Abstract
In Mediterranean intermittent streams, the hydrological fragmentation in summer and the successive water flow re-convergence in autumn allow exploring how local processes shape the microbial community within the same habitat. The objectives of this study were to determine how bacterial community composition responded to hydrological fragmentation in summer, and to evaluate whether the seasonal shifts in community composition predominate over the effects of episodic habitat fragmentation. The bacterial community was assessed along the intermittent stream Fuirosos (Spain), at different levels of phylogenetic resolution by in situ hybridization, fingerprinting, and 16S rRNA gene sequencing. The hydrological fragmentation of the stream network strongly altered the biogeochemical conditions with the depletion of oxidized solutes and caused changes in dissolved organic carbon characteristics. In the isolated ponds, beta-Proteobacteria and Actinobacteria increased their abundance with a gradual reduction of the alpha-diversity as pond isolation time increased. Moreover, fingerprinting analysis clearly showed a shift in community composition between summer and autumn. In the context of a seasonal shift, the temporary stream fragmentation simultaneously reduced the microbial dispersion and affected local environmental conditions (shift in redox regime and quality of the dissolved organic matter) tightly shaping the bacterioplankton community composition.
Introduction
Microbial communities can exhibit spatial variability at scales ranging from millimetres to thousands of kilometres [1]. Understanding the mechanisms governing such spatial distribution is a key issue for elucidating the extent, specificity and stability of microbial associations and the implications for ecosystem functioning [2]–[5]. Microbial ecologists have recently started to examine the role of dispersal in shaping community similarities on large spatial scales [6]–[8]. Moreover, the microbial responses to local environmental stresses have to be carefully considered to properly interpret the dynamics of the microbial world [9].
The temporal and spatial variability found in bacterial community composition is mainly driven by species sorting and fast local growth, which counterbalance cell dispersion [10]–[12]. Both local interactions (e.g. within/between species, between species and the environment) and regional processes (e.g. dispersal) influence local community assembly [4]. High dispersal rates can lead to a continuous, worldwide supply of taxa that can be found even in less suitable habitats [13]. Local environmental characteristics are relatively more important when variation between sites increases [7], [14], [15,]. To gain a better understanding on how the local environment shapes bacterial communities it is necessary to address the proper spatial resolution at which microorganisms assemble into local communities, thus minimising regional effects at larger spatial scale [6], [16].
In the case of freshwater environments, most studies tended to focus on either similar habitats across different spatial scales or interconnected habitats, reaching different conclusions on the importance of environmental factors [11], [17] and geographic distance [8], [18]–[20]. So far, few studies have focused on assessing microbial assemblages when a habitat undergoes a gradual fragmentation, which interrupts the flow of carbon and energy, the dispersion of biota, and causes a marked environmental heterogeneity [21], [22]. In intermittent streams, hydrological fragmentation in summer and water flow re-convergence in autumn could provide the opportunity to explore how local processes shape a microbial community within the same habitat. Episodes of low flow fragment the hydrological stream network into a patched landscape of unconnected standing water bodies. As a consequence, fragmentation increases the environmental heterogeneity and decreases hydrological connectivity and the potential for bacterial cell dispersal.
We designed a field study following a gradient approach (from flowing to stagnant waters, in ponds disconnected at different times) that provides a framework for discussing how bacterial community structure relates to water biogeochemistry during the stream habitat fragmentation episodes. To avoid confounding regional influences [14], we selected a small semi-pristine intermittent Mediterranean stream (Fuirosos, Spain) and microbial assemblages were assessed along the whole stream network. During summer, the streambed was completely dry except in the five locations where water samples were collected. A second sampling campaign was carried out in the same sites when the stream connectivity was re-established in autumn. The hydrology and the water chemistry were characterised and the bacterial assemblages were assessed at different levels of phylogenetic resolution by Fluorescence in situ Hybridization Catalysed Reported Deposition (CARD-FISH) and by Denaturing Gradient Gel Electrophoresis (DGGE) and 16S rRNA gene sequencing.
In this study, we aimed to determine (i) how the abundance and composition of bacterioplankton communities respond to the hydrological and biogeochemical changes when the stream shifts between free-flowing and fragmented non-flowing conditions in summer; and (ii) how the seasonal shift, in the transition from summer to autumn, affects the occurrence of the major bacterial species and predominates over the effects of episodic habitat fragmentation.
Results
Hydrological and chemical dynamics
Hydrological fragmentation enhanced anaerobic conditions due to both depletion of oxidised solutes (O2, N-NO3, SO4) and accumulation of reduced solutes such as dissolved organic carbon (DOC), dissolved organic nitrogen (DON) and N-NH4 in isolated ponds (i.e. sites 2, 3 and 5) (Fig. 1 and 2; Table 1). The lowest values of the Chemical Index (CI<0.3), used to describe the degree of the aerobic/anaerobic conditions in the waters bodies, were found in the isolated ponds, while the highest values (CI>3) were observed in flowing waters (both in summer and in autumn) and in groundwater. An inverse relationship between the CI and the Pond Isolation Time (PIT) was observed (r = 0.92, p<0.05, df = 3).
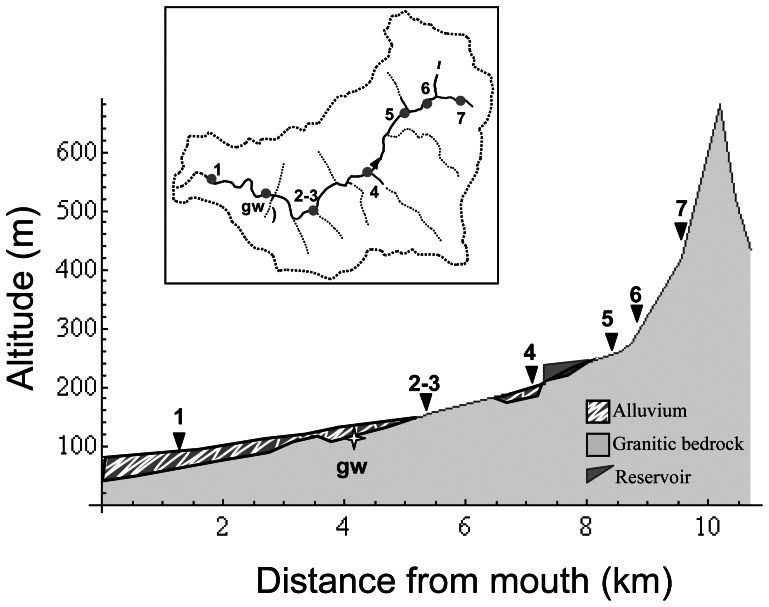
Figure 1. Sampling point locations.
Sampling point locations along the longitudinal-altitudinal profile and the fluvial network (inset). Triangles: surface waters. Star: riparian groundwater.
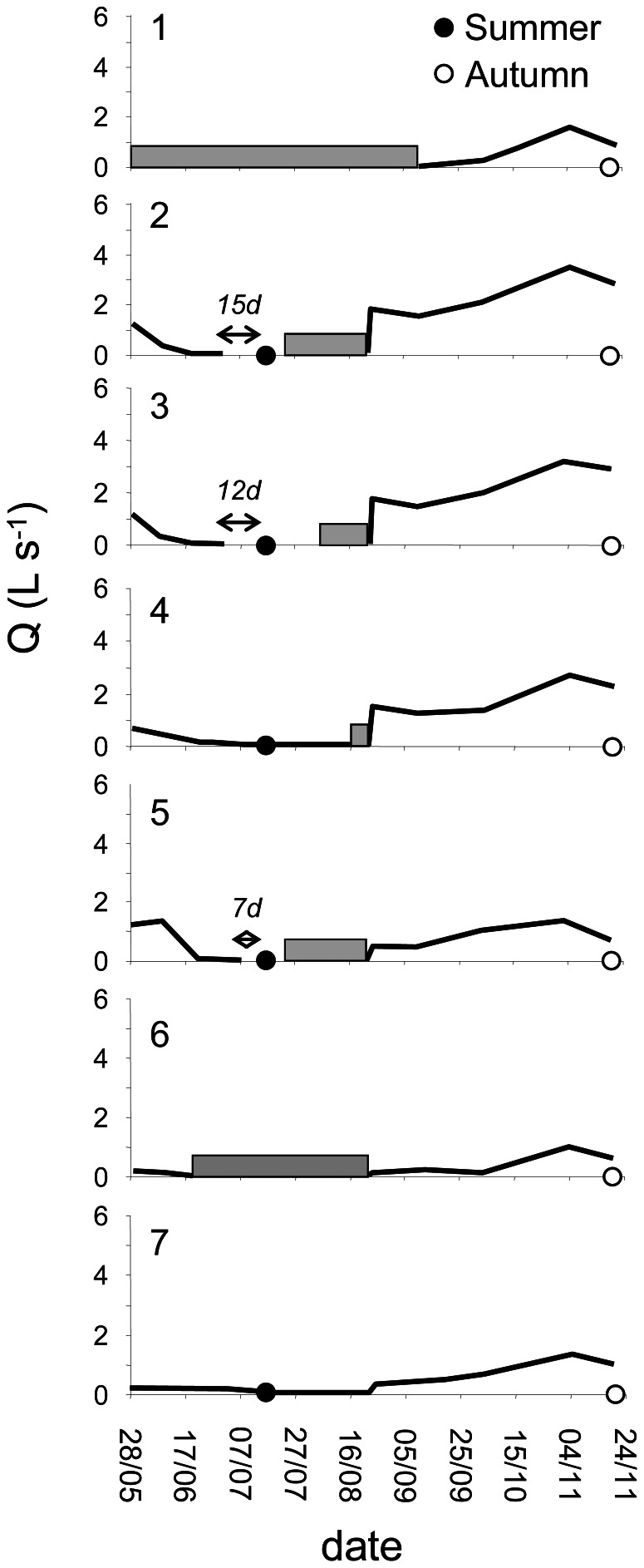
Figure 2. Water discharge in the sampling points.
Discharge from May to November in the sampling points (1–7). (•) = summer sampling; (○) = autumn sampling; (↔) = period between water segregation in the pond (no flow) and sampling time; gray bars indicate the period of complete dryness of the stream reach.
Table 1. Water physical and chemical characteristics in sampling points.
| Summer | Autumn | |||||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | gw | 1 | 2–3 | 4 | 5 | 6 | 7 | gw | ||
| Water physical and chemical characteristics | PIT (d) | dry | 15 | 12 | 0 | 7 | dry | 0 | nm | nm | nm | nm | nm | nm | nm | nm |
| Q (l/s) | dry | 0 | 0 | 0.1 | 0 | dry | 0.2 | nm | 0.9 | 2.9 | 2.3 | 0.7 | 0.6 | 0.6 | nm | |
| Temperature (°C) | dry | 18.2 | 17.1 | 17.5 | 16.7 | dry | 18.0 | 17.0 | 11.3 | 11.3 | 11.4 | 9.5 | 11.4 | 9.6 | 14.8 | |
| O2 (mg/l) | dry | 0.5 | 1.0 | 1.5 | 2.0 | dry | 9.0 | 6.5 | 8.3 | 10.8 | 7.3 | 9.1 | 5.4 | 10.3 | 8.4 | |
| pH | dry | 7.3 | 7.3 | 6.9 | 7.3 | dry | 8.0 | 6.9 | 7.2 | 7.6 | 7.5 | 7.5 | 7.0 | 7.6 | 7.4 | |
| EC (µS/cm) | dry | 463 | 349 | 349 | 408 | dry | 161 | 387 | 265 | 267 | 220 | 258 | 274 | 164 | 419 | |
| Chloride (mg/l) | dry | 33.9 | 29.7 | 18.9 | 21.7 | dry | 14.1 | 20.2 | 26.3 | 23.5 | 20.5 | 21.6 | 23.2 | 14.2 | 23.2 | |
| (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | ||||
| SO4 (mg/l) | dry | 4.7 | 3.6 | 2.8 | 10.3 | dry | 8.4 | 36.5 | 21.6 | 21.9 | 19.2 | 19.1 | 21.5 | 9.3 | 14.4 | |
| (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | ||||
| N-NO3 (mg/l) | dry | 0.08 | 0.01 | 0.02 | 0.08 | dry | 0.23 | 1.31 | 0.25 | 0.01 | 0.12 | 0.34 | 0.74 | 0.04 | 0.02 | |
| (*) | (0.001) | (0.004) | (*) | (0.060) | (*) | (0.050) | (0.010) | (*) | (*) | (*) | (0.030) | (0.060) | ||||
| N-NH4 (mg/l) | dry | 13.00 | 1.89 | 0.10 | 1.55 | dry | 0.02 | 0.04 | 0.04 | 0.04 | 0.02 | 0.01 | 0.03 | 0.01 | 0.01 | |
| (*) | (*) | (0.020) | (0.190) | (0.005) | (0.010) | (0.060) | (*) | (0.004) | (0.003) | (*) | (0.010) | (0.004) | ||||
| P-PO4 (mg/l) | dry | 0.017 | 0.017 | 0.001 | 0.031 | dry | 0.017 | 0.008 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | |
| (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | ||||
| CI | dry | −3.3 | −0.6 | 2.7 | 0.3 | dry | 6.3 | 5.0 | 5.2 | 5.6 | 6.1 | 6.6 | 5.3 | 7.2 | 6.6 | |
| (*) | (*) | (0.1) | (0.1) | (0.3) | (0.3) | (1.3) | (*) | (*) | (0.6) | (0.6) | (*) | (*) | ||||
| DOM characteristics | DOC (mg/l) | dry | 33.1 | 5.9 | 6.0 | 6.1 | dry | 1.8 | 1.7 | 1.7 | 2.8 | 4.1 | 2.0 | 2.2 | 1.9 | 1.1 |
| (*) | (*) | (*) | (*) | (*) | (0.60) | (0.02) | (*) | (*) | (*) | (*) | (0.41) | (*) | ||||
| DON (mg/l) | dry | 1.31 | 0.14 | 0.29 | 0.87 | dry | 0.05 | 0.09 | 0.11 | 0.17 | 0.32 | 0.16 | 0.14 | 0.08 | 0.17 | |
| (*) | (0.05) | (0.03) | (0.21) | (0.03) | (0.01) | (0.02) | (0.03) | (0.06) | (0.02) | (0.02) | (0.03) | (0.14) | ||||
| DOC∶DON | dry | 25 | 41 | 21 | 7 | dry | 37 | 19 | 16 | 17 | 13 | 13 | 16 | 25 | 6 | |
| (*) | (*) | (5) | (*) | (13) | (*) | (*) | (2) | (3) | (*) | (*) | (5) | (3) | ||||
| BDOC (%) | dry | 39.6 | 16.9 | 7.7 | 26.1 | dry | 5.3 | 14.2 | 10.8 | 6.5 | 12.7 | 5.8 | 9.9 | 14.5 | 35.9 | |
| (3.9) | (2.7) | (2.7) | (*) | (3.4) | (10.5) | (1.5) | (5.7) | (5.5) | (2.3) | (11.6) | (*) | (6.4) | ||||
| FI | dry | 1.85 | 1.80 | 1.72 | 1.90 | dry | 1.60 | 1.87 | 1.76 | 1.77 | 1.69 | 1.72 | 1.74 | 1.62 | 1.95 | |
| (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | ||||
| SUVA (l/mg C cm) | dry | 0.88 | 1.38 | 2.42 | 2.04 | dry | 2.28 | 0.69 | 0.50 | 0.75 | 1.11 | 1.29 | 0.36 | 1.40 | 0.30 | |
| (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | (*) | ||||
| IC/IA | dry | 0.91 | 0.75 | 0.71 | 0.89 | dry | 0.55 | 0.63 | 0.42 | 0.54 | 0.38 | 0.38 | 0.49 | 0.58 | 0.63 | |
Standard deviation is reported in brackets; (*) = standard deviation lower than 10%. (gw) = groundwater samples; (PIT) = Pond Isolation Time; (Q) = Discharge; (EC) = Electrical Conductivity; (CI) = Chemical Index; (BDOC) = Biodegradable Dissolved Organic Carbon; (FI) = Fluorescence Index; (SUVA) = Specific UV Absorbance; (IC/IA) = Ratio of intensities of C and A fluorescence peaks. See text for additional details.
On average, the descriptors used to characterize the dissolved organic matter (DOM) such as DOC∶DON ratio, percentage of biodegradable DOC (BDOC), and ratio of intensities of C and A fluorescence peaks (IC/IA, a measure of in situ microbial degradation), showed higher values in summer than in autumn (Table 1). In particular, DOC was more biodegradable in ponds (BDOC>16.9%) than in running waters (BDOC<14.5%). In summer, BDOC was directly related to DON (r = 0.92, df = 4, p<0.01,) and inversely related to CI (r = 0.84, df = 4, p<0.05). Moreover, IC/IA showed higher values (>0.75) in ponds than in running waters and groundwater.
The Fluorescence Index (FI, a descriptor of DOM origin) value around 1.6, which is typically found in soil leachates in the Fuirosos basin [23], indicated a greater contribution of allochthonous DOC in the flowing waters than in isolated ponds. In summer, specific UV absorbance at 254 nm (SUVA, a measure of DOM aromaticity) values ranged from 0.88 to 2.42 with highest values in running water sites 4 and 7 and lowest values in groundwater and sites 2 and 3. In autumn, these values were lower than in summer and tended to increase with respect to CI values.
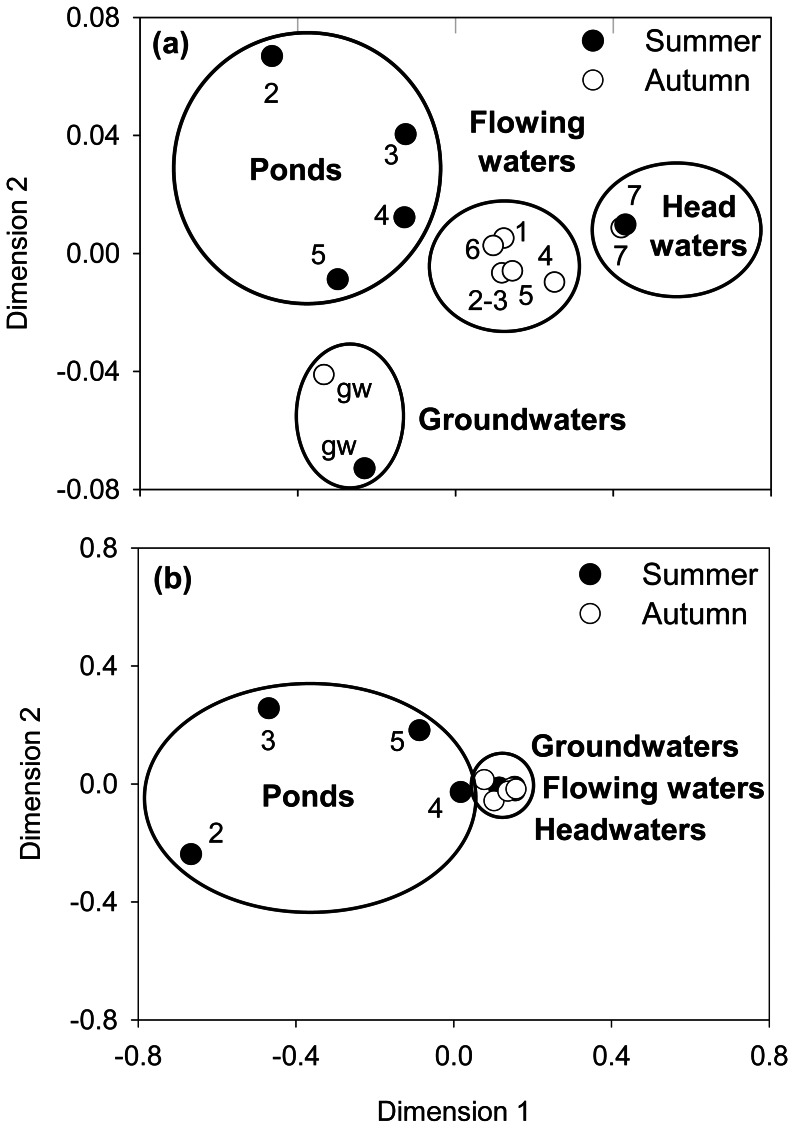
Overall, hydrological fragmentation enhanced biogeochemical heterogeneity as graphically summarised by the non-Metric Multi-Dimensional Scaling analysis (nMDS). In the headwater and groundwater sites (7 and gw) changes between the two hydrological periods (summer and autumn) were minimal compared to the high biogeochemical shift in isolated ponds (Fig. 3 a).
Figure 3. nMDS ordination analysis.
(a) biogeochemical data as in Table 1, and (b) community composition analysed by CARD-FISH from both summer and autumn samplings.
Bacterial community composition by in situ hybridization
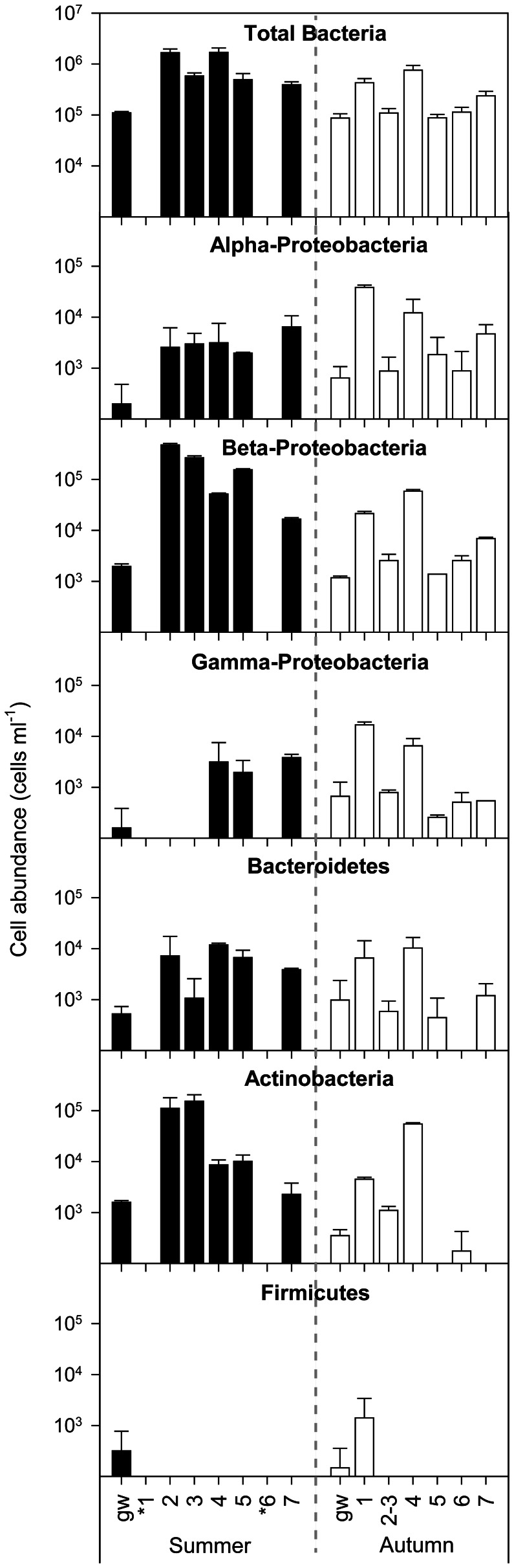
Bacterial cell abundance averaged 9.7×105 cells/ml during summer, with great variability among the sites (CV = 68%). In autumn, the cell abundance was lower (2.9×105 cells/ml) and less variable (CV = 21%). Groundwaters (site gw) had the lowest concentrations at any time (average 9.8×104 cells/ml, CV = 16%) (Fig. 4). We also observed statistically significant differences, both between seasons (two way ANOVA F = 569.03; P<0.001) and among the sites (F = 163.46; P<0.001). Pair-wise multiple comparisons (Student-Newman-Keuls method) revealed no significant differences between sites 2 and 4 or among sites 3, 5 and 7 in the summer. In autumn, cell abundance in site 4 showed the highest value, significantly different from any other site except site 1 (q = 1.709 p>0.05). Cell abundance was positively correlated with DOC concentration (r = 0.72; P<0.004) and water temperature (r = 0.51; P<0.05). The multiple regression analysis highlighted that the seasonal and spatial dynamics of cell abundance could be significantly explained by DOC, Temperature, and NH4 dynamics (r = 0.93; P<0.001).
Figure 4. Bacterial abundance.
Abundance of the selected bacterial phylogenetic groups as analysed by CARD-FISH. Values are expressed as cells per ml. Error bars indicate standard deviation. (*) = dry reach in summer.
When quantifying the occurrence of specific clusters, beta-Proteobacteria was the most abundant among the analysed groups in both the summer (up to 4.7×105±0.4×105 cells/ml in Site 2) and the autumn (up to 5.9×104±0.4×104 cells/ml in site 4). On average, Actinobacteria was the second-most abundant group, reaching the highest values in summer (isolated ponds 2 and 3). The alpha- and gamma-Proteobacteria showed the highest values in autumn at sites 1 and 4. The Bacteroidetes did not display a seasonal trend, and were below 1×104 cells/ml, while the Firmicutes were only detected in groundwater samples and in site 1 during autumn (Fig. 4).
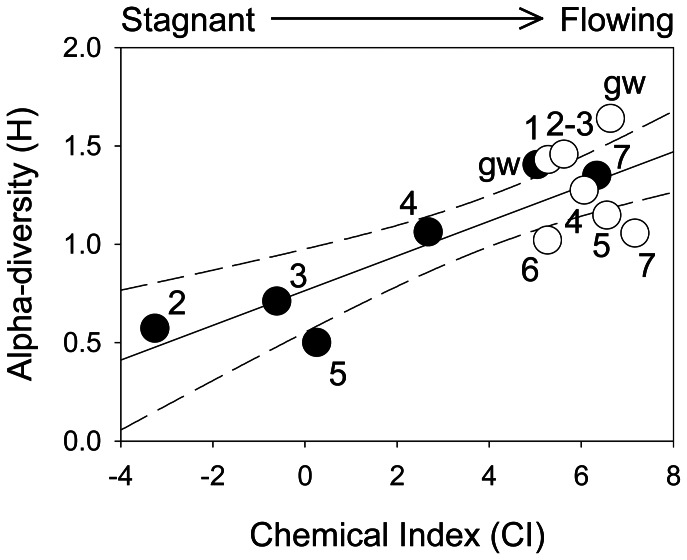
The nMDS analysis revealed a consistent shift of community composition in the isolated ponds (Fig. 3 b). CI was the chemical variable most strongly associated with community composition (Mantel test r = 0.51; p<0.001) followed by qualitative DOM properties (i.e. intensity of fluorescence peaks A and C, r = 0.37 and r = 0.33 respectively; p<0.001). When considering two or more variables together, the explanatory power of the Mantel test did not improve. Overall, changes in the alpha-diversity level of the bacterial community were observed in summer (Shannon entropy, H = 1.35 in headwaters and H = 0.50 in isolated ponds), whereas in the autumn it increased from 1.06 to 1.46 along the stream continuum. We observed a significant linear relationship (r = 0.82; p<0.05) between CI and the alpha-diversity level of the bacterial community (Fig. 5).
Figure 5. Chemical Index vs alpha-diversity.
Relationship between biogeochemical conditions (expressed as Chemical Index – CI = log([O2]/[N-NH4])) and the alpha-diversity index (r = 0.82; p<0.05). s = summer; a = autumn.
Bacterial phylogenetic assessment by fingerprinting and sequencing
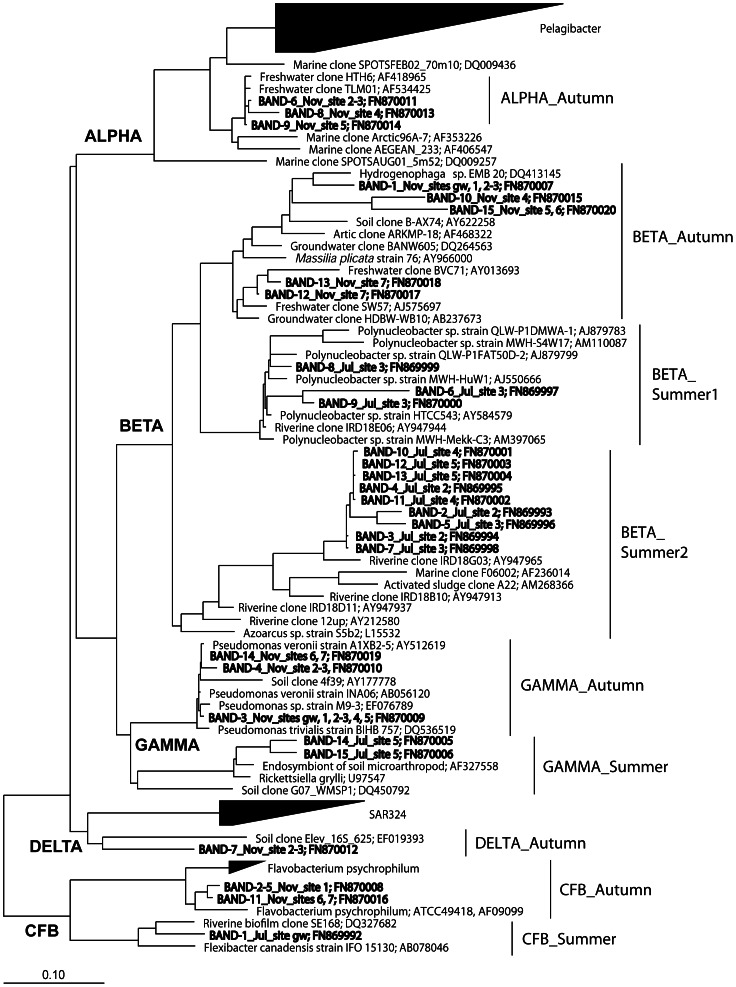
The number of DGGE bands was similar among samples and ranged between 8 and 13. The prominent bands (overall c.a. >80% of total band intensity in the lanes) were sequenced and alpha-, beta-, gamma-, and delta-Proteobacteria and Bacteroidetes were identified (Fig. 6). DGGE detects populations with a relative abundance around 0.1–1% of the total PCR-targeted cells [24], and we cannot, therefore, disregard the possible presence of other populations below this detection limit. It was possible to detect a shift in the retrieved phylotypes between summer and autumn samples for each bacterial phylum by grouping the samples according to the sampling season (Fig. 6).
Figure 6. Maximum parsimony tree of the bacterial 16S rRNA gene sequences.
Partial sequences were inserted into the optimised and validated tree available in the ARB program without changing the initial topology of the consensus tree provided by default. The scale bar represents 10% estimated divergence. Those identical sequences found in different sites are indicated in the band code. ALPHA, BETA, GAMMA, DELTA = Classes of Proteobacteria; CFB = Bacteroidetes.
The alpha-Proteobacteria (freshwater SAR11 cluster) were only detected in autumn as very prominent DGGE bands in sampling sites 2, 3, 4 and 5. The beta-Proteobacteria were abundant in most of the sites and split into three groups for the two different sampling periods: i) beta_Autumn, detected only in the autumn samples and related to Hydrogenophaga and Massilia spp.; ii) beta_ Summer1, detected in the summer samples and closely related to Polynucleobacter sp.; and iii) beta -Summer2, detected in the summer samples and distantly related to Azoarcus sp. (see accession numbers in Fig. 6). Overall, beta-Proteobacteria were heterogeneously distributed both in space and in time for the different sampling points.
We also observed changes in population composition between the two sampling periods for gamma-Proteobacteria (closely related to Pseudomonas sp. in autumn and to Rickettsiella sp. in summer) and Bacteroidetes (closely related to Flavobacterium spp. in autumn and to Flexibacter spp. in summer). The gamma-Proteobacteria were widespread in autumn, but were only detected in site 5 in summer. The delta-Proteobacteria (distantly related to the SAR324 cluster) were only detected in autumn. Bacteroidetes were found in a few sites in autumn (sites 1, 6, and 7) and only in site gw in the summer.
Discussion
In the Mediterranean region, hydrological fragmentation is a common feature of both headwaters and extended reaches of large rivers [25]–[26]. When stream flow is interrupted, the longitudinal lentic water continuum disappears. The water mass remains fragmented and stagnant in ponds and the hydrological connections to the surrounding environment are lost [27]. Our results showed that the qualitative DOM characteristics were extremely sensitive to the hydrological conditions showing a higher contribution of autochthonous organic solutes in the disconnected ponds in summer. Furthermore, the biogeochemical status of each disconnected pond was clearly related to the length of the isolation period. Hence, the generation of a set of transient isolated patches with high rates of biogeochemical processes [28], [29] and with different isolation times, increases the chemical heterogeneity spectrum of the entire Fuirosos stream system during summer [30].
The effect of biogeochemistry on microbial community composition and functioning is well known [31]. In the present study, the aquatic bacterial communities responded promptly to the biogeochemical heterogeneity. We observed that total bacterial abundance was positively related to DOC and N-NH4 concentrations and to temperature, with the highest values in the isolated ponds, where DOM mainly originated from in-situ microbial processes (i.e. high FI and Ic/Ia values). The alpha-, beta- and gamma-Proteobacteria accounted for the largest number of bacteria among the analysed groups, in agreement with the findings of other studies carried out in riverine systems by either 16S rDNA gene sequencing or FISH counts [32]–[34]. The dominance of beta-Proteobacteria in freshwater habitats was highlighted in a variety of other investigations [3], [22]. The alpha and gamma classes were consistently less abundant than the beta-Proteobacteria, and were also associated with more oxic conditions. Members of Bacteroidetes, reported as an autochthonous component of the limnetic habitat [32], were found in lower numbers. As shown by earlier investigations, the proportion of Actinobacteria can vary considerably in different freshwater habitats [35] and may locally belong to the dominant fractions of freshwater bacterial communities. Firmicutes were only locally present in low-stream reaches and groundwater with a low relative abundance, as reported for other stream environments [32], [36], [37]. The distribution of these bacterial classes showed relevant changes between stagnant and flowing waters. The degree of aerobic/anaerobic conditions seemed to be the main driver of the bacterial community composition and it was related to PIT and to autochthonous DOM. In summer, the bacterial communities showed higher alpha diversity in the stream reaches with flowing water. In the pond most recently isolated (site 5, PIT = 7 days), the community was composed by all the groups primarily found in headwaters, although a reduction in the abundance of alpha- and gamma-Proteobacteria was already evident. As isolation time increased (PIT>10 days) and CI decreased (Sites 2, and 3), the abundance of the beta-Proteobacteria and Actinobacteria increased significantly. These groups were probably adapted to the selective conditions that follow the interruption of the flow. Overall, during the summer, the change in community composition was closely related to the chemical variability determined by the isolation time. In autumn, as flow was restored, the alpha diversity of the communities increased downstream. This could be related to the greater contribution of organic substances derived from the allochthonous DOM in the upper reaches and the autochthonous DOM in the downstream reaches, as indicated by the increasing FI values. Therefore, when the river continuum was re-established, organic C transport provides a linkage along the stream that is fundamental to the nature of fluvial systems [38].
Variations in bacterial species composition, characterised by PCR-based community fingerprinting techniques, are often related to physical, chemical, and biological factors (e.g. [11], [39], [40]). Microcosm experiments clearly demonstrated that variations in DOM composition and origin (allochthonous versus autochthonous) could affect community composition [31], [41], [42]. In our study, the analysis of the genetic fingerprinting showed a temporal clustering instead of a spatial grouping. This indicated a seasonal shift of the populations within the same phylogenetic group (e.g. beta-, gamma-Proteobacteria and Bacteroidetes) in the transition from summer to autumn conditions for most of the sites. The sequences of beta-Proteobacteria retrieved from the hypoxic summer ponds were mainly affiliated to two different clusters, which were closely related to the genera Azoarcus spp. and Polynucleobacter spp., while the groups found in autumn were related to Hydrogenophaga and Massilia spp. The genus Azoarcus is reported to degrade aromatic compounds under denitrifying conditions [43], [44], and it might contribute to the removal of nitrogen and aromatic DOM in the isolated ponds. It is likely that Polynucleobacter spp. is ubiquitously distributed in lentic and stagnant habitats worldwide [45]. Recent observations from laboratory studies suggest that they prefer autochthonous rather than allochthonous substrate sources [46], [47]. Our results, therefore, confirm that the source and lability of DOM could drive bacterial community composition [48].
In conclusion, we found that (i) the temporary hydrological fragmentation simultaneously reduced the microbial dispersion and affected local environmental characteristics (i.e. redox regime and DOM quality) prompting the gradual development of selected bacterial groups in isolated water ponds, and (ii) a shift of the populations within the same phylogenetic classes (i.e. beta- and gamma-Proteobacteria) was observed in the transition from summer to autumn conditions. In the context of the seasonal dynamics, the temporary limitation in microbial dispersal and the environmental changes, promoted by pond isolation, gradually revealed local patterns in the community composition.
Materials and Methods
All the sampling sites along the Fuirosos stream are located in the protected area of Natural Park Montnegre-Corredor, under the authority of the Diputació de Barcelona. No specific permissions were required for all these locations to carry out the research activities, reported in this study, by the University of Barcelona. We confirm that the field studies did not involve endangered or protected species.
Site description and sampling
Fuirosos, a tributary of the River Tordera, is a third-order stream that drains a forested granitic catchment area of 16.2 km2 (NE Spain, 41°42′N, 2°34′W, 50–770 m a.s.l.). The climate is Mediterranean, with mean monthly temperatures ranging from 3°C in January to 24°C in August. Precipitation mostly falls in the autumn and spring, with occasional summer storms. The average annual mean rainfall for the region is 750 mm [49], and the catchment is covered mainly by perennial woodland, with agricultural fields representing <10% of the area.
The mean daily flow at the hydrochemical long-term monitoring station (Site 1, Fig. 1) ranged between 0 and 20 L s−1 [50]. During summer, the basal discharge decreased from 15 L s−1 in May to 0 L s−1on June 6th, when the water flow stopped and the water masses started to be confined in ponds. During this period, daily hydrological monitoring along the stream network allowed us to estimate the age of each isolated water pond. Surface flow recovered in the fluvial network on August 24th and the basal discharge gradually increased to 3–4 L s−1 and remained steady at around these values throughout November. In order to cover a wide range of hydrological conditions, samples were collected during summer (July), when the stream was disconnected into a series of isolated ponds, and after autumnal rainfall (November) when the stream connectivity was re-established. During the summer sampling, the entire stream network was completely dry, except in the five ponds where water was collected (ponds 2, 3, 4, 5, 7; Fig. 1 and 2). Ponds 2, 3, 5 were completely isolated, with stagnant water since 15, 12 and 7 days before sampling respectively. Ponds 2 and 3 were 15 meters apart. In ponds 4 and 7 water was still flowing (0.1–0.2 L s−1). During the autumn sampling, when the surface flow was continuous along the fluvial network (discharge 3–4 L s−1), samples were collected in six locations including those previously sampled in summer (Fig. 1 and 2). In autumn, only one sample was collected in correspondence of the summer ponds 2 and 3 as no differences were registered in water physical and chemical in situ measurements in the two contiguous sites.
Samples were also collected from a shallow, perched, riparian aquifer (site gw), which was recharged by stream water, by a peristaltic pump from a well (2 m depth) located outside the stream edge that perforated the sandy-gravel and the weathered granite layers [51]. We decided to sample this aquifer because during dry periods, it loses the hydrological connectivity with surface water, essentially becoming an underground pond with stagnant infiltrated water [27]. It was not possible to determine the PIT value for the groundwater site. However, it can be presumed that it was isolated from the surface's running water for at least 15 days before the sampling date.
Overall, the sampling sites covered a hydrological gradient including: surface stagnant waters in isolated ponds (sampling points 2, 3 and 5 in summer); surface waters under very low-flow (sampling points 4 and 7 in summer) and basal-flow conditions (sampling points 1 to 7 in autumn); and groundwater (gw) (Fig. 1 and 2). Temperature, pH, electrical conductivity, oxygen concentration and, when possible, discharge were measured at each sampling location. Triplicate water samples were collected in clean acid-washed bottles for water chemical characterisation (2 L) and in sterile flasks for microbiological analysis (1 L). The water samples were pre-filtered in the field with pre-combusted GF/F filters (Whatman) and transported in an icebox (4°C).
Water chemistry
Chloride and sulphate content was analysed with liquid chromatography using a Metrohm 76 compact IC, while nitrates and ammonia were determined colorimetrically using a Technicon auto-analyser, by means of the Griess-Ilosvay method [52] and oxidation by salicylate [53], respectively. The relationship between the concentration of dissolved oxygen and nitrogen, in ammonium form (CI = log([O2]/[N-NH4]) was used to describe the degree of the aerobic/anaerobic conditions in the waters bodies [30]. Under anaerobic conditions, CI is expected to have low values as a consequence of the decrease of oxygen and the concomitant accumulation of reduced solutes (i.e. NH4) [54], [55].
DOC and total dissolved nitrogen (TDN) concentrations were determined using a Shimadzu TOC-VCS with a coupled TDN analyser unit. Meanwhile, DON was estimated from the difference between the TDN and the dissolved inorganic nitrogen. Five descriptors were used to characterise the DOM: DOC∶DON ratio, BDOC, SUVA, FI, IC/IA. The BDOC content was determined according to the method proposed by Servais et al. [56], using site 4 GF/F filtered water as inoculum in all samples. SUVA was calculated from the measured absorbance at 254 nm, corrected by the cuvette path length in meters and the DOC concentration. Previous studies have revealed that SUVA is highly correlated to DOM aromaticity [57], [58]. Fluorescence measurements were performed using a Shimadzu RF-5301PC. The FI, calculated from the ratio between the emission intensities at 450 and 500 nm at a fixed excitation of 370 nm, allows discriminating DOM origin [59]. It ranges from 1.2 to 2, where low values indicate allochthonous origin while high values point to an autochthonous origin. The EEMs were obtained by concatenating emission spectra ranging from 280 to 690 at a range of excitation wavelengths from 240 to 420 in steps of 10 nm. Raman scattering was corrected by subtracting the value of ultra pure water blanks, and the EEMs were then corrected by the Raman area. Fluorescence intensity is expressed in Raman units. The determination of fluorescent peaks was performed by visual “peak picking”, using the coordinates estimated by Coble [60] as reference. The fluorescence maximum of peaks C and A were used to calculate IC/IA [30]. Fluorescent peaks A and C are generally associated with substances of terrestrial origin [60]. However, the fluorescence of certain components, which correspond to peak C, increases as a result of the microbial degradation of estuarine DOM of an autochthonous origin [61]. Accordingly, high values in this ratio provide information about the magnitude of the in-situ microbial processing.
Fluorescence in situ hybridization
CARD-FISH was performed following the protocol optimised by Fazi et al. [62], [63]. The following rRNA-target HRP-labelled probes (Biomers, Ulm, Germany) were used: ALF968, targeting sequence types affiliated with alpha-Proteobacteria; BET42a for beta-Proteobacteria; GAM42a for gamma-Proteobacteria; CF319a for Bacteroidetes (formerly Cytophaga-Flavobacterium-Bacteroides); HGC69a for Actinobacteria; LGC354a for Firmicutes [64]. The stained filter sections were inspected on a Leica DM LB 30 epifluorescence microscope (Leica Microsystems GmbH, Wetzlar, Germany) at 1000× magnification. At least 300 cells were counted in >10 microscopic fields randomly selected across the filter sections. The relative abundance of hybridized cells was estimated as the ratio of hybridized cells to total DAPI-stained cells.
DGGE fingerprinting analysis and 16S rRNA gene sequencing
A variable volume (200 ml to 800 ml) of stream water was filtered through a number of 0.2 µm- polycarbonate membranes (Nuclepore) until filter clogging decreased the flow rate to <1 ml per min. Because of the differences in cell abundance among sites, filter clogging provided an indication that we had processed a similar number of cells for each sample. Samples were digested in lysis buffer and phenol extracted [65], [66]. The 16S ribosomal RNA gene was PCR amplified with the universal bacterial primer set 341fGC-907r and run in a DGGE as previously described [67]. Prominent bands were excised from the gel, re-amplified, and sequenced [68]. The sequences were submitted to a BLAST search [69] and inserted into an optimised and validated ARB consensus tree (www.arb-home.de). The 16S rRNA gene sequence accession numbers at EMBL are from FN869992 to FN870020.
Statistical analysis
The abundances of different phylogenetic taxa estimated by CARD-FISH in each sampling site were compared by performing a two-way ANOVA and pair-wise multiple comparisons (Student-Newman-Keuls method). A multiple regression analysis was run to identify the environmental parameters that best explained the variability of the abundance of the bacterial taxa. To explore similarities between the sampling sites along the gradient from flowing to stagnant waters, the nMDS was performed with log transformed data according to the basic Euclidean distance matrix. The analysis was computed with either the environmental variables or the abundances of the bacterial taxa estimated by CARD-FISH. In addition, Mantel tests were run (permutation tests for correlation between Euclidean similarity matrices; 1000 randomised runs) to determine which combinations of environmental variables were more closely related with the similarity patterns of the abundances of the bacterial taxa [70]. Multivariate analyses were performed by the PAST software package (PAlaeontological STatistics, ver. 2.05). The alpha-diversity index was based on the relative abundance of the phylogenetic taxa estimated by CARD-FISH [Shannon entropy, H = −Σ (Pi ln Pi), Pi = relative abundance].
In order to evaluate if seasonal (summer versus autumn) community changes predominate over episodic habitat fragmentation, the primary DGGE bands at the same position in the different lanes of the gel were identified. A binary matrix (1/0) was produced to build a dissimilarity matrix based on the Jaccard coefficient (Sj) and a dendrogram with the un-weighted pair group average linkage method (UPGMA). Relative band intensities were calculated by comparison with the total intensity of all bands in each lane [68].
Funding Statement
Participation of AB and EV was supported by Project CGL2011-30151-C02-02/BTE, funded by Ministerio de Economìa y Competitividad. AB, EOC and EV are members of the Graccie Consortium. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, et al. (2006) Microbial biogeography: putting microorganisms on the map. Nature Reviews Microbiology 4: 102–112. [DOI] [PubMed] [Google Scholar]
- 2. Chaffron S, Rehrauer H, Pernthaler J, von Mering C (2010) A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Research 20: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barberan A, Casamayor EO (2010) Global phylogenetic community structure and beta-diversity patterns in surface bacterioplankton metacommunities. Aquatic Microbial Ecology 59: 1–10. [Google Scholar]
- 4. Logue JB, Mouquet N, Peter H, Hillebrand H, Metacommunity Working G (2011) Empirical approaches to metacommunities: a review and comparison with theory. Trends in Ecology & Evolution 26: 482–491. [DOI] [PubMed] [Google Scholar]
- 5. Portillo MC, Anderson SP, Fierer N (2012) Temporal variability in the diversity and composition of stream bacterioplankton communities. Environmental Microbiology 14: 2417–2428. [DOI] [PubMed] [Google Scholar]
- 6. Besemer K, Hodl I, Singer G, Battin TJ (2009) Architectural differentiation reflects bacterial community structure in stream biofilms. Isme Journal 3: 1318–1324. [DOI] [PubMed] [Google Scholar]
- 7. Östman O, Drakare S, Kritzberg ES, Langenheder S, Logue JB, et al. (2010) Regional invariance among microbial communities. Ecology Letters 13: 118–127. [DOI] [PubMed] [Google Scholar]
- 8. Barberan A, Casamayor EO (2011) Euxinic Freshwater Hypolimnia Promote Bacterial Endemicity in Continental Areas. Microbial Ecology 61: 465–472. [DOI] [PubMed] [Google Scholar]
- 9. Schimel J, Balser TC, Wallenstein M (2007) Microbial stress-response physiology and its implications for ecosystem function. Ecology 88: 1386–1394. [DOI] [PubMed] [Google Scholar]
- 10. Curtis TP, Sloan WT (2005) Exploring microbial diversity - A vast below. Science 309: 1331–1333. [DOI] [PubMed] [Google Scholar]
- 11. Fierer N, Morse JL, Berthrong ST, Bernhardt ES, Jackson RB (2007) Environmental controls on the landscape-scale biogeography of stream bacterial communities. Ecology 88: 2162–2173. [DOI] [PubMed] [Google Scholar]
- 12. Jackson RB, Fierer N, Schimel JP (2007) New directions in microbial ecology. Ecology 88: 1343–1344. [Google Scholar]
- 13. Hervas A, Camarero L, Reche I, Casamayor EO (2009) Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environmental Microbiology 11: 1612–1623. [DOI] [PubMed] [Google Scholar]
- 14. Comte J, del Giorgio PA (2009) Links between resources, C metabolism and the major components of bacterioplankton community structure across a range of freshwater ecosystems. Environmental Microbiology 11: 1704–1716. [DOI] [PubMed] [Google Scholar]
- 15. Sommaruga R, Casamayor EO (2009) Bacterial ‘cosmopolitanism’ and importance of local environmental factors for community composition in remote high-altitude lakes. Freshwater Biology 54: 994–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Logue JB, Lindström ES (2008) Biogeography of bacterioplankton in inland waters. Freshwater Reviews 1: 99–114. [Google Scholar]
- 17. Van der Gucht K, Cottenie K, Muylaert K, Vloemans N, Cousin S, et al. (2007) The power of species sorting: Local factors drive bacterial community composition over a wide range of spatial scales. Proceedings of the National Academy of Sciences of the United States of America 104: 20404–20409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Logue JB, Robinson CT, Meier C, Van der Meer JR (2004) Relationship between sediment organic matter, bacteria composition, and the ecosystem metabolism of alpine streams. Limnology and Oceanography 49: 2001–2010. [Google Scholar]
- 19. Reche I, Pulido-Villena E, Morales-Baquero R, Casamayor EO (2005) Does ecosystem size determine aquatic bacterial richness? Ecology 86: 1715–1722. [Google Scholar]
- 20. Lindstrom ES, Forslund M, Algesten G, Bergstrom AK (2006) External control of bacterial community structure in lakes. Limnology and Oceanography 51: 339–342. [Google Scholar]
- 21. Larned ST, Datry T, Arscott DB, Tockner K (2010) Emerging concepts in temporary-river ecology. Freshwater Biology 55: 717–738. [Google Scholar]
- 22. Romaní AM, Amalfitano S, Artigas J, Fazi S, Sabater S, et al. (2012) Microbial biofilm structure and organic matter use in mediterranean streams. Hydrobiologia 1–16. [Google Scholar]
- 23. Romaní AM, Vazquez E, Butturini A (2006) Microbial availability and size fractionation of dissolved organic carbon after drought in an intermittent stream: biogeochemical link across the stream–riparian interface. Microb Ecol 52: 501–512. [DOI] [PubMed] [Google Scholar]
- 24. Casamayor EO, Schafer H, Baneras L, Pedros-Alio C, Muyzer G (2000) Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: Comparison by microscopy and denaturing gradient gel electrophoresis. Applied and Environmental Microbiology 66: 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gasith A, Resh VH (1999) Streams in Mediterranean climate regions: Abiotic influences and biotic responses to predictable seasonal events. Annual Review of Ecology and Systematics 30: 51–81. [Google Scholar]
- 26. von Schiller D, Acuna V, Graeber D, Marti E, Ribot M, et al. (2011) Contraction, fragmentation and expansion dynamics determine nutrient availability in a Mediterranean forest stream. Aquatic Sciences 73: 485–497. [Google Scholar]
- 27. Butturini A, Bernal S, Nin E, Hellin C, Rivero L, et al. (2003) Influences of the stream groundwater hydrology on nitrate concentration in unsaturated riparian area bounded by an intermittent Mediterranean stream. Water Resources Research 39: 1–13. [Google Scholar]
- 28. McClain ME, Boyer EW, Dent CL, Gergel SE, Grimm NB, et al. (2003) Biogeochemical hot spots and hot moments at the interface of terrestrial and aquatic ecosystems. Ecosystems 6: 301–312. [Google Scholar]
- 29. Benstead JP, Leigh DS (2012) An expanded role for river networks. Nature Geosci 5: 678–679. [Google Scholar]
- 30. Vazquez E, Amalfitano S, Fazi S, Butturini A (2011) Dissolved organic matter composition in a fragmented Mediterranean fluvial system under severe drought conditions. Biogeochemistry 102: 59–72. [Google Scholar]
- 31. Judd KE, Crump BC, Kling GW (2006) Variation in dissolved organic matter controls bacterial production and community composition. Ecology 87: 2068–2079. [DOI] [PubMed] [Google Scholar]
- 32. Zwart G, Crump BC, Agterveld M, Hagen F, Han SK (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquatic Microbial Ecology 28: 141–155. [Google Scholar]
- 33. Amalfitano S, Fazi S, Zoppini A, Caracciolo AB, Grenni P, et al. (2008) Responses of benthic bacteria to experimental drying in sediments from Mediterranean temporary rivers. Microbial Ecology 55: 270–279. [DOI] [PubMed] [Google Scholar]
- 34. Zoppini A, Amalfitano S, Fazi S, Puddu A (2010) Dynamics of a benthic microbial community in a riverine environment subject to hydrological fluctuations (Mulargia River, Italy). Hydrobiologia 657: 37–51. [Google Scholar]
- 35. Allgaier M, Grossart HP (2006) Diversity and seasonal dynamics of Actinobacteria populations in four lakes in northeastern Germany. Applied and Environmental Microbiology 72: 3489–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fazi S, Amalfitano S, Piccini C, Zoppini A, Puddu A, et al. (2008) Colonization of overlaying water by bacteria from dry river sediments. Environmental Microbiology 10: 2760–2772. [DOI] [PubMed] [Google Scholar]
- 37. Gao XQ, Olapade OA, Leff LG (2005) Comparison of benthic bacterial community composition in nine streams. Aquatic Microbial Ecology 40: 51–60. [Google Scholar]
- 38. Webster JR (2007) Spiraling down the river continuum: stream ecology and the U-shaped curve. Journal of the North American Benthological Society 26: 375–389. [Google Scholar]
- 39. Muylaert K, Van der Gucht K, Vloemans N, De Meester L, Gillis M, et al. (2002) Relationship between bacterial community composition and bottom-up versus top-down variables in four eutrophic shallow lakes. Applied and Environmental Microbiology 68: 4740–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yannarell AC, Triplett EW (2005) Geographic and environmental sources of variation in lake bacterial community composition. Applied and Environmental Microbiology 71: 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Findlay SEG, Sinsabaugh RL, Sobczak WV, Hoostal M (2003) Metabolic and structural response of hyporheic microbial communities to variations in supply of dissolved organic matter. Limnology and Oceanography 48: 1608–1617. [Google Scholar]
- 42. Kirchman DL, Dittel AI, Findlay SEG, Fischer D (2004) Changes in bacterial activity and community structure in response to dissolved organic matter in the Hudson River, New York. Aquatic Microbial Ecology 35: 243–257. [Google Scholar]
- 43. Anders HJ, Kaetzke A, Kampfer P, Ludwig W, Fuchs G (1995) Taxonomic position of aromatic-degrading denitrifying Pseudomonad strains K-172 and KB-740 and their description as new members of the genera Thauera, as Thauera-aromatica sp-nov, and Azoarcus, as Azoarcus-evansii sp-nov, respectively, members of the beta-subclass of the proteobacteria. International Journal of Systematic Bacteriology 45: 327–333. [DOI] [PubMed] [Google Scholar]
- 44. Song B, Haggblom MM, Zhou JZ, Tiedje JM, Palleroni NJ (1999) Taxonomic characterization of denitrifying bacteria that degrade aromatic compounds and description of Azoarcus toluvorans sp nov and Azoarcus toluclasticus sp nov. International Journal of Systematic Bacteriology 49: 1129–1140. [DOI] [PubMed] [Google Scholar]
- 45. Jezberova J, Jezbera J, Brandt U, Lindstrom ES, Langenheder S, et al. (2010) Ubiquity of Polynucleobacter necessarius ssp asymbioticus in lentic freshwater habitats of a heterogenous 2000 km2 area. Environmental Microbiology 12: 658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hahn MW (2006) The microbial diversity of inland waters. Current Opinion in Biotechnology 17: 256–261. [DOI] [PubMed] [Google Scholar]
- 47. Wu QLL, Hahn MW (2006) High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environmental Microbiology 8: 1660–1666. [DOI] [PubMed] [Google Scholar]
- 48. Crump BC, Kling GW, Bahr M, Hobbie JE (2003) Bacterioplankton community shifts in an arctic lake correlate with seasonal changes in organic matter source. Applied and Environmental Microbiology 69: 2253–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ninyerola M, Pons X, Roure JM (2000) A methodological approach of climatological modelling of air temperature and precipitation through GIS techniques. International Journal of Climatology 20: 1823–1841. [Google Scholar]
- 50. Butturini A, Alvarez M, Bernal S, Vazquez E, Sabater F (2008) Diversity and temporal sequences of forms of DOC and NO3-discharge responses in an intermittent stream: Predictable or random succession? Journal of Geophysical Research-Biogeosciences 113. [Google Scholar]
- 51. Medici C, Butturini A, Bernal S, Vazquez E, Sabater F, et al. (2008) Modelling the non-linear hydrological behaviour of a small Mediterranean forested catchment. Hydrological Processes 22: 3814–3828. [Google Scholar]
- 52.Keeney DR, Nelson DW (1982) Nitrogen-inorganic forms. In: Methods of Soil Analysis, Part 2. 2nd Edn., Page, A. L., D. R Keeney, D. E Baker, R. H Miller, Jr. R Ellis and J. D Rhoades (Eds.). Madison, W. I. Agron. Monogr. 9. ASA and SSSA., ISBN: 0891180729.
- 53.Hach Company (1992) Hach water analysis handbook, 2nd edition. Hach Company, Loveland, Colorado.
- 54. Bleich ME, Silveira RML, Nogueira FMB (2009) Limnological Patterns in Northern Pantanal Lagoons. Brazilian Archives of Biology and Technology 52: 755–764. [Google Scholar]
- 55. Stanhope JW, Anderson IC, Reay WG (2009) Base Flow Nutrient Discharges from Lower Delmarva Peninsula Watersheds of Virginia, USA. Journal of Environmental Quality 38: 2070–2083. [DOI] [PubMed] [Google Scholar]
- 56. Servais P, Anzil A, Ventresque C (1989) Simple method for determination of biodegradable dissolved organic carbon in water. Applied and Environmental Microbiology 55: 2732–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weishaar JL, Aiken GR, Bergamaschi BA, Fram MS, Fujii R, et al. (2003) Evaluation of specific ultraviolet absorbance as an indicator of the chemical composition and reactivity of dissolved organic carbon. Environmental Science & Technology 37: 4702–4708. [DOI] [PubMed] [Google Scholar]
- 58. Hood E, Williams MW, McKnight DM (2005) Sources of dissolved organic matter (DOM) in a Rocky Mountain stream using chemical fractionation and stable isotopes. Biogeochemistry 74: 231–255. [Google Scholar]
- 59. McKnight DM, Boyer EW, Westerhoff PK, Doran PT, Kulbe T, et al. (2001) Spectrofluorometric characterization of dissolved organic matter for indication of precursor organic material and aromaticity. Limnology and Oceanography 46: 38–48. [Google Scholar]
- 60. Coble PG (1996) Characterization of marine and terrestrial DOM in seawater using excitation emission matrix spectroscopy. Marine Chemistry 51: 325–346. [Google Scholar]
- 61. Stedmon CA, Markager S (2005) Tracing the production and degradation of autochthonous fractions of dissolved organic matter by fluorescence analysis. Limnology and Oceanography 50: 1415–1426. [Google Scholar]
- 62. Fazi S, Amalfitano S, Pernthaler J, Puddu A (2005) Bacterial communities associated with benthic organic matter in headwater stream microhabitats. Environmental Microbiology 7: 1633–1640. [DOI] [PubMed] [Google Scholar]
- 63. Fazi S, Amalfitano S, Pizzetti H, Pernthaler J (2007) Efficiency of fluorescence in situ hybridization for bacterial cell identification in temporary river sediments with contrasting water content. Systematic and Applied Microbiology 30: 463–470. [DOI] [PubMed] [Google Scholar]
- 64. Loy A, Maixner F, Wagner M, Horn M (2007) probeBase - an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Research 35: D800–D804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Demergasso C, Escudero L, Casamayor EO, Chong G, Balagué V, Pedros-Alio C (2008) Novelty and spatio-temporal heterogeneity in the bacterial diversity of hypersaline Lake Tebenquiche (Salar de Atacama). Extremophiles 12: 491–504. [DOI] [PubMed] [Google Scholar]
- 66. Ferrera I, Massana R, Casamayor EO, Balague V, Sanchez O, et al. (2004) High-diversity biofilm for the oxidation of sulfide-containing effluents. Applied Microbiology and Biotechnology 64: 726–734. [DOI] [PubMed] [Google Scholar]
- 67. Casamayor EO, Pedros-Alio C, Muyzer G, Amann R (2002) Microheterogeneity in 16S ribosomal DNA-defined bacterial populations from a stratified planktonic environment is related to temporal changes and to ecological adaptations. Applied and Environmental Microbiology 68: 1706–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Casamayor EO, Muyzer G, Pedros-Alio C (2001) Composition and temporal dynamics of planktonic archaeal assemblages from anaerobic sulfurous environments studied by 16S rDNA denaturing gradient gel electrophoresis and sequencing. Aquatic Microbial Ecology 25: 237–246. [Google Scholar]
- 69. Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clarke KR, Warwick RM (2001) Changes in marine communities: an approach to statistical analysis and interpretation, 2nd edition. PRIMER-E, Plymouth. 172 pp.