lower than in a high cell density condition like plate grown cells. This suggests that Esp surface exposure is dependent on cell density, which may indicate involvement of quorum sensing systems. In contrast to E. faecalis where an fsr quorum sensing system has been studied in detail [38], quorum sensing systems have never been described in E. faecium [39]. Because of this and the fact that only small fluctuations were observed for ebrB expression during different growth-phases, it is still unclear how exactly cell density dependent expression of Esp expression is regulated. EbrB is, however, necessary for basal expression while Esp is completely absent in all growth phases and in plate grown cells in E1162▵ebrB. It would be of interest to further investigate the existence of quorum sensing systems in E. faecium as they, like in E. faecalis, often play a role in virulence [38]–[43].
In the semi-static biofilm model and the flow cell system, differences were observed in biofilm formation between wild-type, mutants and complemented strains. In the semi-static model differences were more pronounced after 72 h of growth. In the flow cell model biofilm formation was observed in the wild-type strain and to lesser extent also in mutant strains. Furthermore, biofilm development in the mutant strains was delayed compared to wild-type. These data demonstrate a role for EbrB in early and later biofilm. From the Comstat analysis, including total biomass and average thickness, it is clear that biofilm formation in E1162ΔebrB is more affected than in E1162Δesp. A similar effect was observed using the flow cells. In addition, in the complemented strains more biofilm formation was observed in the ebrB complemented strain compared to the esp complemented strain. An explanation for this more pronounced effect of EbrB on E. faecium biofilm formation, relative to Esp, may reside in the fact that EbrB also controls expression of additional three genes located just downstream of esp on the esp containing ICEEfm1 of E. faecium. Two of these genes encoding a putative NADH oxidase and a putative muramidase and it has been shown previously that proteins belonging to these classes are implicated in biofilm formation [9], [44]–[47].
In a mouse intestinal colonization model the ebrB mutant was clearly attenuated. Given the fact that in the ebrB mutant esp expression is affected resulting in the absence of Esp surface exposure, this observation seems to contradict with previous findings of Heikens et al. that based on a comparison of colonization capacities of an insertion-deletion E1162Δesp:cat mutant with wild-type E1162 concluded that Esp is not essential in intestinal colonization of mice [30]. In fact an unexplained higher number of E1162Δesp:cat was found in the small intestine. An explanation for these contradicting results might be that in the current study a different decolonization regime was used, resulting in higher colonization rates. However, another plausible explanation is that additional genes located downstream of esp and also controlled by EbrB contribute to colonization. The exact role of the genes located downstream of esp and contained on ICEEfm1 in biofilm formation and colonization remains to be investigated.
In conclusion, we identified EbrB as a regulator of esp in E. faecium and its downstream encoded genes. Based on our current and previous [15] observations that Esp expression is growth condition dependent, indicates that expression of esp is regulated by environmental signals likely through EbrB. Regulation of AraC-type regulators by environmental signals and the effect of this on virulence gene expression is well documented and recently nicely reviewed [48]. Furthermore, we determined that during growth Esp is increasingly expressed at RNA and protein level, with highest expression in stationary phase which suggests involvement of an unidentified quorum sensing like system in regulation of esp expression in E. faecium. Understanding how virulence gene expression is regulated in E. faecium may foster the development of compounds that block the action of virulence gene regulators, like ebrB, which would impact on gut intestinal colonization and biofilm formation of specific multidrug resistance E. faecium hospital lineages that are enriched in the esp containing ICEEfm1.
Supporting Information
The effect of targeted mutations of esp and ebrB on growth of E. faecium . Overnight cultures of wild-type, mutants and complemented E. faecium were inoculated at an initial cell density of OD660 0.0025 in BHI or TSBg. Growth curves of strain E1162, the different mutant strains (panel A: BHI Δesp; panel B: BHI ΔebrB; panel C: TSBg Δesp; panel D: TSBg ΔebrB) and in trans complemented strains are shown. Growth curves are mean data of three independent experiments.
(TIF)
Aggregation of esp and ebrB complemented strains. Pictures of overnight in TSBg (panel A) and BHI (panel B) grown E1162Δesp complemented with the empty vector pEF25 and pEF25+esp and E1162ΔebrB complemented with the empty vector pEF25 and pEF25+ebrB. In the esp complemented strain E1162▵esp+esp grown in TSBg and to a lesser extent in the ebrB complemented strain E1162▵ebrB+ebrB cells have aggregated and form a sediment on the bottom of the tube. Mutants complemented with the empty vector grown in TSBG and all strains grown in BHI produced a more turbid, planktonic growth pattern.
(TIF)
Acknowledgments
The authors like to thank Joost Daalhuizen and Marieke S. ten Brink for their expert technical assistance.
Funding Statement
J.T.'s, W.v.S.'s and R.J.L.W.'s research leading to these results has received funding from the European Union Seventh Framework Programme (FP7-HEALTH-2011-single-stage) under grant agreement no. 282004, EvoTAR. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Willems RJ, van Schaik W (2009) Transition of Enterococcus faecium from commensal organism to nosocomial pathogen. Future Microbiol 4: 1125–1135. [DOI] [PubMed] [Google Scholar]
- 2. van Schaik W, Willems RJ (2010) Genome-based insights into the evolution of enterococci. Clin Microbiol Infect 16: 527–532. [DOI] [PubMed] [Google Scholar]
- 3. Willems RJ, Top J, van Santen M, Robinson DA, Coque TM, et al. (2005) Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis 11: 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Willems RJ, Top J, van Schaik W, Leavis H, Bonten M, et al. (2012) Restricted gene flow among hospital subpopulations of Enterococcus faecium . MBio 3: e00151–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hendrickx AP, van Wamel WJ, Posthuma G, Bonten MJ, Willems RJ (2007) Five genes encoding surface exposed LPXTG proteins are enriched in hospital-adapted Enterococcus faecium Clonal Complex-17 isolates. J Bacteriol 189: 8321–8332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hendrickx AP, Bonten MJ, Luit-Asbroek M, Schapendonk CM, Kragten AH, et al. (2008) Expression of two distinct types of pili by a hospital-acquired Enterococcus faecium isolate. Microbiology 154: 3212–3223. [DOI] [PubMed] [Google Scholar]
- 7. Hendrickx AP, van Luit-Asbroek M, Schapendonk CM, van Wamel WJ, Braat JC, et al. (2009) SgrA, a nidogen-binding LPXTG surface adhesin implicated in biofilm formation, and EcbA, a collagen binding MSCRAMM, are two novel adhesins of hospital-acquired Enterococcus faecium . Infect Immun 77: 5097–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sava IG, Heikens E, Huebner J (2010) Pathogenesis and immunity in enterococcal infections. Clin Microbiol Infect 16: 533–540. [DOI] [PubMed] [Google Scholar]
- 9. Leavis H, Top J, Shankar N, Borgen K, Bonten M, et al. (2004) A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J Bacteriol 186: 672–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Top J, Sinnige JC, Majoor EA, Bonten MJ, Willems RJ, et al. (2011) The recombinase IntA is required for excision of esp-containing ICEEfm1 in Enterococcus faecium . J Bacteriol 193: 1003–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Schaik W, Top J, Riley DR, Boekhorst J, Vrijenhoek JE, et al. (2010) Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heikens E, Bonten MJ, Willems RJ (2007) Enterococcal surface protein Esp is important for biofilm formation of Enterococcus faecium E1162. J Bacteriol 189: 8233–8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leendertse M, Heikens E, Wijnands LM, van Luit-Asbroek M, Teske GJ, et al. (2009) Enterococcal surface protein transiently aggravates Enterococcus faecium-induced urinary tract infection in mice. J Infect Dis 200: 1162–1165. [DOI] [PubMed] [Google Scholar]
- 14. Heikens E, Singh KV, Jacques-Palaz KD, van Luit-Asbroek M, Oostdijk EA, et al. (2011) Contribution of the enterococcal surface protein Esp to pathogenesis of Enterococcus faecium endocarditis. Microbes Infect 13: 1185–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Wamel WJ, Hendrickx AP, Bonten MJ, Top J, Posthuma G, et al. (2007) Growth condition-dependent Esp expression by Enterococcus faecium affects initial adherence and biofilm formation. Infect Immun 75: 924–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leenhouts K, Buist G, Bolhuis A, ten Berge A, Kiel J, et al. (1996) A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet 253: 217–224. [DOI] [PubMed] [Google Scholar]
- 17.Muscholl-Silberhorn A (2000) Electrotransformation of enterococci. In: Eynard, Natalie, editors. Electrotransformation of Bacteria. Berlin: Springer. pp. 134–140.
- 18. Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL (2008) The Vienna RNA websuite. Nucleic Acids Res 36: W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X, Paganelli FL, Bierschenk D, Kuipers A, Bonten MJ, et al. (2012) Genome-wide identification of ampicillin resistance determinants in Enterococcus faecium . PLoS Genet 8: e1002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang X, Vrijenhoek JE, Bonten MJ, Willems RJ, van Schaik W (2011) A genetic element present on megaplasmids allows Enterococcus faecium to use raffinose as carbon source. Environ Microbiol 13: 518–528. [DOI] [PubMed] [Google Scholar]
- 22. Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P (1991) Shuttle vectors containing a multiple cloning site and a lacZα gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102: 99–104. [DOI] [PubMed] [Google Scholar]
- 23. Wilson AC, Perego M, Hoch JA (2007) New transposon delivery plasmids for insertional mutagenesis in Bacillus anthracis . J Microbiol Methods 71: 332–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dunne WM, Wang W (1997) Clonal dissemination and colony morphotype variation of vancomycin-resistant Enterococcus faecium isolates in metropolitan Detroit, Michigan. J Clin Microbiol 35: 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wästfelt M, Stalhammar-Carlemalm M, Delisse AM, Cabezon T, Lindahl G (1996) Identification of a family of streptococcal surface proteins with extremely repetitive structure. J Biol Chem 271: 18892–18897. [DOI] [PubMed] [Google Scholar]
- 26. Hsiao A, Worrall DS, Olefsky JM, Subramaniam S (2004) Variance-modeled posterior inference of microarray data: detecting gene-expression changes in 3T3-L1 adipocytes. Bioinformatics 20: 3108–3127. [DOI] [PubMed] [Google Scholar]
- 27. Hsiao A, Ideker T, Olefsky JM, Subramaniam S (2005) VAMPIRE microarray suite: a web-based platform for the interpretation of gene expression data. Nucleic Acids Res 33: W627–W632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lebreton F, van Schaik W, Sanguinetti M, Posteraro B, Torelli R, et al. (2012) AsrR is an oxidative stress sensing regulator modulating Enterococcus faecium opportunistic traits, antimicrobial resistance, and pathogenicity. PLoS Pathog 8: e1002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Heikens E, Leendertse M, Wijnands LM, van Luit-Asbroek M, Bonten MJ, et al. (2009) Enterococcal surface protein Esp is not essential for cell adhesion and intestinal colonization of Enterococcus faecium in mice. BMC Microbiol 9: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang X, Top J, de Been M, Bierschenk D, Rogers M, et al. (2013) Identification of a genetic determinant in clinical Enterococcus faecium strains which contributes to intestinal colonization during antibiotic treatment. J Infect Dis 207: 1780–1786. [DOI] [PubMed] [Google Scholar]
- 32. Egan SM (2002) Growing repertoire of AraC/XylS activators. J Bacteriol 184: 5529–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huffman JL, Brennan RG (2002) Prokaryotic transcription regulators: more than just the helix-turn-helix motif. Curr Opin Struct Biol 12: 98–106. [DOI] [PubMed] [Google Scholar]
- 34. Manzanera M, Marques S, Ramos JL (2000) Mutational analysis of the highly conserved C-terminal residues of the XylS protein, a member of the AraC family of transcriptional regulators. FEBS Lett 476: 312–317. [DOI] [PubMed] [Google Scholar]
- 35. Michel JL, Madoff LC, Olson K, Kling DE, Kasper DL, et al. (1992) Large, identical, tandem repeating units in the C protein alpha antigen gene, bca, of group B streptococci. Proc Natl Acad Sci U S A 89: 10060–10064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lindahl G, Stalhammar-Carlemalm M, Areschoug T (2005) Surface proteins of Streptococcus agalactiae and related proteins in other bacterial pathogens. Clin Microbiol Rev 18: 102–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maddox SM, Coburn PS, Shankar N, Conway T (2012) Transcriptional regulator PerA influences biofilm-associated, platelet binding, and metabolic gene expression in Enterococcus faecalis . PLoS One 7: e34398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qin X, Singh KV, Weinstock GM, Murray BE (2001) Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J Bacteriol 183: 3372–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paganelli FL, Willems RJ, Leavis HL (2012) Optimizing future treatment of enterococcal infections: attacking the biofilm? Trends Microbiol 20: 40–49. [DOI] [PubMed] [Google Scholar]
- 40. Hancock LE, Perego M (2004) The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J Bacteriol 186: 5629–5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sifri CD, Mylonakis E, Singh KV, Qin X, Garsin DA, et al. (2002) Virulence effect of Enterococcus faecalis protease genes and the quorum-sensing locus fsr in Caenorhabditis elegans and mice. Infect Immun 70: 5647–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Singh KV, Nallapareddy SR, Nannini EC, Murray BE (2005) Fsr-independent production of protease(s) may explain the lack of attenuation of an Enterococcus faecalis fsr mutant versus a gelE-sprE mutant in induction of endocarditis. Infect Immun 73: 4888–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thurlow LR, Thomas VC, Narayanan S, Olson S, Fleming SD, et al. (2010) Gelatinase contributes to the pathogenesis of endocarditis caused by Enterococcus faecalis . Infect Immun 78: 4936–4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arciola CR, Campoccia D, Speziale P, Montanaro L, Costerton JW (2012) Biofilm formation in Staphylococcus implant infections. A review of molecular mechanisms and implications for biofilm-resistant materials. Biomaterials 33: 5967–5982. [DOI] [PubMed] [Google Scholar]
- 45. Loo CY, Mitrakul K, Jaafar S, Gyurko C, Hughes CV, et al. (2004) Role of a nosX homolog in Streptococcus gordonii in aerobic growth and biofilm formation. J Bacteriol 186: 8193–8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Montanaro L, Poggi A, Visai L, Ravaioli S, Campoccia D, et al. (2011) Extracellular DNA in biofilms. Int J Artif Organs 34: 824–831. [DOI] [PubMed] [Google Scholar]
- 47. Thomas VC, Hiromasa Y, Harms N, Thurlow L, Tomich J, et al. (2009) A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis . Mol Microbiol 72: 1022–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yang J, Tauschek M, Robins-Browne RM (2011) Control of bacterial virulence by AraC-like regulators that respond to chemical signals. Trends Microbiol 19: 128–135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The effect of targeted mutations of esp and ebrB on growth of E. faecium . Overnight cultures of wild-type, mutants and complemented E. faecium were inoculated at an initial cell density of OD660 0.0025 in BHI or TSBg. Growth curves of strain E1162, the different mutant strains (panel A: BHI Δesp; panel B: BHI ΔebrB; panel C: TSBg Δesp; panel D: TSBg ΔebrB) and in trans complemented strains are shown. Growth curves are mean data of three independent experiments.
(TIF)
Aggregation of esp and ebrB complemented strains. Pictures of overnight in TSBg (panel A) and BHI (panel B) grown E1162Δesp complemented with the empty vector pEF25 and pEF25+esp and E1162ΔebrB complemented with the empty vector pEF25 and pEF25+ebrB. In the esp complemented strain E1162▵esp+esp grown in TSBg and to a lesser extent in the ebrB complemented strain E1162▵ebrB+ebrB cells have aggregated and form a sediment on the bottom of the tube. Mutants complemented with the empty vector grown in TSBG and all strains grown in BHI produced a more turbid, planktonic growth pattern.
(TIF)