Abstract
Understanding the tissue-specific pattern of gene expression is critical in elucidating the molecular mechanisms of tissue development, gene function, and transcriptional regulations of biological processes. Although tissue-specific gene expression information is available in several databases, follow-up strategies to integrate and use these data are limited. The objective of the current study was to identify and evaluate novel tissue-specific genes in human and mouse tissues by performing comparative microarray database analysis and semi-quantitative PCR analysis. We developed a powerful approach to predict tissue-specific genes by analyzing existing microarray data from the NCBI′s Gene Expression Omnibus (GEO) public repository. We investigated and confirmed tissue-specific gene expression in the human and mouse kidney, liver, lung, heart, muscle, and adipose tissue. Applying our novel comparative microarray approach, we confirmed 10 kidney, 11 liver, 11 lung, 11 heart, 8 muscle, and 8 adipose specific genes. The accuracy of this approach was further verified by employing semi-quantitative PCR reaction and by searching for gene function information in existing publications. Three novel tissue-specific genes were discovered by this approach including AMDHD1 (amidohydrolase domain containing 1) in the liver, PRUNE2 (prune homolog 2) in the heart, and ACVR1C (activin A receptor, type IC) in adipose tissue. We further confirmed the tissue-specific expression of these 3 novel genes by real-time PCR. Among them, ACVR1C is adipose tissue-specific and adipocyte-specific in adipose tissue, and can be used as an adipocyte developmental marker. From GEO profiles, we predicted the processes in which AMDHD1 and PRUNE2 may participate. Our approach provides a novel way to identify new sets of tissue-specific genes and to predict functions in which they may be involved.
Introduction
Tissue-specific gene expression plays a fundamental role in multi-cellular biology. In general, about 100 to 200 signature genes are expressed in a specific tissue. A detailed understanding of the tissue-specific pattern of gene expression can help elucidate the molecular mechanisms of tissue development, gene function, and transcriptional regulation of biological processes [1]. Tissue-specific transcript analysis can indicate novel functions of known and unknown genes. The expression of tissue-specific genes can also be used as an indicator for many complex diseases. Examples include the tissue-specific expression of insulin signaling-related genes in diabetes, the stroma-tumor interaction-related genes in cancer, and the tissue-specific expression of mutant IKBKAP (inhibitor of kappa light polypeptide enhancer in B cells, kinase complex-associated protein) gene in Familial Dysautonomia [2].
Microarrays are established technologies that can provide large-scale gene expression data through measurements of transcript abundance in various tissues. Various tissue-specific expression information is available in many databases including GEO [3], ArrayExpress [4], TiGER [5], BODYMAP [6] and BioGPS [7]. The Gene Expression Omnibus (GEO) database contains gene expression profiles derived from curated GEO DataSets (GDS), which store originally submitted records obtained from common commercial arrays (Affymetrix, Agilent, Illumina, or Nimblegen). The GDS contains several thousand gene expression profiles with 4 to 70 microarrays per profile and 12,000 to 30,000 genes per microarray, comparing diverse tissues and cells of human and mouse origins under various experimental conditions.
The GeneAtlas data on the website (http://biogps.org) provide baseline expression data for the expression patterns of thousands of predicted genes, as well as known and poorly characterized genes, across more than 60 murine tissues, and over 100 human tissues. However, the data from microarray experiments represent only a starting point toward understanding the microarray-derived measurements of differential gene expression. Although huge amounts of useful data are available to scientists, there is a lack of a follow-up strategy to integrate and use these data to identify novel sets of genes that are important for each field of study. There have been no attempts to integrate these valuable databases to identify novel sets of tissue-specific genes that might have important functions in tissue growth and development.
The objective of the current study was to identify and evaluate novel tissue-specific genes across the human and mouse by performing an analysis of microarray databases and semi-quantitative PCR analysis. In the current study, we developed a unique approach to generate accurate predictions of tissue-specific genes by comparing expression profiles for various tissues across the human and mouse. The semi-quantitative PCR analysis confirmed the accuracy of our predictions. We identified 59 genes across 6 human and mouse adult tissues: 10 kidney-specific, 11 liver-specific, 11 lung-specific, 11 heart-specific, 8 muscle-specific, and 8 adipose-specific. Among them we discovered 3 novel tissue-specific genes: AMDHD1 (amidohydrolase domain containing 1) in the liver, PRUNE2 (prune homolog 2) in the heart, and ACVR1C (activin A receptor, type IC) in the adipose tissue. The processes in which PRUNE2 and AMDHD1 may participate were predicted according to the GEO profiles. Further studies have shown that ACVR1C is adipose tissue-specific and adipocyte-specific in adipose tissue, and can be used as an adipocyte developmental marker. Our approach provides a novel method for identifying novel tissue-specific genes and predicting functions in which they may be involved.
Methods
Data Sources and Processing
The microarray expression profiles from 6 tissues (kidney, liver, lung, heart, muscle, and adipose) were derived from the GEO DataSet (GDS) in the NCBI web site: GDS3142 for mouse and GDS596 for human. Each tissue was represented by GEO samples (GSMs) from 2 to 4 subjects (human kidney: GSM18955 and GSM18956; human liver: GSM18953 and GSM18954; human lung: GSM18949 and GSM18950; human heart: GSM18951 and GSM18952; human muscle: GSM19013 and GSM19014; human adipose: GSM18975 and GSM18976; mouse kidney: GSM252083, GSM252084, and GSM252085; mouse liver: GSM252074, GSM252075, and GSM252076; mouse lung: GSM252080, GSM252081, and GSM252082; mouse heart: GSM252113, GSM252114, and GSM252115; mouse muscle: GSM252070, GSM252071, GSM252072, and GSM252073; mouse adipose: GSM252093, GSM252094, and GSM252095).
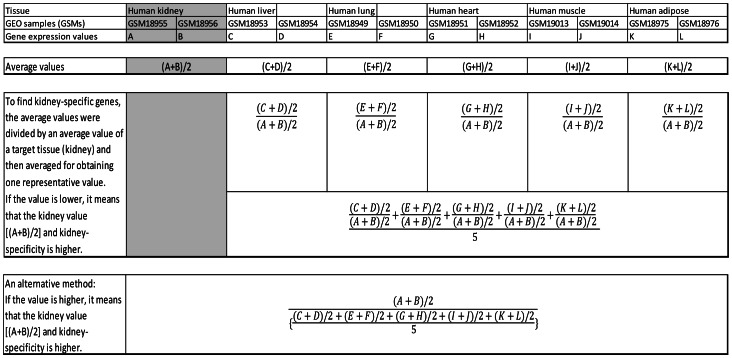
Tissue-specific genes were determined as follows: i) Gene expression values for each tissue in the GSM data (e.g., A and B in human kidney, C and D in human liver, and E and F in human lung) were averaged to obtain an average value [e.g., (A+B)/2, (C+D)/2, and (E+F)/2]; ii) To find tissue-specific genes (e.g., kidney-specific genes), the average values were divided by an average value of a target tissue {e.g., [(C+D)/2]/[(A+B)/2] and [(E+F)/2]/[(A+B)/2] }and then averaged to obtain one representative value <e.g., {[(C+D)/2]/[(A+B)/2]+[(E+F)/2]/[(A+B)/2]}/2>. If the value is lower, it means that the kidney value [(A+B)/2] and kidney-specificity is higher; iii) Averaged values were sorted in ascending order representing a lower value with a higher tissue-specific expression; iv) This method also shows relative gene expression ratios in other non-target tissues {e.g., [(C+D)/2]/[(A+B)/2] and [(E+F)/2]/[(A+B)/2]};v) An alternative method for finding tissue-specificity is to divide an average gene expression value for a target tissue [e.g., (A+B)/2 for kidney] by an average of averages of gene expression values in other tissues <e.g., {[(C+D)/2]+[(E+F)/2]}/2>, and sort the resulting values in descending order. Highly ranked genes shown in both human and mouse were selected for further analysis (Figure 1 shows the process of selecting kidney-specific genes; Table S1 shows the Excel spreadsheets of tissue-specific genes in selected tissues). The rank for each gene in each tissue is shown in Table S1.
Figure 1. Steps for selecting kidney-specific genes.
Tissue-specific genes were determined as follows: i) Gene expression values for each tissue were averaged; ii) The averaged values were divided by an averaged value for kidney; iii) The results were averaged and sorted in ascending order with a lower value representing higher tissue-specific expression. Highly ranked genes with lower values are candidates for kidney-specific genes; iv) An alternative method for finding tissue-specificity is to divide an average gene expression value for kidney by an average of averages of gene expression values in other tissues.
Animal Use and Ethics Statement
All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at The Ohio State University. All experiments were performed in accordance with the Prevention of Cruelty to Animals Act (1986). All mice were raised in a mouse housing facility at the Ohio State University and fed ad libitum. Mice were euthanized by CO2 inhalation followed by cervical dislocation. White adipose tissue (WAT), brown adipose tissue (BAT), liver, muscle, heart, lung, spleen, and kidney were harvested from 3-month-old mice to isolate total RNAs (n = 3). Mouse inguinal adipose tissue was collected from 1-month-old FVB mice [8].
Differentiation of Preadipocytes
The 3T3-L1 preadipocytes were differentiated to adipocytes as previously described [8]. The 3T3-L1 preadipocytes (American Type Culture Collection, Manassas, VA, USA) were cultured in DMEM culture media (Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (Invitrogen) and the mixture solution of penicillin and streptomycin (Pen Strep; Invitrogen). The preadipocytes were maintained and grown to confluence at 37°C in 5% CO2. After 2 d post-confluence (Day 0), differentiation of 3T3-L1 preadipocytes to adipocytes was induced by treating the preadipocytes for 2 d with a differentiation media, which contains 1 µM dexamethasone, 0.25 mM isobutylmethylxanthine, and 1 µg/ml insulin (Sigma-Aldrich Co., St. Louis, MO, USA) additionally in the culture media. Two days after induction (Day 2), the differentiation media was changed to the insulin media, which was composed of 1 µg/ml insulin in the culture media, for the next 2 d. Two days later (Day 4), the insulin media was changed to the DMEM culture media for another 4 d and the media was changed every 2 d. Total RNA was isolated from the 3T3L1 adipocytes at d 0, 2, 4, 6, and 8 post-differentiation.
cDNA Synthesis and PCR Analysis
Mouse total RNA was isolated from kidney, liver, lung, heart, muscle, and adipose tissue of adult mice using Trizol reagent (Invitrogen; [8]). Adult human RNAs from kidney, liver, lung, heart and muscle were purchased from Agilent Technologies (Santa Clara, CA, USA) and adult human RNA from adipose tissue was bought from Clontech Laboratories (Mountain View, CA, USA). Stromal vascular (SV) and fat cell (FC) fractionation were isolated according to procedures described previously [8], [9], [10]. In brief, the adipose tissue was incubated with 3.2 mg/ml collagenase II (Sigma-Aldrich) in DMEM media for 1 h at 37°C in a vigorous shaker to separate each cell. The digested adipose tissue was filtered to remove large cell masses, and then centrifuged for 5 min at 500×g to isolate the floating FC fraction from the pellet of the SV fraction. Both SV and FC fractions were gathered for RNA isolation (n = 3).
RNA was reverse-transcribed to cDNA using moloney murine leukemia virus reverse transcriptase (Invitrogen). The PCR reaction consisted of 1 µL of cDNA, 0.5 µL of 10 mM deoxynucleoside triphosphate mix (dNTP), 2.5 µL of 10×ThermopolII (Mg-free) reaction buffer, 0.5 µL of 100 mM MgSO4, 0.5 nM of each of the forward and reverse primers, 0.125 µL of Taq DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA), and nuclease-free water up to 25 µL. The cycling parameters were 95°C for 5 min, followed by 25 to 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 40 s with a final elongation at 72°C for 10 min. A 1% agarose gel was used to check PCR amplification.
For cDNA reverse transcription, 1 µg of total RNA, oligo dT, and M-MLV reverse transcriptase (Invitrogen) were used. The conditions for reverse transcription were 65°C for 5 min, 37°C for 50 min, and 70°C for 15 min. Quantitative real-time PCR was performed as described previously [8], [11] using AmpliTaq Gold polymerase (Applied Biosystems, Foster City, CA, USA) and SYBR green I as a detection dye. The sequences of primers for real-time PCR of cyclophilin (cyc), delta-like 1 (DLK1), fatty acid binding protein 4 (FABP4) and stearoyl-CoA desaturase-1 (SCD1) were as described previously [8], [11]. All other primers used are listed in Table 1. The mRNA expression of each gene was normalized to mRNA expression of cyclophilin, which was used as an internal control.
Table 1. Primer sequences for PCR amplification.
| Gene name | Primer sequence (5′–3′) | Gene name | Primer sequence (5′–3′) |
| SCGB1A1 | h-f: CCACCAGACTCAGAGACGGAAC | SLC22A2 | h-m-f: GTCAGCAAAGCAGGCTGGTTA |
| h-r: TGGGCGTGGACTCAAAGCA | h-m-r: GCATGATGAGGCCCTGGTA | ||
| m-f: ACAACATCACCCCACATCTACAGAC | PDZK1 | h-m-f: CTGGTCAGAAAGAGTGGGAATTCA | |
| m-r: CAAAGAGGAAGGAGGGGTTGG | h-m-r: TTTCAACCACTTCCTCATGGCT | ||
| SFTPB | h-m-f: ACCCTCTGCTGGACAAGCT | SLC12A3 | h-m-f: ATCATTTCCAACTTCTTCCTCTGC |
| h-m-r: AGCCAGGCACTGGCAGAT | h-m-r: GGTAGTTCTTGATGTGGTCTTCCAC | ||
| SFTPC | h-m-f: AGCAAAGAGGTCCTGATGGAGA | SPP1 | h-f: GAAAGCCATGACCACATGGA |
| h-m-r: CCAGTGGAGCCGATGGA | h-r: TGGGTTTCAGCACTCTGGTCA | ||
| CLIC3 | h-m-f: GCGGCTCTTCATGGTCCT | m-f: GAAAGCCATGACCACATGGA | |
| h-m-r: CAGGTAGCTGTCCAGCCTGG | m-r: ACTATCGATCACATCCGACTGATC | ||
| SLC34A2 | h-m-f: GCAGGCATGACCTTCATCGT | SLC34A1 | h-m-f: ATGCTCAACTCCCTGCTCAAGG |
| h-m-r: CGAACCAGCGATACTTGGCA | h-m-r: GCAAACCAGCGGTACTTGGC | ||
| AGER | h-m-f: AACATCACAGCCCGGATTG | FXYD2 | h-f: GGACGTGGACCCGTTCTACTATG |
| h-m-r: GTCTCAGGGTGTCTCCTGGTC | h-r: GGAGCTTCCTTCAGCCCAAG | ||
| TNNT2 | h-m-f: ATCCCCGATGGAGAGAGAGTG | m-f: CTATGAAACCGTCCGCAAAGG | |
| GCCACAGCTCCTTGGCCTTC | m-r: GATGAGGCTACACATGCTCCTCA | ||
| FHOD3 | h-m-f: AGACCAGAGGGAAGATGATCAC | AMDHD1 | h-m-f: CCTCGAGCCAGGAAGATGTTAG |
| h-m-r: CAGGGCTCTGGCTTCTTCTGG | h-m-r: TCAATTAATTCATGATGGCCTCC | ||
| NPPA | h-m-f: TAGAAGATGAGGTCATGCCCC | AMBP | h-m-f: CCTATGTGGTCCACACCAACTATG |
| GTCCTTGGTGCTGAAGTTTATTC | h-m-r: GACTATGGGGAGATTGCAGGC | ||
| PLN | h-m-f: AAAGTCCAATACCTCACTCGCTC | GNMT | h-m-f: CTTTGATGCTGTCATCTGCCTTG |
| GAAATGCCTCAGCAAGCACGTC | h-m-r: CAGGACGCTGTGCTGGCAC | ||
| MYH6 | h-m-f: TCCGCAAACAGCTGGAGGTG | HPX | h-m-f: CGCTACTACTGCTTCCAGGGTAAC |
| h-m-r: CTTCTGGTTGATGAGGCTGGTG | h-m-r: GCCTCCCTTTGTCAGGAAGAC | ||
| CSRP3 | h-m-f: CAAAATGTGGAGCCTGTGAAAAG | ALB | h-m-f: TGCAACACAAAGATGACAACCC |
| h-m-r: CTCTTCCCACAGATGGCACAG | h-m-r: TGCCCAGGAAGACATCCTT | ||
| RYR2 | h-m-f: AGGTCTCCACTTCTTCTGTGG | APOA1 | h-m-f: CGGCAGAGACTATGTGTCCCAG |
| h-m-r: CCAGGCTAGGTAGAGGAAGGA | h-m-r: CTTCTGGCGGTAGAGCTCCA | ||
| ACTN3 | h-f: GGCTCTCTGGAGGAGCAGATG | SLC27A5 | h-m-f: TCCTGCGGTACTTGTGTAAC |
| h-r: CTCGGGTCAGTACCTGGTTCTC | h-m-r: TCGAACTGCACCAGCTCAAAG | ||
| m-f: CCCAGCCGTGACCAGACACTG | FGG | h-m-f: GGCTGGGAAATGATGAGAAGAT | |
| m-r: TTGGGGTCCACCATGGTCATG | h-m-r: CACAGTTGCCTTCAAACTTATC | ||
| PRUNE2 | h-m-f: CTACCAGATGATTGACAGACGG | CYC | h-f: CTCCTTTGAGCTGTTTGCAG |
| h-m-r: GATGATGCTCTCTGGAATGTGG | h-r: CACCACATGCTTGCCATCC | ||
| ACVR1C | h-m-f: GTTTGCCTCCTGTCCATAGC | m-f: AGCACTGGAGAGAAAGGATTTGG | |
| h-m-r: GGTTCCCACTTTAGGATTCTG | m-r: TCTTCTTGCTGGTCTTGCCATT |
Statistical Analysis
Statistical analysis for the tissue distribution of gene expression was performed by a mixed ANOVA model (tissues showing significant expression at α = 0.05 vs. other tissues) followed by a Fisher’s protected least significant difference test. Analysis of SV and FC fraction was performed using the Student’s t test at P<0.05. Differences among the developmental time points were compared by one-way ANOVA followed by the Tukey’s post hoc test (P<0.05). To compare the difference between a control and an experimental group from GEO DataSets (GDS), Student’s t test was conducted (P<0.05). In addition, one-way ANOVA followed by Tukey’s post hoc test (P<0.05) was performed to compare multiple treatments from GDS. All statistical analyses were conducted using SAS software (version 9.2, SAS Institute Inc., Cary, NC, USA).
Results
Discovery of Tissue-Specific Genes
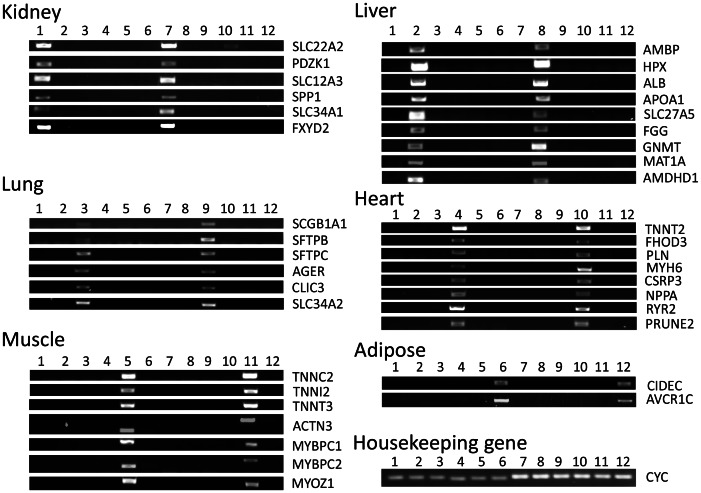
Twelve Excel spreadsheet files were generated for the kidney, liver, lung, heart, muscle, and adipose tissue specific expression in the human and mouse, respectively. By comparing top-rated genes across the human and mouse, 10 kidney, 11 liver, 11 lung, 11 heart, 8 muscle, and 8 adipose genes were selected. Names and ranks of selected genes, and ratios to an average of other tissues are provided in Table S1. These genes are significantly more highly expressed in certain tissues than in other tissues (P<0.001) with few exceptions (Table 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13). The tissue-specific expressions of those genes were confirmed either by existing publications or by semi-quantitative PCR and gel-electrophoresis (Figure 2). Three novel tissue-specific genes discovered by our method included AMDHD1 in the liver, PRUNE2 in the heart and ACVR1C in the adipose tissue. Their tissue-specific expressions were confirmed by both semi-quantitative PCR and real-time PCR (Figures 2, 3A, 4A, and 5A).
Table 2. Human kidney-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| GALNT11 | 1768650±25854 | 209850±77262 | 295450±8851 | 244800±68610 | 155500±30505 | 271400±4801 | <.0001 |
| SLC22A6 | 2441850±177177 | 41900±13602 | 8250±350 | 56900±43807 | 73550±12452 | 8250±150 | <.0001 |
| SLC22A8 | 4959550±860580 | 70850±13252 | 38800±10702 | 69900±29504 | 223350±60559 | 61700±29905 | <.0001 |
| SLC22A2 | 1041350±175076 | 136550±57659 | 82700±32805 | 41450±4651 | 375600±112217 | 31350±11652 | <.0001 |
| KL | 1565900±114717 | 74350±53258 | 59600±7101 | 38500±27004 | 576700±214132 | 24950±19053 | <.0001 |
| PDZK1 | 2576450±82362 | 90950±68260 | 9000±1500 | 72200±35005 | 451500±61209 | 48250±1850 | <.0001 |
| SLC12A3 | 1107100±29805 | 45800±4801 | 19200±1600 | 50100±20903 | 130300±900 | 23800±10302 | <.0001 |
| SPP1 | 6054550±271291 | 112100±35005 | 105000±2200 | 59100±10902 | 365750±275592 | 140150±550 | <.0001 |
| SLC34A1 | 1566950±94764 | 268150±53958 | 90150±51358 | 142850±116768 | 129300±38306 | 60300±8701 | <.0001 |
| FXYD2 | 11852250±272891 | 155850±39056 | 62500±33105 | 265050±171576 | 642700±59909 | 99750±46257 | <.0001 |
P value represents the significance of gene expression in the human kidney compared to other tissues.
Table 3. Mouse kidney-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| GALNT11 | 3362±149 | 170±2 | 198±21 | 156±2 | 157±5 | 215±15 | <.0001 |
| SLC22A6 | 1986±129 | 105±3 | 110±6 | 157±7 | 118±4 | 103±6 | <.0001 |
| SLC22A8 | 1741±130 | 86±6 | 88±4 | 120±3 | 80±1 | 85±3 | <.0001 |
| SLC22A2 | 1872±59 | 109±5 | 83±3 | 99±4 | 97±3 | 92±6 | <.0001 |
| KL | 1914±65 | 76±1 | 76±6 | 93±3 | 85±1 | 72±3 | <.0001 |
| PDZK1 | 4070±129 | 530±49 | 98±2 | 106±3 | 106±4 | 111±5 | <.0001 |
| SLC12A3 | 1987±249 | 136±2 | 120±3 | 143±11 | 120±4 | 115±6 | <.0001 |
| SPP1 | 4933±273 | 435±67 | 1037±210 | 104±4 | 145±12 | 104±18 | <.0001 |
| SLC34A1 | 7269±107 | 102±3 | 94±8 | 101±10 | 92±3 | 89±4 | <.0001 |
| FXYD2 | 9786±153 | 76±2 | 99±15 | 113±4 | 106±2 | 215±12 | <.0001 |
P value represents the significance of gene expression in the mouse kidney compared to other tissues.
Table 4. Human liver-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| HAMP | 171900±42806 | 11845400±1589440 | 25500±20103 | 690200±445667 | 370050±119868 | 54150±15252 | <.0001 |
| AHSG | 77800±44407 | 24651250±4147376 | 10550±150 | 189200±33805 | 318450±76662 | 33700±13702 | <.0001 |
| AMBP | 29800±14102 | 44128150±5392864 | 9100±3601 | 16750±4151 | 65750±7251 | 21050±10552 | <.0001 |
| HPX | 196050±100465 | 21364500±2278144 | 110450±33855 | 102750±33455 | 403000±131520 | 56350±3050 | <.0001 |
| ALB | 511500±6101 | 32913750±61559 | 50450±9351 | 91800±88613 | 114300±38006 | 53000±11602 | <.0001 |
| APOA1 | 74300±2600 | 50972350±10240696 | 45900±11902 | 349250±121768 | 309150±136971 | 52800±25004 | <.0001 |
| SLC27A5 | 58150±12552 | 11243300±360854 | 65400±25504 | 38300±9101 | 105450±13052 | 35000±7401 | <.0001 |
| FGG | 53400±49207 | 10354250±496925 | 74250±24254 | 135350±28454 | 333450±64160 | 54100±21403 | <.0001 |
| GNMT | 89750±70861 | 4526150±933091 | 12350±1750 | 44200±30205 | 243850±135070 | 20000±8101 | <.0001 |
| MAT1A | 46600±12902 | 8485900±318148 | 26350±2950 | 38750±14052 | 301400±41106 | 43650±8751 | <.0001 |
P value represents the significance of gene expression in the human liver compared to other tissues.
Table 5. Mouse liver-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| HAMP | 82±2 | 6092±1249 | 122±4 | 121±16 | 83±3 | 79±4 | <.0001 |
| AHSG | 93±1 | 8121±414 | 92±8 | 124±3 | 96±2 | 94±4 | <.0001 |
| AMBP | 94±5 | 7279±204 | 79±4 | 101±2 | 89±1 | 82±4 | <.0001 |
| HPX | 84±3 | 9046±350 | 93±5 | 100±1 | 98±3 | 84±1 | <.0001 |
| ALB | 183±3 | 16668±614 | 106±7 | 139±10 | 120±8 | 142±7 | <.0001 |
| APOA1 | 91±4 | 14910±361 | 86±9 | 127±4 | 103±3 | 88±1 | <.0001 |
| SLC27A5 | 84±4 | 5640±335 | 87±8 | 115±3 | 94±5 | 80±3 | <.0001 |
| FGG | 191±5 | 9792±373 | 81±1 | 96±4 | 87±4 | 75±4 | <.0001 |
| GNMT | 485±6 | 9337±214 | 181±6 | 216±10 | 182±11 | 115±9 | <.0001 |
| MAT1A | 76±2 | 4738±705 | 116±5 | 89±2 | 78±2 | 68±2 | <.0001 |
| AMDHD1 | 82±2 | 1612±126 | 68±6 | 82±3 | 69±3 | 67±4 | <.0001 |
P value represents the significance of gene expression in the mouse liver compared to other tissues.
Table 6. Human lung-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| CLDN5 | 247000±52608 | 245800±18503 | 3249550±98165 | 634400±200330 | 572600±375857 | 54650±8451 | <.0001 |
| CLDN18 | 158500±80912 | 248750±25154 | 2896050±202881 | 520000±331950 | 643050±209982 | 113450±19153 | <.0001 |
| LPCAT1 | 212500±131420 | 49550±19953 | 1304850±440417 | 27050±1650 | 105200±5201 | 90550±64060 | <.001 |
| MUC1 | 1190450±33755 | 34900±1400 | 3937250±418013 | 48050±30255 | 314650±164475 | 47250±14852 | <.0001 |
| SCGB1A1 | 157000±97015 | 131700±80312 | 8461200±606492 | 192650±121068 | 459000±116518 | 92700±16402 | <.0001 |
| SMAD6 | 130100±40306 | 48700±18203 | 770000±236136 | 193750±67260 | 85350±12552 | 23200±3901 | <.001 |
| SFTPB | 130800±16002 | 112700±76812 | 18122550±2916290 | 128500±49708 | 335300±18903 | 143050±29955 | <.0001 |
| SFTPC | 71700±3601 | 72400±9501 | 28720600±2420766 | 43900±8501 | 112150±8951 | 24450±3651 | <.0001 |
| AGER | 54700±15702 | 17150±5751 | 1179650±770166 | 52950±19353 | 87900±21303 | 70850±55658 | <.05 |
| CLIC3 | 32700±16302 | 23050±5351 | 1270200±311047 | 40600±14802 | 108600±71411 | 81950±13252 | <.0001 |
| SLC34A2 | 252100±20203 | 219050±89864 | 2808200±206331 | 247100±171326 | 711800±399460 | 61950±5451 | <.0001 |
P value represents the significance of gene expression in the human lung compared to other tissues.
Table 7. Mouse lung-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| CLDN5 | 196±8 | 186±24 | 3131±327 | 259±30 | 227±5 | 721±98 | <.0001 |
| CLDN18 | 92±2 | 100±3 | 2162±188 | 113±10 | 88±3 | 82±2 | <.0001 |
| LPCAT1 | 179±7 | 147±9 | 2681±141 | 190±7 | 159±5 | 289±31 | <.0001 |
| MUC1 | 198±23 | 102±4 | 916±33 | 127±4 | 102±7 | 95±6 | <.0001 |
| SCGB1A1 | 92±1 | 73±3 | 16192±411 | 107±4 | 79±2 | 86±9 | <.0001 |
| SMAD6 | 165±5 | 134±5 | 1725±207 | 185±5 | 150±11 | 242±43 | <.0001 |
| SFTPB | 146±6 | 148±14 | 8236±493 | 222±4 | 147±14 | 148±16 | <.0001 |
| SFTPC | 100±5 | 105±4 | 17295±341 | 142±4 | 98±1 | 94±2 | <.0001 |
| AGER | 108±4 | 99±3 | 10701±129 | 218±8 | 140±7 | 118±5 | <.0001 |
| CLIC3 | 154±4 | 95±3 | 1355±23 | 93±2 | 96±8 | 98±4 | <.0001 |
| SLC34A2 | 147±7 | 134±9 | 5913±238 | 265±17 | 137±11 | 104±9 | <.0001 |
P value represents the significance of gene expression in the mouse lung compared to other tissues.
Table 8. Human heart-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| FHL2 | 448350±147472 | 149000±91214 | 270350±7151 | 10829800±196430 | 996650±208782 | 545000±8801 | <.0001 |
| HSPB7 | 619350±7951 | 218500±41206 | 349200±7201 | 16034200±279242 | 4024050±402711 | 776550±73761 | <.0001 |
| MYOZ2 | 10800±5901 | 10900±3501 | 7650±550 | 1594350±58059 | 1238400±3501 | 6000±2200 | <.0001 |
| TNNT2 | 156200±14402 | 108600±35405 | 46200±3601 | 9868550±1550184 | 423300±216733 | 113600±47107 | <.0001 |
| FHOD3 | 285850±26454 | 36500±11702 | 34850±17253 | 817700±2100 | 171100±65210 | 47300±36405 | <.0001 |
| PLN | 37900±16302 | 40200±14302 | 14900±7701 | 1052750±251388 | 503800±348953 | 15950±2550 | <.005 |
| MYH6 | 17600±2700 | 28700±4801 | 53050±15452 | 5613450±320798 | 1098450±264690 | 12700±7001 | <.0001 |
| CSRP3 | 12150±4151 | 11600±2600 | 5300±400 | 5745250±515928 | 2571250±283193 | 8150±1850 | <.0001 |
| NPPA | 872650±221483 | 182900±79512 | 138900±74211 | 5658450±205681 | 2447950±762865 | 336900±52408 | <.0001 |
| RYR2 | 72050±61359 | 28600±5401 | 20250±12952 | 291400±22103 | 99800±10502 | 10450±3951 | <.0005 |
| PRUNE2 | 126700±42006 | 71650±20053 | 16850±12052 | 127800±46207 | 86450±36956 | 255800±8601 | <.5 |
P value represents the significance of gene expression in the human heart compared to other tissues.
Table 9. Mouse heart-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| FHL2 | 279±7 | 140±5 | 173±6 | 8829±429 | 161±2 | 146±7 | <.0001 |
| HSPB7 | 94±1 | 97±7 | 154±16 | 4441±118 | 1016±181 | 116±1 | <.0001 |
| MYOZ2 | 80±3 | 96±2 | 193±14 | 8934±199 | 2157±145 | 82±1 | <.0001 |
| TNNT2 | 74±4 | 79±4 | 422±27 | 16112±248 | 113±5 | 66±2 | <.0001 |
| FHOD3 | 346±22 | 82±2 | 81±4 | 1353±58 | 421±36 | 75±10 | <.0001 |
| PLN | 102±3 | 75±8 | 185±8 | 14565±615 | 112±8 | 86±9 | <.0001 |
| MYH6 | 71±2 | 73±2 | 839±124 | 19317±1430 | 83±2 | 87±12 | <.0001 |
| CSRP3 | 86±2 | 411±5 | 247±8 | 8180±256 | 1294±176 | 86±0 | <.0001 |
| NPPA | 77±4 | 88±7 | 261±19 | 4431±361 | 83±3 | 80±2 | <.0001 |
| RYR2 | 70±1 | 69±3 | 128±6 | 2889±51 | 74±3 | 68±2 | <.0001 |
| PRUNE2 | 72±1 | 77±3 | 74±0 | 215±26 | 85±1 | 83±2 | <.0001 |
P value represents the significance of gene expression in the mouse heart compared to other tissues.
Table 10. Human muscle-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| MYOT | 12900±6001 | 19500±7601 | 17250±12752 | 92450±35755 | 5427150±568736 | 14050±4651 | <.0001 |
| TNNC2 | 200500±7901 | 85800±14002 | 139350±32055 | 81450±13252 | 72503600±4693109 | 64650±21753 | <.0001 |
| TNNI2 | 40300±8301 | 25100±5901 | 28150±7951 | 43850±450 | 25906100±111517 | 22500±9601 | <.0001 |
| TNNT3 | 220450±83863 | 228450±94964 | 161100±14102 | 565100±373356 | 11062700±366455 | 149900±32805 | <.0001 |
| ACTN3 | 221850±24654 | 198850±90564 | 166450±15552 | 256300±87213 | 7269400±844828 | 163550±40656 | <.0001 |
| MYBPC1 | 179500±64710 | 158700±64710 | 57750±10352 | 279550±101765 | 20410650±63360 | 113800±3100 | <.0001 |
| MYBPC2 | 16600±3701 | 24550±8851 | 6800±100 | 50450±5851 | 12523950±251088 | 14650±6751 | <.0001 |
| MYOZ1 | 311100±176027 | 238550±67760 | 180600±6901 | 372150±141771 | 15847500±217533 | 148900±12202 | <.0001 |
P value represents the significance of gene expression in the human muscle compared to other tissues.
Table 11. Mouse muscle-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| MYOT | 66±1 | 65±2 | 63±0 | 836±14 | 7300±174 | 64±3 | <.0001 |
| TNNC2 | 82±3 | 94±7 | 80±3 | 135±3 | 15148±298 | 90±2 | <.0001 |
| TNNI2 | 89±4 | 94±8 | 105±13 | 109±4 | 15552±388 | 92±4 | <.0001 |
| TNNT3 | 79±4 | 101±11 | 90±4 | 83±4 | 14045±248 | 83±3 | <.0001 |
| ACTN3 | 92±3 | 107±5 | 101±20 | 119±4 | 11726±449 | 150±8 | <.0001 |
| MYBPC1 | 89±2 | 95±3 | 85±2 | 97±3 | 3414±295 | 87±4 | <.0001 |
| MYBPC2 | 83±2 | 82±4 | 100±2 | 183±13 | 9194±118 | 94±5 | <.0001 |
| MYOZ1 | 139±6 | 161±6 | 157±50 | 214±14 | 7523±84 | 145±11 | <.0001 |
P value represents the significance of gene expression in the mouse muscle compared to other tissues.
Table 12. Human adipose-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| RETN | 59850±32055 | 86150±42056 | 437450±156374 | 174450±100065 | 211200±89313 | 73500±32705 | <.5 |
| ADIPOQ | 173600±64710 | 141750±17753 | 34000±17103 | 393850±11152 | 299050±41256 | 10250050±1283044 | <.0001 |
| LEP | 13250±3351 | 8900±400 | 8950±1250 | 26550±5151 | 78650±15952 | 24350±2450 | <1 |
| PPARG | 42850±7951 | 35750±14352 | 80400±21503 | 22600±7001 | 84600±1400 | 917150±14252 | <.0001 |
| CIDEC | 546300±145022 | 787350±256989 | 400000±153723 | 894150±271191 | 1235150±349403 | 10546600±390859 | <.0001 |
P value represents the significance of gene expression in the human adipose tissue compared to other tissues.
Table 13. Mouse adipose-specific gene expression values.
| Gene | Kidney | Liver | Lung | Heart | Muscle | Adipose | P valuea |
| RETN | 168±13 | 136±2 | 240±8 | 190±18 | 254±44 | 11518±177 | <.0001 |
| ADIPOQ | 177±39 | 67±0 | 355±30 | 123±10 | 830±177 | 15125±409 | <.0001 |
| LEP | 74±2 | 80±1 | 75±2 | 88±4 | 105±10 | 1719±137 | <.0001 |
| PPARG | 108±3 | 168±24 | 148±4 | 172±16 | 135±8 | 2016±159 | <.0001 |
| CIDEC | 83±6 | 88±7 | 132±4 | 82±5 | 123±12 | 5175±484 | <.0001 |
| CCDC80 | 146±2 | 191±10 | 362±2 | 734±34 | 409±46 | 6447±167 | <.0001 |
| DGAT2 | 739±27 | 1252±158 | 213±8 | 727±29 | 330±16 | 4503±922 | <.0001 |
| ACVR1C | 58±2 | 61±2 | 63±2 | 60±2 | 68±2 | 776±208 | <.0001 |
P value represents the significance of gene expression in the mouse adipose tissue compared to other tissues.
Figure 2. Expression of adult human and mouse gene transcripts detected by PCR reaction and agarose gel electrophoresis.
Lanes 1–6 contain PCR products from human and lanes 7–12 contain PCR products from mouse. Lanes 1 and 7: kidney, lanes 2 and 8: liver, lanes 3 and 9: lung, lanes 4 and 10: heart, lanes 5 and 11: muscle, and lanes 6 and 12: adipose. Housekeeping genes, human and mouse cyclophilin (cyc), serve as a loading control.
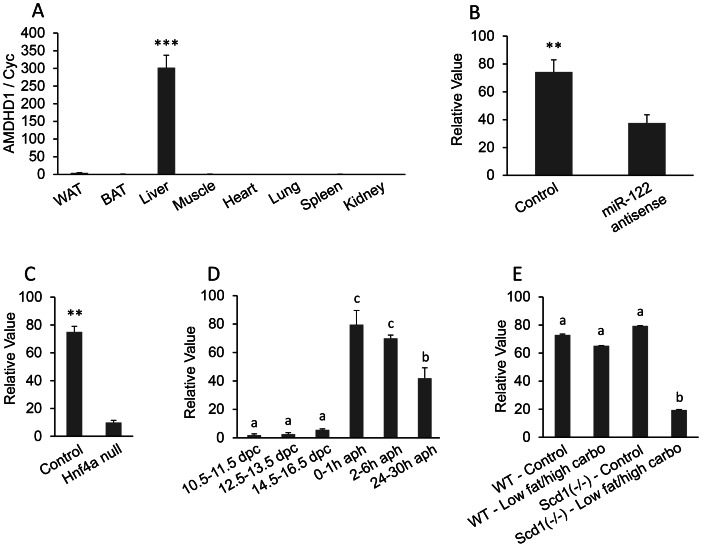
Figure 3. AMDHD1 (amidohydrolase domain containing 1) mRNA expression.
A, Real-time PCR for AMDHD1 mRNA tissue distribution. Total RNA were isolated from the white adipose tissue (WAT), brown adipose tissue (BAT), liver, muscle, heart, lung, spleen, and kidney of adult mice. The mRNA expression was measured by quantitative real-time reverse transcription PCR (qRT-PCR) (n = 3). The bar represents mean ± SEM. Statistical significance is indicated by ***(P<0.001). Housekeeping gene cyclophilin (cyc) was used to normalize the mRNA expression. B–E, Analysis of microarray DataSets obtained from the NCBI website, which contains expression profiles for AMDHD1. B: GDS1729 (n = 5 per group), C: GDS1916 (n = 3 per group), D: GDS2577 (n = 3–4 per group), and E: GDS1517 (n = 5 per group). HNF4α: hepatocyte nuclear factor 4 alpha; dpc: days post conception; aph: after partial hepatectomy.
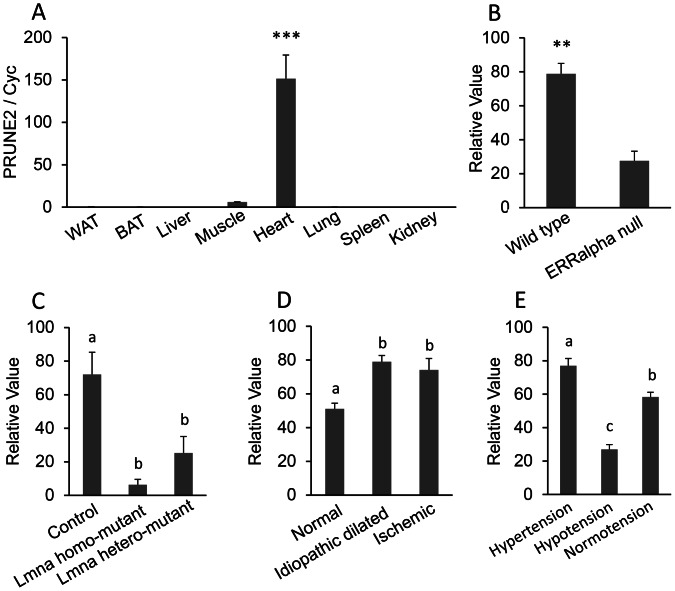
Figure 4. PRUNE2 (prune homolog 2) mRNA expression.
A, Real-time PCR for PRUNE2 mRNA tissue distribution. Total RNA were isolated from the white adipose tissue (WAT), brown adipose tissue (BAT), liver, muscle, heart, lung, spleen, and kidney of adult mice. The mRNA expression was measured by quantitative real-time reverse transcription PCR (qRT-PCR) (n = 3). The bar represents mean ± SEM. Statistical significance is indicated by ***(P<0.001). Housekeeping gene cyclophilin (cyc) was used to normalize the mRNA expression. B–E, Analysis of microarray DataSets obtained from the NCBI website that contains expression profiles for PRUNE2. B: GDS2727 (n = 3 per group), C: GDS2746 (n = 6–8 per group), D: GDS651 (n = 11–15 per group), and E: GDS3673 (n = 5 per group). ERRα: estrogen-related receptor alpha; Lmna: lamin A/C.
Figure 5. ACVR1C (activin A receptor, type IC) mRNA expression.
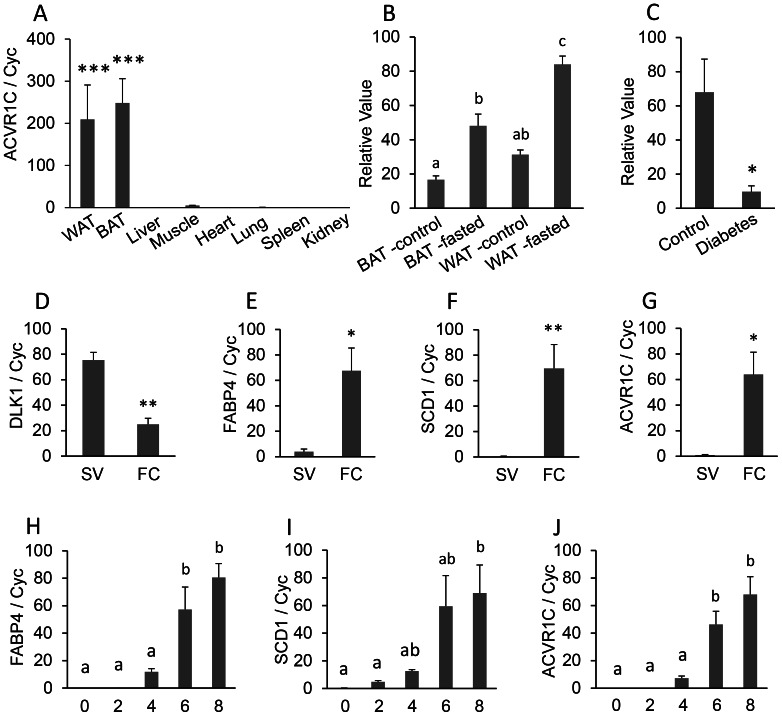
A, Real-time PCR for ACVR1C mRNA tissue distribution. Total RNA were isolated from the white adipose tissue (WAT), brown adipose tissue (BAT), liver, muscle, heart, lung, spleen, and kidney of adult mice. The mRNA expression was measured by quantitative real-time reverse transcription PCR (qRT-PCR) (n = 3). The bar represents mean ± SEM. Statistical significance is indicated by ***(P<0.001). Housekeeping gene cyclophilin (cyc) was used to normalize the mRNA expression. B and C, Analysis of microarray DataSets obtained from the NCBI website containing expression profiles for ACVR1C. B: GDS3135 (n = 4 per group) and C: GDS3665 (n = 5 per group). D–G, Relative expression of DLK1, FABP4, SCD1, and ACVR1C in the stromal-vascular (SV) and fat cell (FC) fractions from mouse inguinal adipose tissue. Each bar indicates mean and SEM (n = 5). Statistical significance by Student’s t test is shown: *, P<0.05; **, P<0.01. The gene expression was normalized to cyclophilin (cyc) mRNA expression. H–J, Developmental regulation of ACVR1C during adipogenic differentiation of 3T3-L1 cells. The bar represents mean ± SEM (n = 3). Letters a and b show significant differences in gene expression among several time-points (day 0, 2, 4, 6, and 8) in adipocyte differentiation at P<0.05. The mRNA abundance was measured by quantitative real-time reverse transcription PCR (qRT-PCR) and normalized to cyclophilin (cyc) mRNA.
Regulation of Novel Gene Expression Under Different Physiological Conditions and Disease Models
Gene expression under different physiological conditions and disease models was investigated using GEO profiles to help understand potential functions of the novel genes. The GEO profiles provided information to predict the function of AMDHD1 in the liver. GDS1729 in the GEO Profiles shows that microRNA miR-122 antisense inhibits the expression of AMDHD1 in the liver (Figure 3B, P <0.01). The miR-122 down-regulates target mRNAs to establish tissue-specific gene expression patterns. The decreased expression of AMDHD1 in miR-122 treated liver indicated that AMDHD1 is one of the target genes of miR-122 and is involved in liver development and formation. GDS1916 shows that the expression of AMDHD1 is decreased significantly when hepatocyte nuclear factor 4 alpha (HNF4α) is deleted (Figure 3C, P <0.01). HNF4α is a promoter for the expression of hundreds of metabolism-related genes in the liver. The co-expression of AMDHD1 and HNF4α indicates that AMDHD1 may be involved in hepatogenesis. The relatively greater expression of AMDHD1 in the regenerating liver compared to the developing liver indicates the role of AMDHD1 in renewal and repair of the liver (GDS2577; Figure 3D, P <0.05). Expression of AMDHD1 in control mice is not influenced by a low fat, high carbohydrate diet, whereas SCD1 null mice treated with a low fat, high carbohydrate diet have significantly decreased expression of AMDHD1 (GDS-1517; Figure 3E, P <0.05). Because SCD1 is the rate-limiting enzyme that catalyzes the synthesis of monounsaturated fatty acids [12], this result indicates that AMDHD1 may be involved in fatty acid metabolism, which is one of the main functions of the liver.
PRUNE2 was previously studied in the central nervous system and in cancer patients, but there are no reports related to the function of PRUNE2 in the heart. GEO profiles provide some information to predict the function of PRUNE2 in the heart. The expression of PRUNE2 is decreased in ERRα (estrogen-related receptor alpha) deficient mouse hearts (GDS2727; Figure 4B; P<0.01). ERRα regulates cellular energy metabolism, which is pivotal in high energy demand tissues such as the heart. The decreased expression of PRUNE2 in the heart of ERRα deficient mice indicates the function of PRUNE2 may be related to energy metabolism in the heart. Bonne et al. (1999) identified Lmna (lamin A/C) mutations in the autosomal dominant form of Emery–Dreifuss muscular dystrophy (AD-EDMD) [13]. The decreased expression of PRUNE2 in the Lmna H222P mutants (GDS 2746; Figure 4C; P<0.05) suggests a relationship between PRUNE2 and Emery–Dreifuss muscular dystrophy. Patients with idiopathic dilated heart failure and ischemic heart failure have increased expression of PRUNE2, which also indicates a role of PRUNE2 in heart function (GDS651, Figure 4D; P<0.05). PRUNE2 is also related to heart tension with higher expression in the heart of the hypertensive blood pressure high (BPH) inbred strains of mice and lower expression in genetically hypotensive blood pressure low (BPL) inbred strains compared to the normotensive blood pressure normal (BPN) inbred strains (GDS3673; Figure 4E; P<0.05). All of these data suggest that PRUEN2 is related to heart function and heart disease.
ACVR1C, also named activin receptor-like kinase 7 (ALK7), has a known ligand, Nodal, and is one of the type I transforming growth factor-β (TGF-β) receptors [14]. GDS 3135 shows that, in fasted rats, the expression of ACVR1C increased significantly in both WAT and BAT (Figure 5B), suggesting a role for ACVR1C in releasing fats in both WAT and BAT. GDS3665 shows that the expression of ACVR1C is decreased significantly (P<0.05; Figure 5C) in the adipose tissue from obese diabetic women compared to control women, which indicates that ACVR1C may be a contributing factor to obese diabetic symptoms.
ACVR1C is BAT and WAT-Specific
Given that average relative expression in other tissues such as liver, muscle, heart, lung, spleen, and kidney was approximately 1, the mRNA expression of ACVR1C in BAT and WAT was about 250-fold and 210-fold higher than in other tissues, respectively (Figure 5A). Therefore, ACVR1C showed significantly higher expression in both BAT and WAT. In addition, fractionation of SV cells and fat cells was verified by predominant expression of DLK1 (a preadipocyte marker) in SV cells (P<0.01), and FABP4 and SCD1 (adipocyte markers) in fat cells (P<0.05 and P<0.01, respectively). In SV cells, mRNA expression of ACVR1C was significantly reduced. However, in fat cells, mRNA expression of ACVR1C was enhanced approximately 60-fold compared to expression in SV cells. Taken together, these results suggest that ACVR1C expression is specific to adipose tissue and adipocytes in the fat cell fraction.
Developmental Regulation of ACVR1C in 3T3-L1 Cells
Developmental regulation of gene expression of ACVR1C has been evaluated during adipogenic differentiation of 3T3-L1 preadipocytes (Figure 5J). The development of adipocytes was demonstrated by gradual increases in the expression of adipocyte markers, FABP4 and SCD1, during differentiation. In addition, expression of both FABP4 and SCD1 increased significantly after d 6 (P<0.05). During 3T3-L1 preadipocyte differentiation, expression of ACVR1C showed a highly correlated pattern of expression to that of FABP4, with a significant increase at d 6 (P<0.05).
Discussion
We have established a novel and powerful approach to predict tissue-specific genes. By comparing one human and one mouse GEO DataSet (GDS) from a microarray, we identified a total of 59 tissue-specific or tissue-related genes in the kidney, liver, lung, heart, muscle, and adipose tissue. After confirmation of the tissue-specific expression by semi-quantitative PCR, we searched for the functions of these genes in specific tissues using NCBI PubMed and GEO profiles to further support our approach. The following tissue-specific genes selected from a microarray were categorized as follows: i) Genes that are verified as tissue-specific in human in previous publications; ii) Genes that are verified as tissue-specific in mouse in previous publications; iii) Genes that are verified as tissue-specific in both human and mouse in previous publications; iv) Genes that are not confirmed as tissue-specific, but are said to contain tissue-related functions in previous publications; and v) Novel genes that have not been previously reported as tissue-specific or tissue-related, but are verified as tissue-specific by our PCR (Table 14, 15, 16, 17, 18, 19).
Table 14. Kidney-specific expression confirmed by publications and/or semi-quantitative PCR.
Table 15. Liver-specific expression confirmed by publications and/or semi-quantitative PCR.
Table 16. Lung-specific expression confirmed by publications and/or semi-quantitative PCR.
Table 17. Heart-specific expression confirmed by publications and/or semi-quantitative PCR.
Table 18. Muscle-specific expression confirmed by publications and/or semi-quantitative PCR.
Table 19. Adipose-specific expression confirmed by publications and/or semi-quantitative PCR.
Kidney-Specific Genes
The 10 kidney-specific genes that we identified are GALNT11 ((UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11), SLC22A6 [solute carrier family 22 (organic anion transporter), member 6], SLC22A8 [solute carrier family 22 (organic anion transporter), member 8], SLC22A2 [solute carrier family 22 (organic cation transporter), member 2], KL (klotho), PDZK1 (PDZ domain containing 1), SLC12A3 [solute carrier family 12 (organic cation transporter), member 3], SPP1 (secreted phosphoprotein 1), SLC34A1 [solute carrier family 34 (sodium phosphate), member 1], and FXYD2 (FXYD domain containing ion transport regulator 2). Among them, genes that have been verified by publications for the human and/or mouse include GALNT11 [15], SLC22A6 [16], [17], SLC22A8 [18], [19], SLC22A2 [20], [21], KL [22], [23], PDZK1 [24], and SLC12A3 [25]. Genes confirmed by PCR include SLC22A2, PDZK1, SLC12A3, SPP1, SLC34A1, and FXYD2.
The SLC genes belong to the solute carrier family. The SLC22A2, SLC22A6, and SLC22A8 genes are members of the organic anion transporter SLC22 gene family [26]. SLC12A3 belongs to an electroneutral cation-chloride-coupled cotransporter gene family. SLC34A1 is a member of the type II sodium-phosphate co-transporter family. All of these genes are responsible for solute (organic anion, sodium/chloride, and phosphate, respectively) transportation in the kidney, which is the key aspect for kidney function.
KL is a single transmembrane protein that is mainly produced in the kidney and brain. Severely reduced production of KL can induce chronic renal failure in the human kidney [27]. FXYD2 is the gamma subunit of the Na,K-ATPase and functions in regulating the enzyme’s activity by inducing ion channel activity. Mutations in this gene have been associated with renal hypomagnesaemia [28]. SPP1 is synthesized by the kidney and secreted into the urine by epithelial cells [29]. It functions to inhibit the nucleation and aggregation of calcium oxalate crystals [30]. PDZK1 is a scaffold protein that is located in brush borders of proximal tubular cells [24]. PDZK1 binds to and mediates the localization of cell surface proteins and plays an important role in cholesterol metabolism [31]. GALNT11 is an enzyme that is highly expressed in the kidney and catalyzes O-linked oligosaccharide biosynthesis and transfers N-acetyl-D-galactosamine residue to a serine or threonine residue on the protein receptor.
Liver-Specific Genes
HAMP (hepcidin antimicrobial peptide), AHSG (alpha-2-HS-glycoprotein), AMBP (alpha-1-microglobulin/bikunin precursor), HPX (hemopexin), ALB(albumin), APOA1 (apolipoprotein A-I), SLC27A5 [solute carrier family 27 (fatty acid transporter), member 5], FGG (fibrinogen gamma chain), GNMT (glycine N-methyltransferase), MAT1A (methionine adenosyltransferase I, alpha), and AMDHD1 (amidohydrolase domain containing 1) exhibit significantly higher expression in liver than in other tissues. The liver-specific expressions of HAMP [32], [33], AHSG [34], and AMBP [35] in human and/or mouse were confirmed by previous publications. AMBP, HPX, ALB, APOA1, SLC27A5, FGG, GNMT, and MAT1A were confirmed as liver-specific by semi-quantitative PCR.
HAMP functions in the maintenance of iron homeostasis, and is required for intestinal iron absorption and iron storage in macrophages. Mutations in this gene may cause hemochromatosis type 2B, which is an endocrine liver disease. HPX is a plasma glycoprotein that can bind and transport heme from the plasma to the liver for iron recovery to prevent heme-mediated oxidative damage and heme-bound iron loss [36]. AHSG and ALB are both synthesized by hepatocytes and secreted to the serum. AHSG is involved in ectopic calcium deposition [37], insulin resistance [38], [39], and fat accumulation in the liver [40]. ALB composes about half of the blood serum protein. It serves as a carrier for steroids, fatty acids, thyroid hormones, and drugs and as a regulator for the colloidal osmotic pressure of blood. ALB levels are decreased in chronic liver disease and nephrotic syndrome.
AMBP is a liver-specific precursor protein of alpha-1-microglobulin and bikunin. Alpha-1-microglobulin belongs to the lipocalin transport protein superfamily and functions in inflammatory processes, whereas bikunin is a urinary trypsin inhibitor. FGG is a blood-borne glycoprotein that can be cleaved by thrombin to form fibrin and act as a co-factor in platelet aggregation. Defects in this gene lead to several disorders, including dysfibrinogenemia, hypofibrinogenemia, and thrombophilia. APOA1 is the main component of high density lipoprotein (HDL) in blood plasma. It promotes reverse transport of cholesterol from tissues to the liver for excretion by promoting cholesterol efflux from tissues [41]. SLC27A5, GNMT, and MATA1A have enzymatic activities. SLC27A5 is a fatty acid transporter, and encodes Acyl-CoA synthetase, which is involved in bile acid metabolism in the liver. GNMT catalyzes the conversion of S-adenosyl-methionine (SAM) to S-adenosylhomocysteine and sarcosine. MAT1A catalyzes the formation of S-adenosylmethionine from methionine and ATP. All of these activities occur predominantly in the liver and are important for normal liver functions.
AMDHD1 is only contained in mouse GDS and is highly ranked, indicating its liver-specific expression. Human GDS596 only contains data for about 22,000 spots, whereas the mouse GDS3142 has data for more than 45,000 spots. Searching for “AMDHD1 liver” in NCBI PubMed (http://www.ncbi.nlm.nih.gov/pubmed) returns no results. Searching GEO profiles provides indications about the function of AMDHD1 in the liver. AMDHD1 protein contains 426 amino acids and has been reported to be involved in the histidine metabolism pathway [42]. The expression of AMDHD1 in the liver is inhibited by microRNA miR-122 antisense. The miR-122 makes up 70% of all microRNA in the adult liver. It is highly expressed in the developing and adult liver [43]. It negatively regulates target mRNAs and is thought to be important for establishing tissue-specific gene expression patterns. The expression of AMDHD1 is negatively regulated by miR-122 in the liver, suggesting that AMDHD1 may be involved in liver development and formation. AMDHD1 does not have miR-122 binding sites (www.microrna.org), which indicates that miR-122 is a trans-acting factor for AMDHD1. HNF4α is a nuclear receptor that can activate the expression of hundreds of genes in the liver, especially metabolism-related genes in glucose, fatty acid, cholesterol, and drug metabolism [44], [45]. The HNF4α null mice are embryonic lethal [46]. The decreased expression of AMDHD1 in HNF4α deleted liver suggests that AMDHD1 is involved in hepatogenesis. GDS2577 shows that expression of AMDHD1 is significantly higher in the regenerating liver than in the developing liver, which suggests a function for AMDHD1 in renewal and repair of the liver.
SCD1 is an enzyme that is responsible for forming a double bond in stearoyl-CoA to form monounsaturated fatty acid from saturated fatty acid [12]. A low fat, high carbohydrate diet may cause SCD1 null mice to develop severe hypercholesterolemia. The significantly lower expression of AMDHD1 in the low fat, high carbohydrate treated SCD1 null mice (GDS-1517) indicates that AMDHD1 is involved in fatty acid metabolism in the liver.
Lung-Specific Genes
Lung-specific genes identified in the current study include LPCAT1 (lysophosphatidylcholine acyltransferase 1), MUC1 (mucin 1, cell surface associated), SCGB1A1 [secretoglobin, family 1A, member 1 (uteroglobin)], SFTPB (surfactant protein B), SFTPC (surfactant protein C), AGER (advanced glycosylation end product-specific receptor), CLDN5 (claudin 5), CLDN18 (claudin 18), SLC34A2 [solute carrier family 34 (sodium phosphate), member 2], SMAD6 (SMAD family member 6), and CLIC3 (chloride intracellular channel 3). Among them, lung-specific expression in human and/or mouse was either confirmed by publications {CLDN5 [47], [48], CLDN18 [49], LPCAT1 [50], [51], MUC1 [52], [53], SCGB1A1 [54], and SMAD6 [55]} or PCR (SCGB1A1, SFTPB, SFTPC, AGER, SLC34A2, and CLIC3).
Bridges et al. (2010) reported that LPCAT1 functions in surfactant phospholipid synthesis and is essential for transitioning to air breathing in neonatal mice [56]. MUC1 serves as a protective layer in the airway against bacterial and enzyme attack. SCGB1A1 is an anti-inflammatory agent that decreases systemic inflammation and increases surfactant protein and vascular endothelial growth factor expression. It functions in reducing lung injury, improves pulmonary compliance and oxygenation. SFTPB and SFTPC are both expressed on the pulmonary surfactant to promote alveolar stability by reducing air-liquid interface tension.
The CLDNs are located on tight junction strands on the cell membrane of the lung and serve as a physical barrier for solutions and water. Mutations in CLDN5 may cause velocardiofacial syndrome [47], whereas mutations in CLDN18 are related to lung adenocarcinomas [57]. AGER is highly expressed in the embryonic brain and adult lung. AGER expression is significantly decreased in human lung carcinomas, which suggests that AGER may function in suppressing lung cancer. SLC34A2 is a phosphate transport protein. Mutations in SLC34A2 may cause pulmonary alveolar microlithiasis [58].
SMAD6 inhibits transforming growth factor-beta (TGF-β) superfamily-regulated cell growth and development. CLIC3 is a component of chloride ion channels. The functions of these two genes are understudied in the lung. By searching the GEO profile, we predict that SMAD6 and CLIC3 could be related to idiopathic pulmonary fibrosis and pulmonary adenocarcinomas. Compared to the normal lung tissue, the expressions of SMAD6 and CLIC3 are lower in tissues with idiopathic pulmonary fibrosis (GDS1252) and pulmonary adenocarcinoma (GDS1650 and GDS 3257).
Heart-Specific Genes
In the heart, FHL2 (four and a half LIM domains 2), HSPB7 [heat shock 27 kDa protein family, member 7 (cardiovascular)], MYOZ2 (myozenin 2), FHOD3 (formin homology 2 domain containing 3), PLN (phospholamban), MYH6 (myosin, heavy chain 6, cardiac muscle, alpha), CSRP3 [cysteine and glycine-rich protein 3 (cardiac LIM protein)], NPPA (natriuretic peptide A), RYR2 (ryanodine receptor 2, cardiac), TNNT2 (troponin T type 2, cardiac), and PRUNE2 have significantly higher expressions compared to other tissues. Publications confirmed the human and/or mouse heart-specific expression of FHL2 [59], [60], HSPB7 [61], [62], and MYOZ2 [63], and PCR confirmed the heart specific expression of TNNT2, FHOD3, PLN, MYH6, CSRP3, NPPA, RYR2, and PRUNE2.
FHL2 functions in many fundamental processes by interacting with a variety of types of proteins including structural proteins, kinases, and transcription factors. HSPB7 interacts with alpha filamin, and is potentially involved in chaperone activity and maintenance of the cytoskeletal network in the cardiac muscle. MYOZ may serve as intracellular binding proteins involved in linking Z line proteins and localizing calcineurin signaling to the sarcomere. FHOD3 is an actin-organizing protein that may regulate stress fiber formation. PLN has been postulated to regulate the activity of the calcium pump of the cardiac sarcoplasmic reticulum. CSRP3 is an organizer of cytosolic structures in cardiomyocytes. Mutations in this gene may cause hypertrophic cardiomyopathy and dilated cardiomyopathy in humans [64]. NPPA is a hormone playing a key role in cardiovascular homeostasis. Both MYH6 and RYR2 are responsible for cardiac muscle contraction. RYR2 mutations may cause catecholaminergic polymorphic ventricular tachycardia (CPVT) and arrhythmogenic right ventricular dysplasia (ARVD) [65], [66].
Full length PRUNE2 contains 3,088 amino acids, which can be divided into 19 exons. PRUNE2 has 5 isoforms: BMCC1 [Bcl-2/adenovirusE1B 19 kDa-interacting protein (BNIP) 2 and Cdc42 GAP homology (BCH) motif-containing molecule at the C-terminal region1], BNIPXL (BNIP2 extralong), C9orf65 (Chromosome 9 Open Reading Frame 65), PRUNE2, and Olfaxin. BMCC1 is associated with neuronal apoptosis [67], BNIPXL is an N-terminal truncated form of BMCC1 and is related to cellular transformation [68], C9orf65 is a biomarker that distinguishes leiomyosarcomas from gastrointestinal stromal tumors [69], PRUNE2 is a binding protein of 8-oxo-GTP that contains C9orf65 and BMCC1 [70], [71]. These four PRUNE2 isoforms may play crucial roles in Alzheimer’s disease and cancer. Olfaxin is the most recently discovered PRUNE2 isoform that is located in the olfactory systems [72].
Neuronal tissues were not included in our samples. The expression of PRUNE2 was significantly greater in the heart compared to other tissues in the mouse (P<0.001) but not human (P<0.5) Searching NCBI PubMed returned no publications about PRUNE2 in the heart. However, our semi-quantitative and real-time quantitative PCR confirmed the high expression of PRUNE2 in both human and mouse heart.
From GEO profiles we found that expression of PRUNE2 is decreased in ERRα-deficient mouse hearts. Considering ERRα is an orphan nuclear receptor that plays a critical role in regulating cellular energy metabolism, the decreased expression of PRUNE2 in ERRα-deficient mouse hearts suggests that PRUNE2 may be involved in energy metabolism in the mouse heart.
PRUNE2 is likely to be related to heart diseases. The expression of PRUNE2 is decreased in Lmna H222P mutants, which in turn are related to Emery–Dreifuss muscular dystrophy [13]. These results suggest an association between PRUNE2 and Emery–Dreifuss muscular dystrophy.
The function of PRUNE2 in cardiomyocyte diseases is linked to serum response factor (SRF). SRF plays a critical role in mesodermal development [73] and the deletion of SRF causes embryonic lethal cardiovascular phenotypes. Expression of PRUNE2 is upregulated in SRF-null mutants, which indicates that PRUNE2 may be associated with cardiomyocyte diseases. Increased expression of PRUNE2 is also found in patients with idopathic dilated heart failure and ischemic heart failure, which supports the role of PRUNE2 in heart function. PRUNE2 expression is the highest in the hearts of genetically hypertensive BPH inbred strains of mice compared to the normotensive BPN and hypotensive BPL inbred strains. All of these data suggest an important role for PRUNE2 in heart function and heart diseases.
Muscle-Specific Genes
All 8 of the muscle specific genes we identified are well-studied for their roles in muscle function: MYOT (myotilin), TNNC2 (troponin C type 2, fast), TNNI2 (troponin I type 2, skeletal, fast), TNNT3 (troponin T type 3, skeletal, fast), MYBPC1 (myosin binding protein C, slow type), MYOZ1 (myozenin 1), ACTN3 (actinin, alpha 3), and MYBPC2 (myosin binding protein C, fast type). We confirmed the muscle-specific expression of TNNC2, TNNI2, TNNT3, ACTN3, MYBPC1, MYBPC2, and MYOZ1 by PCR and two publications supported the muscle-specific expression of MYOT [74], [75].
MYOT is located within the Z-disc of sarcomeres. Mutations in the MYOT gene cause various forms of muscular dystrophy [76], [77], [78], [79]. Troponin is a key protein controlling striated muscle contraction. It is composed of 3 subunits: the TNNI subunit inhibits actomyosin ATPase, the TNNC subunit binds to calcium and overcomes the inhibitory action of the troponin complex on actin filaments, and the TNNT subunit binds to tropomyosin and TNNC. TNNI2, TNNC2, and TNNT3 are specifically expressed in muscle, whereas TNNT2 is mainly expressed in cardiac muscle. MYBPC1 and MYBPC2 are skeletal muscle slow-twitch and fast-twitch myosin binding proteins, which can regulate the activity of actin-activated myosin ATPase to modulate muscle contraction. MYOZ1 functions in modulating calcineurin signaling in skeletal muscle.
Adipose-Specific Genes
Six genes that were confirmed by publications to be highly expressed in human and mouse adipose tissue are: RETN (resistin) [80], [81], [82], ADIPOQ (adiponectin, C1Q and collagen domain containing) [83], LEP (leptin) [84], [85], PPARγ (peroxisome proliferator-activated receptor gamma) [86], [87], CIDEC (Cell death–inducing DFF45-like effector C) [88], CCDC80 (coiled-coil domain containing 80) [89], and DGAT2 (diacylglycerol O-acyltransferase 2) [90]. Our PCR data showed the adipose specific expression of CIDEC (cell death-inducing DFFA-like effector c) and ACVR1C.
Both RETN and ADIPOQ are adipokines that control insulin sensitivity and fat metabolism. LEP controls the size of adipose depots by affecting food intake and energy expenditure. PPARγ is a well-known regulator of adipocyte differentiation, glucose hemeostasis, and blood pressure. CIDEC is localized around the lipid droplet in adipocytes and regulates lipid droplet formation [91], [92]. It can regulate energy balance and obesity [93] and induce cell apoptosis [94]. CCDC80 is a secreted protein that regulates adipocyte differentiation [95], whereas DGAT2 is an enzyme that catalyzes the final step of mammalian triglyceride synthesis [96] and may be involved in the mechanisms of obesity, insulin resistance, and leptin resistance.
Our SV and fat cell fractionation studies showed that ACVR1C is adipocyte-specific, but not preadipocyte-specific. ACVR1C was further confirmed to be both WAT-specific and BAT-specific. When fasted, the expression of ACVR1C increases significantly in rat WAT and BAT, which suggests a role for ACVR1C in fat metabolism. In the adipose tissue from obese diabetic women, the expression of ACVR1C is decreased significantly, which suggests a role for ACVR1C in obese diabetic symptoms.
In summary, our approach provides a new and powerful procedure to discover novel tissue-specific genes and predict the processes or pathways in which they may be involved. With this method, we discovered novel tissue-specific genes: AMDHD1 in the liver, PRUNE2 in the heart, and ACVR1C in the adipose tissue. Our procedure also can be extended to other tissues in other species. This approach is an efficient way of integrating valuable databases to identify novel sets of tissue-specific genes that are related to tissue growth and development, and diseases.
Supporting Information
Tissue-specific gene expression values based on GEO DataSet(GDS)596 for the human and GDS3142 for the mouse. The microarray gene expression profiles for the six tissues (kidney, liver, lung, heart, muscle, and adipose tissue) of the human and mouse were collected from GEO DataSet(GDS) in the National Center for Biotechnology Information (NCBI) web page. *Average gene expression values for each tissue including a target tissue were obtained. **Those average values for each tissue were divided by an average value of a target tissue and then averaged for obtaining one representative value. If the representative value is lower, then tissue-specificity for a target tissue is higher because it was divided by an average value of a target tissue. ***An alternative approach shows that an average value of a target tissue was divided by average of averages for each tissue. If the result value is higher, then tissue-specificity for a target tissue is higher because an average value of a target tissue was divided. Figure 1 shows the schematic diagram for the above procedures.
(XLSX)
Funding Statement
This project was supported by Agriculture and Food Research Initiative Competitive grant number 2010-65206-20716 from the USDA National Institute of Food and Agriculture. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Niehrs C, Pollet N (1999) Synexpression groups in eukaryotes. Nature 402: 483–487. [DOI] [PubMed] [Google Scholar]
- 2. Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, et al. (2001) Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet 68: 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, et al. (2007) NCBI GEO: mining tens of millions of expression profiles. Nucleic Acids Res 35: D760–D765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parkinson H, Sarkans U, Shojatalab M, Abeygunawardena N, Contrino S, et al. (2005) ArrayExpress–a public repository for microarray gene expression data at the EBI. Nucleic Acids Res 33: D553–D555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu X, Yu X, Zack DJ, Zhu H, Qian J (2008) TiGER: a database for tissue-specific gene expression and regulation. BMC Bioinformatics 9: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ogasawara O, Otsuji M, Watanabe K, Iizuka T, Tamura T, et al. (2006) BodyMap-Xs: anatomical breakdown of 17 million animal ESTs for cross-species comparison of gene expression. Nucleic Acids Res 34: D628–D631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, et al. (2002) Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA 99: 4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li B, Shin J, Lee K (2009) Interferon-stimulated gene ISG12b1 inhibits adipogenic differentiation and mitochondrial biogenesis in 3T3-L1 cells. Endocrinology 150: 1217–1224. [DOI] [PubMed] [Google Scholar]
- 9. Deiuliis JA, Li B, Lyvers-Peffer PA, Moeller SJ, Lee K (2006) Alternative splicing of delta-like 1 homolog (DLK1) in the pig and human. Comp Biochem Physiol Part B 145: 50–59. [DOI] [PubMed] [Google Scholar]
- 10. Deiuliis JA, Shin J, Bae D, Azain MJ, Barb R, et al. (2008) Developmental, hormonal, and nutritional regulation of porcine adipose triglyceride lipase (ATGL). Lipids 43: 215–225. [DOI] [PubMed] [Google Scholar]
- 11. Li B, Zerby HN, Lee K (2007) Heart fatty acid binding protein is up-regulated during porcine adipocyte development. J Anim Sci 85: 1651–1659. [DOI] [PubMed] [Google Scholar]
- 12. Dobrzyn P, Ntambi JM, Dobrzyn A (2008) Stearoyl-CoA desaturase: A novel control point of lipid metabolism and insulin sensitivity. Eur J Lipid Sci Technol 110: 93–100. [Google Scholar]
- 13. Bonne G, Di Barletta MR, Varnous S, Bécane HM, Hammouda EH, et al. (1999) Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy. Nat Genet 21: 285–288. [DOI] [PubMed] [Google Scholar]
- 14. Roberts HJ, Hu S, Qiu Q, Leung PC, Caniggia I, et al. (2003) Identification of novel isoforms of activin receptor-like kinase 7 (ALK7) generated by alternative splicing and expression of ALK7 and its ligand, Nodal, in human placenta. Biol Reprod 68: 1719–1726. [DOI] [PubMed] [Google Scholar]
- 15. Schwientek T, Bennett EP, Flores C, Thacker J, Hollmann M, et al. (2002) Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in Drosophila. J Biol Chem 277: 22623–22638. [DOI] [PubMed] [Google Scholar]
- 16. Lopez-Nieto CE, You G, Bush KT, Barros EJ, Beier DR, et al. (1997) Molecular cloning and characterization of NKT, a gene product related to the organic cation transporter family that is almost exclusively expressed in the kidney. J Biol Chem 272: 6471–6478. [DOI] [PubMed] [Google Scholar]
- 17. Hosoyamada M, Sekine T, Kanai Y, Endou H (1999) Molecular cloning and functional expression of a multispecific organic anion transporter from human kidney. Am J Physiol 276: F122–128. [DOI] [PubMed] [Google Scholar]
- 18. Cha SH, Sekine T, Fukushima JI, Kanai Y, Kobayashi Y, et al. (2001) Identification and characterization of human organic anion transporter 3 expressing predominantly in the kidney. Mol Pharmacol 59: 1277–1286. [DOI] [PubMed] [Google Scholar]
- 19. Buist SC, Klaassen CD (2004) Rat and mouse differences in gender-predominant expression of organic anion transporter (Oat1–3; Slc22a6–8) mRNA levels. Drug Metab Dispos 32: 620–625. [DOI] [PubMed] [Google Scholar]
- 20. Aoki M, Terada T, Kajiwara M, Ogasawara K, Ikai I, et al. (2008) Kidney-specific expression of human organic cation transporter 2 (OCT2/SLC22A2) is regulated by DNA methylation. Am J Physiol Renal Physiol 295: F165–F170. [DOI] [PubMed] [Google Scholar]
- 21. Mooslehner KA, Allen ND (1999) Cloning of the mouse organic cation transporter 2 gene, Slc22a2, from an enhancer-trap transgene integration locus. Mamm Genome 10: 218–224. [DOI] [PubMed] [Google Scholar]
- 22. Matsumura Y, Aizawa H, Shiraki-Iida T, Nagai R, Kuro-o M, et al. (1998) Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem Biophys Res Commun 242: 626–630. [DOI] [PubMed] [Google Scholar]
- 23. Shiraki-Iida T, Aizawa H, Matsumura Y, Sekine S, Iida A, et al. (1998) Structure of the mouse klotho gene and its two transcripts encoding membrane and secreted protein. FEBS Lett 424: 6–10. [DOI] [PubMed] [Google Scholar]
- 24. Gisler SM, Pribanic S, Bacic D, Forrer P, Gantenbein A, et al. (2003) PDZK1: I. a major scaffolder in brush borders of proximal tubular cells. Kidney Int 64: 1733–1745. [DOI] [PubMed] [Google Scholar]
- 25. Mastroianni N, De Fusco M, Zollo M, Arrigo G, Zuffardi O, et al. (1996) Molecular cloning, expression pattern, and chromosomal localization of the human Na-Cl thiazide-sensitive cotransporter (SLC12A3). Genomics 35: 486–493. [DOI] [PubMed] [Google Scholar]
- 26. Koepsell H, Endou H (2004) The SLC22 drug transporter family. Pflugers Arch 447: 666–676. [DOI] [PubMed] [Google Scholar]
- 27. Koh N, Fujimori T, Nishiguchi S, Tamori A, Shiomi S, et al. (2001) Severely reduced production of klotho in human chronic renal failure kidney. Biochem Biophys Res Commun 280: 1015–1020. [DOI] [PubMed] [Google Scholar]
- 28. Sha Q, Pearson W, Burcea LC, Wigfall DA, Schlesinger PH, et al. (2008) Human FXYD2 G41R mutation responsible for renal hypomagnesemia behaves as an inward-rectifying cation channel. Am J Physiol Renal Physiol 295: F91–F99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hudkins KL, Giachelli CM, Cui Y, Couser WG, Johnson RJ, et al. (1999) Osteopontin expression in fetal and mature human kidney. J Am Soc Nephrol 10: 444–457. [DOI] [PubMed] [Google Scholar]
- 30. Grover PK, Ryall RL (1999) Inhibition of calcium oxalate crystal growth and aggregation by prothrombin and its fragments in vitro: relationship between protein structure and inhibitory activity. Eur J Biochem 263: 50–56. [DOI] [PubMed] [Google Scholar]
- 31. Komori H, Arai H, Kashima T, Huby T, Kita T, et al. (2008) Coexpression of CLA-1 and human PDZK1 in murine liver modulates HDL cholesterol metabolism. Arterioscler Thromb Vasc Biol 28: 1298–1303. [DOI] [PubMed] [Google Scholar]
- 32. Park CH, Valore EV, Waring AJ, Ganz T (2001) Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem 276: 7806–7810. [DOI] [PubMed] [Google Scholar]
- 33. Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, et al. (2001) A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem 276: 7811–7819. [DOI] [PubMed] [Google Scholar]
- 34. Denecke B, Gräber S, Schäfer C, Heiss A, Wöltje M, et al. (2003) Tissue distribution and activity testing suggest a similar but not identical function of fetuin-B and fetuin-A. Biochem J 376: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Salier JP, Chan P, Raguenez G, Zwingman T, Erickson RP (1993) Developmentally regulated transcription of the four liver-specific genes for inter-alpha-inhibitor family in mouse. Biochem J 296: 85–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tolosano E, Altruda F (2002) Hemopexin: structure, function, and regulation. DNA Cell Biol 21: 297–306. [DOI] [PubMed] [Google Scholar]
- 37. Wang AY, Woo J, Lam CW, Wang M, Chan IH, et al. (2005) Associations of serum fetuin-A with malnutrition, inflammation, atherosclerosis and valvular calcification syndrome and outcome in peritoneal dialysis patients. Nephrol Dial Transplant 20: 1676–1685. [DOI] [PubMed] [Google Scholar]
- 38. Srinivas PR, Wagner AS, Reddy LV, Deutsch DD, Leon MA, et al. (1993) Serum alpha 2-HS-glycoprotein is an inhibitor of the human insulin receptor at the tyrosine kinase level. Mol Endocrinol 7: 1445–1455. [DOI] [PubMed] [Google Scholar]
- 39. Rauth G, Pöschke O, Fink E, Eulitz M, Tippmer S, et al. (1992) The nucleotide and partial amino acid sequences of rat fetuin. Identity with the natural tyrosine kinase inhibitor of the rat insulin receptor. Eur J Biochem 204: 523–529. [DOI] [PubMed] [Google Scholar]
- 40. Stefan N, Hennige AM, Staiger H, Machann J, Schick F, et al. (2006) Alpha2-Heremans-Schmid glycoprotein/fetuin-A is associated with insulin resistance and fat accumulation in the liver in humans. Diabetes Care 29: 853–857. [DOI] [PubMed] [Google Scholar]
- 41. Johnson WJ, Kilsdonk EP, van Tol A, Phillips MC, Rothblat GH (1991) Cholesterol efflux from cells to immunopurified subfractions of human high density lipoprotein: LP-AI and LP-AI/AII. J Lipid Res 32: 1993–2000. [PubMed] [Google Scholar]
- 42. Assié G, Guillaud-Bataille M, Ragazzon B, Bertagna X, Bertherat J, et al. (2010) The pathophysiology, diagnosis and prognosis of adrenocortical tumors revisited by transcriptome analyses. Trends Endocrinol Metab 21: 325–334. [DOI] [PubMed] [Google Scholar]
- 43. Chang J, Nicolas E, Marks D, Sander C, Lerro A, et al. (2004) miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol 1: 17–24. [DOI] [PubMed] [Google Scholar]
- 44. Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, et al. (2004) Control of pancreas and liver gene expression by HNF transcription factors. Science 303: 1378–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Waxman DJ, Holloway MG (2009) Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol 76: 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, et al. (1994) Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes Dev 8: 2466–2477. [DOI] [PubMed] [Google Scholar]
- 47. Sirotkin H, Morrow B, Saint-Jore B, Puech A, Das Gupta R, et al. (1997) Identification, characterization, and precise mapping of a human gene encoding a novel membrane-spanning protein from the 22q11 region deleted in velo-cardio-facial syndrome. Genomics 42: 245–251. [DOI] [PubMed] [Google Scholar]
- 48. Morita K, Furuse M, Fujimoto K, Tsukita S (1999) Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA 96: 511–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Niimi T, Nagashima K, Ward JM, Minoo P, Zimonjic DB, et al. (2001) claudin-18, a novel downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor, encodes lung- and stomach-specific isoforms through alternative splicing. Mol Cell Biol 21: 7380–7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harayama T, Shindou H, Shimizu T (2009) Biosynthesis of phosphatidylcholine by human lysophosphatidylcholine acyltransferase 1. J Lipid Res 50: 1824–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nakanishi H, Shindou H, Hishikawa D, Harayama T, Ogasawara R, et al. (2006) Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J Biol Chem 281: 20140–20147. [DOI] [PubMed] [Google Scholar]
- 52. Ho SB, Niehans GA, Lyftogt C, Yan PS, Cherwitz DL, et al. (1993) Heterogeneity of mucin gene expression in normal and neoplastic tissues. Cancer Res 53: 641–651. [PubMed] [Google Scholar]
- 53. Braga VM, Pemberton LF, Duhig T, Gendler SJ (1992) Spatial and temporal expression of an epithelial mucin, Muc-1, during mouse development. Development 115: 427–437. [DOI] [PubMed] [Google Scholar]
- 54. Peri A, Cordella-Miele E, Miele L, Mukherjee AB (1993) Tissue-specific expression of the gene coding for human Clara cell 10-kD protein, a phospholipase A2-inhibitory protein. J Clin Invest 92: 2099–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, et al. (1997) Smad6 inhibits signalling by the TGF-beta superfamily. Nature 389: 622–626. [DOI] [PubMed] [Google Scholar]
- 56. Bridges JP, Ikegami M, Brilli LL, Chen X, Mason RJ, et al. (2010) LPCAT1 regulates surfactant phospholipid synthesis and is required for transitioning to air breathing in mice. J Clin Invest 120: 1736–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Merikallio H, Pääkkö P, Harju T, Soini Y (2011) Claudins 10 and 18 are predominantly expressed in lung adenocarcinomas and in tumors of nonsmokers. Int J Clin Exp Pathol 4: 667–673. [PMC free article] [PubMed] [Google Scholar]
- 58. Tachibana T, Hagiwara K, Johkoh T (2009) Pulmonary alveolar microlithiasis: review and management. Curr Opin Pulm Med 15: 486–490. [DOI] [PubMed] [Google Scholar]
- 59. Chan KK, Tsui SK, Lee SM, Luk SC, Liew CC, et al. (1998) Molecular cloning and characterization of FHL2, a novel LIM domain protein preferentially expressed in human heart. Gene 210: 345–350. [DOI] [PubMed] [Google Scholar]
- 60. Chu PH, Ruiz-Lozano P, Zhou Q, Cai C, Chen J (2000) Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech Dev 95: 259–265. [DOI] [PubMed] [Google Scholar]
- 61. Vos MJ, Kanon B, Kampinga HH (2009) HSPB7 is a SC35 speckle resident small heat shock protein. Biochim Biophys Acta 1793: 1343–1353. [DOI] [PubMed] [Google Scholar]
- 62. Quraishe S, Asuni A, Boelens WC, O’Connor V, Wyttenbach A (2008) Expression of the small heat shock protein family in the mouse CNS: differential anatomical and biochemical compartmentalization. Neuroscience 153: 483–491. [DOI] [PubMed] [Google Scholar]
- 63. Frey N, Richardson JA, Olson EN (2000) Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc Natl Acad Sci USA 97: 14632–14637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Geier C, Perrot A, Ozcelik C, Binner P, Counsell D, et al. (2003) Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation 107: 1390–1395. [DOI] [PubMed] [Google Scholar]
- 65. Priori SG, Napolitano C, Tiso N, Memmi M, Vignati G, et al. (2001) Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation 103: 196–200. [DOI] [PubMed] [Google Scholar]
- 66. Tiso N, Stephan DA, Nava A, Bagattin A, Devaney JM, et al. (2001) Identification of mutations in the cardiac ryanodine receptor gene in families affected with arrhythmogenic right ventricular cardiomyopathy type 2 (ARVD2). Hum Mol Genet 10: 189–194. [DOI] [PubMed] [Google Scholar]
- 67. Machida T, Fujita T, Ooo ML, Ohira M, Isogai E, et al. (2006) Increased expression of proapoptotic BMCC1, a novel gene with the BNIP2 and Cdc42GAP homology (BCH) domain, is associated with favorable prognosis in human neuroblastomas. Oncogene 25: 1931–1942. [DOI] [PubMed] [Google Scholar]
- 68. Soh UJ, Low BC (2008) BNIP2 extra long inhibits RhoA and cellular transformation by Lbc RhoGEF via its BCH domain. J Cell Sci 121: 1739–1749. [DOI] [PubMed] [Google Scholar]
- 69. Price ND, Trent J, El-Naggar AK, Cogdell D, Taylor E, et al. (2007) Highly accurate two-gene classifier for differentiating gastrointestinal stromal tumors and leiomyosarcomas. Proc Natl Acad Sci USA 104: 3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Clarke RA, Zhao Z, Guo AY, Roper K, Teng L, et al. (2009) New genomic structure for prostate cancer specific gene PCA3 within BMCC1: implications for prostate cancer detection and progression. PloS ONE 4: e4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Iwama E, Tsuchimoto D, Iyama T, Sakumi K, Nakagawara A, et al. (2011) Cancer-related PRUNE2 protein is associated with nucleotides and is highly expressed in mature nerve tissues. J Mol Neurosci 44: 103–114. [DOI] [PubMed] [Google Scholar]
- 72. Li S, Hayakawa-Yano Y, Itoh M, Ueda M, Ohta K, et al. (2012) Olfaxin as a novel Prune2 isoform predominantly expressed in olfactory system. Brain Res 1488: 1–13. [DOI] [PubMed] [Google Scholar]
- 73. Arsenian S, Weinhold B, Oelgeschläger M, Rüther U, Nordheim A (1998) Serum response factor is essential for mesoderm formation during mouse embryogenesis. Embo J 17: 6289–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Salmikangas P, Mykkänen OM, Grönholm M, Heiska L, Kere J, et al. (1999) Myotilin, a novel sarcomeric protein with two Ig-like domains, is encoded by a candidate gene for limb-girdle muscular dystrophy. Hum Mol Genet 8: 1329–1336. [DOI] [PubMed] [Google Scholar]
- 75. Mologni L, Moza M, Lalowski MM, Carpén O (2005) Characterization of mouse myotilin and its promoter. Biochem Biophys Res Commun 329: 1001–1009. [DOI] [PubMed] [Google Scholar]
- 76. Hauser MA, Horrigan SK, Salmikangas P, Torian UM, Viles KD, et al. (2000) Myotilin is mutated in limb girdle muscular dystrophy 1A. Hum Mol Genet 9: 2141–2147. [DOI] [PubMed] [Google Scholar]
- 77. Selcen D, Engel AG (2004) Mutations in myotilin cause myofibrillar myopathy. Neurology 62: 1363–1371. [DOI] [PubMed] [Google Scholar]
- 78. Foroud T, Pankratz N, Batchman AP, Pauciulo MW, Vidal R, et al. (2005) A mutation in myotilin causes spheroid body myopathy. Neurology 65: 1936–1940. [DOI] [PubMed] [Google Scholar]
- 79. Pénisson-Besnier I, Talvinen K, Dumez C, Vihola A, Dubas F, et al. (2006) Myotilinopathyin a family with late onset myopathy. Neuromuscul Disord 16: 427–431. [DOI] [PubMed] [Google Scholar]
- 80. Yang RZ, Huang Q, Xu A, McLenithan JC, Eisen JA, et al. (2003) Comparative studies of resistin expression and phylogenomics in human and mouse. Biochem Biophys Res Commun 310: 927–935. [DOI] [PubMed] [Google Scholar]
- 81. Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, et al. (2001) The hormone resistin links obesity to diabetes. Nature 409: 307–312. [DOI] [PubMed] [Google Scholar]
- 82. Kim KH, Lee K, Moon YS, Sul HS (2001) A cysteine-rich adipose tissue-specific secretory factor inhibits adipocyte differentiation. J Biol Chem 276: 11252–11256. [DOI] [PubMed] [Google Scholar]
- 83. Hu E, Liang P, Spiegelman BM (1996) AdipoQ is a novel adipose-specific gene dysregulated in obesity. J Biol Chem 271: 10697–10703. [DOI] [PubMed] [Google Scholar]
- 84. Bi S, Gavrilova O, Gong DW, Mason MM, Reitman M (1997) Identification of a placental enhancer for the human leptin gene. J Biol Chem 272: 30583–30588. [DOI] [PubMed] [Google Scholar]
- 85. Zhao J, Kunz TH, Tumba N, Schulz LC, Li C, et al. (2003) Comparative analysis of expression and secretion of placental leptin in mammals. Am J Physiol Regul Integr Comp Physiol 285: R438–R446. [DOI] [PubMed] [Google Scholar]
- 86. Vidal-Puig AJ, Considine RV, Jimenez-Liñan M, Werman A, Pories WJ, et al. (1997) Peroxisome proliferator-activated receptor gene expression in human tissues. Effects of obesity, weight loss, and regulation by insulin and glucocorticoids. J Clin Invest 99: 2416–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shin J, Li B, Davis ME, Suh Y, Lee K (2009) Comparative analysis of fatty acid-binding protein 4 promoters: conservation of peroxisome proliferator-activated receptor binding sites. J Anim Sci 87: 3923–3934. [DOI] [PubMed] [Google Scholar]
- 88. Magnusson B, Gummesson A, Glad CA, Goedecke JH, Jernås M, et al. (2008) Cell death–inducing DFF45-like effector C is reduced by caloric restriction and regulates adipocyte lipid metabolism. Met Clin Exp 57: 1307–1313. [DOI] [PubMed] [Google Scholar]
- 89. Okada T, Nishizawa H, Kurata A, Tamba S, Sonoda M, et al. (2008) URB is abundantly expressed in adipose tissue and dysregulated in obesity. Biochem Biophys Res Commun 367: 370–376. [DOI] [PubMed] [Google Scholar]
- 90. Cases S, Stone SJ, Zhou P, Yen E, Tow B, et al. (2001) Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem 276: 38870–38876. [DOI] [PubMed] [Google Scholar]
- 91. Puri V, Konda S, Ranjit S, Aouadi M, Chawla A, et al. (2007) Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J Biol Chem 282: 34213–34218. [DOI] [PubMed] [Google Scholar]
- 92. Keller P, Petrie JT, De Rose P, Gerin I, Wright WS, et al. (2008) Fat-specific protein 27 regulates storage of triacylglycerol. J Biol Chem 283: 14355–14365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nishino N, Tamori Y, Tateya S, Kawaguchi T, Shibakusa T, et al. (2008) FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets. J Clin Invest 118: 2808–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liang L, Zhao M, Xu Z, Yokoyama KK, Li T (2003) Molecular cloning and characterization of CIDE-3, a novel member of the cell-death-inducing DNA-fragmentation-factor (DFF45)-like effector family. Biochem J 370: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tremblay F, Revett T, Huard C, Zhang Y, Tobin JF, et al. (2009) Bidirectional modulation of adipogenesis by the secreted protein Ccdc80/DRO1/URB. J Biol Chem 284: 8136–8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chen HC, Smith SJ, Tow B, Elias PM, Farese RV Jr (2002) Leptin modulates the effects of acyl CoA:diacylglycerol acyltransferase deficiency on murine fur and sebaceous glands. J Clin Invest 109: 175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tissue-specific gene expression values based on GEO DataSet(GDS)596 for the human and GDS3142 for the mouse. The microarray gene expression profiles for the six tissues (kidney, liver, lung, heart, muscle, and adipose tissue) of the human and mouse were collected from GEO DataSet(GDS) in the National Center for Biotechnology Information (NCBI) web page. *Average gene expression values for each tissue including a target tissue were obtained. **Those average values for each tissue were divided by an average value of a target tissue and then averaged for obtaining one representative value. If the representative value is lower, then tissue-specificity for a target tissue is higher because it was divided by an average value of a target tissue. ***An alternative approach shows that an average value of a target tissue was divided by average of averages for each tissue. If the result value is higher, then tissue-specificity for a target tissue is higher because an average value of a target tissue was divided. Figure 1 shows the schematic diagram for the above procedures.
(XLSX)