Abstract
The past 10 years have witnessed a dramatic proliferation in the availability of protein interaction data. However, for interaction mapping based on affinity purification coupled with mass spectrometry (AP-MS), there is a wealth of information present in the datasets that often goes unrecorded in public repositories, and as such remains largely unexplored. Further, how this type of data is represented and used by bioinformaticians has not been well established. Here, we point out some common mistakes in how AP-MS data are handled, and describe how protein complex organization and interaction dynamics can be inferred using quantitative AP-MS approaches.
Keywords: Interaction networks, affinity purification coupled to mass spectrometry, protein-protein interactions, quantitative proteomics, regulated interactions
The hairball: representation of protein-protein interactions
The availability of cDNA and Open Reading Frame (ORF) collections [1–7] and yeast strains engineered to express epitope-tagged proteins [8] first allowed us to begin to characterize at a global level how proteins associate with one another. In 1989, Field and Song published the first yeast two hybrid (Y2H) manuscript [9], introducing an approach which has now been employed to generate large-scale interaction maps in multiple organisms, including yeast [10–13], worms [14, 15], flies [16–18], humans [18–21] and plants [22, 23]. Y2H maps ushered in a new era in the field of protein-protein interactions, and changed the type of question that we can pose: instead of asking “Does protein A interact with protein B?”, or even “What does protein A interact with?”, it has become “How is the cell wired?”.
Y2H primarily detects direct protein-protein interactions (here referred to as binary interactions), and a simple representation of such an interaction between two proteins consists of drawing two circles (or nodes) linked by a line (or edge; Fig. 1A). Each detected interaction can be displayed in the same fashion, and combined to generate a map of the protein-protein interaction network (or interactome; Fig. 1B). These types of representations – and their analysis by computational biologists – are extremely useful, allowing for the study of the organization of any given system, and such “hairballs” also allow for hypothesis generation regarding the biological function of the proteins under analysis. While Y2H is probably the most cost-efficient binary approach for proteome-wide surveys, other techniques optimized for the detection of direct interactions also exist (for review, see [24, 25]). Data from these methods can be depicted and analyzed using the same type of graphical representation.
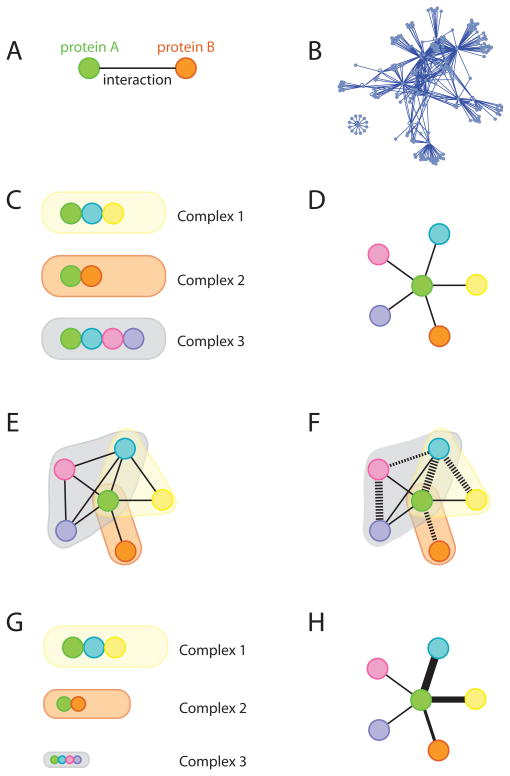
Figure 1. Graphical representations of protein interactions.
A) Graphical representation of a direct protein-protein interaction. The two circles (referred to as “nodes”) represent each of the proteins engaged in an interaction, and the line linking them (the “edge”) represents the interaction. B) Interaction network (or “hairball”) representing ~ 500 interactions amongst ~100 proteins (generated by Cytoscape [103]). C) Protein complexes in a cell. Here, the green protein is found in three different biochemically defined complexes (direct interactions are depicted by contact between the nodes). Not shown here is the relative abundance of these three complexes. D) Unweighted graphical representation (spoke expansion) of the interactions established by the green protein after affinity-purification coupled to mass spectrometry. The organization in different complexes is lost (from this single AP-MS analysis) and direct and indirect interactions are represented in the same manner, as they are indistinguishable in the mass spectrometer. E) Iterative AP-MS helps to resolve complex organization surrounding a central bait. After identification of each of the interaction partners for the green protein, these can be in turned cloned, and analyzed by mass spectrometry. This recapitulates the complex organization shown in (C), though it does not indicates direct or direct interactors. F) Adding binary data to AP-MS data is beneficial to reconstitute the assembly of individual complexes. The dashed lines represent demonstrated (thickest lines) or predicted (thinner lines) direct interactions (the likelihood of a direct interaction is proportional to edge thickness). G) Complexes are not always present in the cell in the same abundances; here, complex 1 is more abundant than complex 2, itself more abundant than complex 3. Most of the green protein will reside in complex 1. H) Quantitative mass spectrometry data provides the relative abundance of each of the interactors for the green protein. This information is shown here as the thickness of the edges.
Parallel to the development of Y2H and other types of binary approaches, dramatic improvements in instrumentation have enabled the efficient coupling of affinity purification to mass spectrometry (AP-MS) for the identification of protein-protein interactions. Proteome-wide surveys of the interactome are still largely limited to S. cerevisiae [26–29], though a growing number of medium-scale AP-MS studies in mammals, insects, plants and various pathogens [30–46] indicate that reconstitution of near “complete” AP-MS interactome maps is not only possible, but likely, in the near future. Importantly, however, the interactions detected by AP-MS differ from those obtained via Y2H, in that they represent a mix of direct and indirect binding relationships. For proteins that take part in multiple alternative complexes (a very common occurrence), the interactors identified in such an analysis thus represent a mixture of multiple protein machines (Fig. 1C; 1D). While techniques such as high-density iterative mapping of protein complexes, the use of quantitative mass spectrometry tools, or binary approaches such as Y2H, can be used to decipher this information (see below), how these types of interactions are depicted and analyzed remain as important challenges to be solved, as it is a priori not always possible to distinguish direct versus indirect interactions in MS data.
In most cases, the same type of network representation used for Y2H (i.e. nodes linked by edges) has been utilized to depict interactions discovered using AP-MS approaches. However, the meaning of edges in AP-MS data is not always clear, as both direct and indirect interactions are similarly represented. Computationally, there has also been much confusion regarding whether to simply draw edges between a bait and all of the interactors detected in the mass spectrometer (referred to as a “spoke” expansion), or to assume that all identified components of an affinity purification are part of a single complex, and draw edges between all prey proteins associated with a given bait (a “matrix expansion”). The matrix expansion model is particularly problematic, in that it completely ignores the partitioning of a bait into mutually-exclusive protein complexes (which may have completely different biological roles; Fig. 1C, E), and improperly implies a series of relationships that may never exist in a cell. Fortunately, this type of expansion method is used less and less.
There is also significant confusion in protein interaction databases regarding how to record, annotate and display AP-MS data. For example, IntAct [47] (currently the largest primary repository of mammalian AP-MS experiments, to our knowledge) records AP-MS data by indicating a single “interaction number”, which encompasses the bait and its interactors as reported by the authors of individual studies. To display this data in a consistent manner, a spoke expansion method is used to record bait-prey relationships, and to display them for a single query (see Fig. 1D). The IntAct site however warns that “most interactions generated by spoke and matrix expansion result in false positives”, and offers a convenient option to “filter” them. This – sadly – only leads to more confusion. For example, in our own dataset on the interactions established by the Ser/Thr phosphatase PPP4C, we deposited both AP-MS data (which is filtered out by the spoke expansion filter), and a confirmation of these interactions by immunoprecipitation followed by immunoblotting (IP/Western) on the same samples [48, 49]. Surprisingly, the IP/Western data survive the filtering process, and are considered to be “binary” data. This is highly problematic because biochemically, the IP/Western data are just as likely as the AP-MS data to be mediated by bridging proteins, yet because the detection method is actually more biased (in that here we only queried for the presence of a single prey with a specific antibody), the interactions are treated differently. This is clearly not the best way to think about interaction data. Importantly, this problem is not limited to IntAct, which actually provides very careful curation of experimental data, enabling us to track down such issues (and we have worked with IntAct to properly annotate our own experiments). Adding to the confusion, some AP-MS data have been deposited by the authors as a set of “binary” interactions (i.e. they were pre-expanded using the spoke model [43]), and are therefore not filtered out by the spoke and matrix expansion filters in IntAct. Other repositories use different rules for annotation and display of interaction data [50]; e.g. BioGRID annotates all interactions in a binary spoke-expanded [51] manner, while HPRD sometimes just reports “complexes” [52]. Given that database aggregators and computational biologists often download entire datasets from public repositories without being aware of the underlying nature of the data, this confusion can lead to spurious conclusions regarding protein-protein interactions.
It is important to note that several of the commonly employed “binary” approaches can also detect both direct and indirect interactions, yet because the detection method is “single channel” (that is, we blind ourselves to everything but the protein for which we have a reagent for detection), the methods are optimistically thought to be “binary”. For example, any experiment in which proteins are expressed in their host of origin (or a closely related species) is susceptible to recovering both direct and indirect interactions, but this fact tends to be ignored.
Simply put, spoke expansion of AP-MS data does not generate false-positives, if the data are handled correctly. If the mass spectrometry and data analysis have been conducted properly, these types of protein identifications are actually of very high quality: what they do not tell you is that an interaction is direct. A better understanding by computational biologists and experimentalists alike of what the edges in AP-MS actually represent is thus critical moving forward. Rather than debating whether an indirect interaction is a false positive, we suggest that it would be more useful to clearly highlight those interactions that have been demonstrated to be direct (using one or more methods outlined below), and to make this data more easily available for interactome analysis. Alternatively, calculating the probability of a direct interaction (based, for example, on future benchmarking of “binary” methods such as Y2H) and overlaying this information on AP-MS data would allow for a much better understanding of the molecular organization of protein complexes. Visualization of the AP-MS interactions amongst all nodes of a network superimposed onto proven direct binding interactions (Fig. 1F and see below) would provide much higher information content to interactome maps.
In summary, while true binary approaches are easily represented by a node-edge-node relationship (and annotated as such in interaction databases), how data generated by AP-MS are recorded, visualized and distributed to the research community remains somewhat problematic. As MS instrumentation increases in speed and sensitivity, the use of AP-MS is also increasing apace. A concerted effort by biologists, curators and bioinformatics experts will be required to address this important issue.
The use of quantitative data in interaction mapping
Most graphic representations of Y2H binary data tend to be unweighted; i.e. all edges possess the same value. If value is added to these types of edges, it is most often based on confidence in the detection of the interaction (e.g. signal strength in a screen). These scores can be very useful (see in particular a confidence score developed by Braun et al. based on reproducibility of the detection of an interaction across several orthogonal binary assays [25, 53]), but they do not directly translate to a likelihood of interaction in a physiological context. Another important issue in our field is that, similar to most Y2H maps, AP-MS network edges are often also represented as being of equal weight, with little consideration for the confidence in each putative interaction or the relative abundance of the interaction partners. We and others have developed new methods to use quantitative information embedded in mass spectrometry data to assist in the identification of true positives in interaction maps [30, 33, 38, 54–58], and such information can very effectively be used to calculate absolute or relative differences in the abundance of proteins across multiple samples, and to better understand protein complex topology (Fig. 1G, H). Excellent reviews on quantitative mass spectrometry applied to protein complexes have been published recently [59–61]; here we will refer only to quantification as it applies to topology and stoichiometry, with a short discussion of interaction dynamics.
Absolute quantification of proteins in a given sample can be determined using isotopically-labeled “heavy” peptide or protein standards. Such peptides are commercially available [62], and can be spiked into any sample of interest prior to MS analysis (Fig. 2A). Since the mass spectrometer measures mass/charge (m/z) ratios, these standards are easily distinguished from the “light” endogenous counterparts in the sample (Fig. 2B). Alternatively, recombinant proteins can be expressed and isotopically labeled (e.g. with 15N or heavy amino acids [63]) in-house, then spiked into a sample prior to proteolysis. A third variation of this approach involves a recombinant, isotopically labeled concatenated polypeptide sequence derived from multiple proteins of interest (qConCat [64, 65]). Ideally, several standard peptides derived from each protein of interest should be used for quantification (to prevent hidden biases that can arise from, e.g. post-translationally modified peptides in one condition and not another). While most researchers would agree that using isotopically-labeled standards is ideal for accurate quantification, this may not be practical for large-scale AP-MS studies, both due to the cost of large numbers of standards, and various technical difficulties, especially in determining the quantity of each standard to be added to each sample to cover a broad dynamic range of protein concentrations across multiple experiments. For example, when a given protein is used as a bait, its abundance in the AP may be several hundred-fold higher than when it is isolated as an interactor with another bait. The proper concentration of each standard peptide must be tuned in each case to ensure that it is present at amounts within the linear range of the mass analyzer.
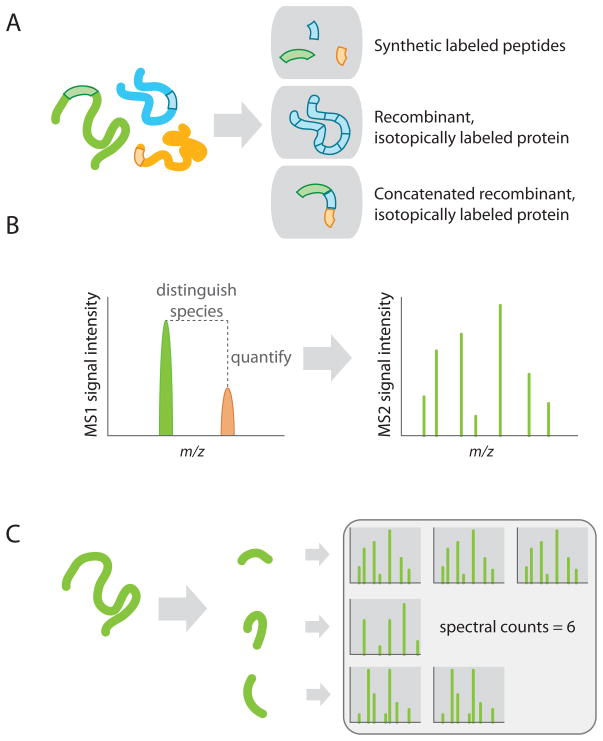
Figure 2. Strategies for quantification of AP-MS data.
A) Absolute quantification with isotopes; alternative sources of isotopically labeled peptides are indicated. In all cases, the absolute concentration of the standards must be determined prior to use in mass spectrometry. B) General principle behind the use of isotopic labels in quantitative proteomics. In the precursor (MS1), the mass to charge ratios (m/z) of all co-eluting peptides are monitored, and their intensity recorded. Since isotopically labeled peptides have different m/z, they are distinguished from each other in the MS1 scan: Relative differences in abundance are proportional to their intensities. Identification (here of the light, green, species) is performed in the MS/MS (or MS2) spectrum. C) Quantification based on spectral counting. Different unique peptides from the same protein may be sequenced; spectral counts refers to the sum of all spectra mapped to a given protein.
A number of alternative approaches have been developed to assess protein abundance. One simple, yet surprisingly effective strategy, is to monitor spectral counts (simply the number of mass spectra assigned to each protein; Fig. 2C) to model the abundance of interactors across parallel purifications [38, 66–68]. Spectral counts are most often normalized to protein length (since larger proteins yield more peptides, they tend to generate more spectra at the same molarity), and sometimes to the expression levels of the bait itself. Spectral counts can be used for filtering out noise in AP-MS experiments, but also to compare the recovery of the same prey across samples [67]. Importantly, spectral counts are more reliable for proteins in the medium to high abundance range in a sample, but are not as useful for low abundance polypeptides. More accurate quantification that does not require isotopes can be performed by analyzing the intensity of the signal in the precursor scan of the mass spectrometer (here referred to as the MS1 scan), or the intensity of the product ions after fragmentation (MS/MS or MS2 scan). Similar to spectral counts, MS1 quantification has been used to identify true positives in AP-MS data, and in some cases to compare the samples quantitatively [58, 69, 70]. However, since different peptides ionize differently in the mass spectrometer (i.e. ion intensities for different peptides at equimolar concentrations can vary widely), these methods can only provide an estimation of abundance (although these issues decrease as the counts or intensities of more peptides from the same protein are averaged; see e.g. [71]). To circumvent this problem, while keeping overall costs of the experiment more manageable, Wepf and colleagues devised an approach in which the recombinantly expressed “bait” protein is fused to an epitope tag that can be used both for isolation and quantification [72]. A single heavy isotopic standard corresponding to a peptide in the epitope tag is spiked into samples to establish a quantitative reference point for the bait in each experiment. Computational analysis can then be used to quantify each protein previously used as a bait across multiple experiments. This approach is more useful when looking at interconnected networks, such that each prey in the dataset is also analyzed as a bait. An extension of this type of approach could consist of spiking a general mixture of heavy peptides into each AP, where some correspond to the epitope tag, some to common contaminants, and others correspond to various components of the network under study, and using these as beacons for quantification of the entire interaction network. While this has not (to our knowledge) been used for interaction proteomics, similar strategies have been applied in the field of biomarker detection [73].
Unfortunately, at present much of this type of data in proteomics experiments is essentially ignored. For example, abundance measures are stripped out of interaction data recorded in the major interaction databases, and in most cases, confidence values are also not tracked. As such, major and minor interactors are given equal weight in such datasets. This is problematic because it enhances the disconnect between small scale and large scale studies, and prevents access to new types of information for modeling by computational biologists. This being said, since abundance levels may vary depending on the experimental set-up, it will be challenging to harmonize quantitative data deposited from different sources.
From interactor lists to complexes
A single AP-MS analysis reveals little regarding the supramolecular architecture of individual protein complexes, but this technique can be harnessed in multiple ways to reveal how protein machines are assembled. For example, the composition of a given complex, and multiple mutually-exclusive assemblies, can often be deduced by performing iterative “high density” AP-MS [74], in which each of the preys from one round of analysis become baits in the next round (Fig. 1E). This is clearly somewhat labor-intensive, but the use of incomplete data (e.g. when not all proteins in a complex are analyzed as baits, or if any of the preys fall below the detection limit) can result in the over-fitting of complex composition and a loss of biologically important information. For example, when we characterized the STRIPAK (STRiatin Interacting Phosphatase And Kinase) complex, 10 different protein families were identified as bona fide components. Only after performing AP-MS on each of the components were we able to define two independent molecular entities in the pulldowns: one complex associated with the cortactin binding protein 2 (CTTNBP2), and a second complex containing the proteins SLMAP and SIKE [34].
An alternative to reciprocal AP-MS (which to date has been used only in smaller scale studies) is to combine the standard AP step with an orthogonal approach to separate multiple bait-containing complexes; this may be accomplished e.g. via gel filtration chromatography or other standard chromatographic steps followed by AP-MS [75]. Despite obvious advantages, this approach has not generally been applied to large-scale AP-MS analysis, most likely due to the additional analytical steps required (e.g. tracking down the fractions in which the bait partitions) and increased analysis time. Approaches such as Blue Native gels have been combined effectively with AP-MS for the analysis of membrane-associated protein complexes [76, 77], and it is likely that such studies will be expanded in the near future. In recent years, parallel (and still largely unpublished) efforts from several groups have attempted to forego the AP step completely, and to systematically analyze protein complexes by chromatographic fractionation coupled to mass spectrometry (L Foster, pers. comm.). Although the dynamic range and limitations of this approach are not entirely clear at present, it could represent a very useful companion to AP-MS analysis to enable the detection of mutually exclusive complexes containing a given protein. Furthermore, as discussed below, this type of approach could be very useful in mapping global changes in interactomes imparted by a stimulus, drug or other perturbation.
An obvious limitation to the use of AP-MS to identify and characterize protein complexes is that the complex must be soluble in the buffer used for affinity purification and the interactions must withstand the affinity purification step. Simply put, if a bait protein and its interacting partners are not extracted efficiently during lysis, they will not be observed by the mass spectrometer. For example, proteins associated with chromatin are often found in the pellet after centrifugation of the crude lysate, unless steps to shear the DNA (such as sonication or treatment with nucleases) are included in the lysis protocol [78–80]. Similarly, membrane proteins are typically poorly recovered in standard extraction buffers, though employing different detergents for their extraction has recently enabled the recovery of multiple complexes associated with different membranes [81–85]. Systematic studies in S. cerevisiae to define the chromatin-associated interactome [79] and the interactome of all membrane-localized proteins (J. Greenblatt, pers. comm.) indicate that these types of approaches will lead to a greatly expanded view of the interactomes for proteins previously thought to be inaccessible to AP-MS analysis. To better understand interactions that do not withstand the affinity purification step (often referred to as “transient” interactions, but more accurately defined as interactions that have a fast “OFF” rate in solution) a variety of different strategies will most likely be required. That these types of interactors do in fact exist has been defined by quantitative proteomics with SILAC, in which combining samples at different times (prior to lysis, after lysis, or after affinity purification) revealed interactions that are stable in solution, and interactors that exchange rapidly [86–88]. The simplest approach to capture rapidly-dissociating interactors is to decrease the chances for the interactions to be lost in the first place. For example, in a dual purification protocol such as Tandem Affinity Purification (TAP), a protein with a fast off rate has the chance to dissociate from its interactors in each of the two purification steps (and during the proteolysis and washes steps). Using a single step purification method, accompanied by shorter incubation times and limited washes, can help to maintain interactors that would otherwise be lost [48, 89]. While these types of samples are likely to contain a larger numbers of contaminants, the use of improved software for statistical analysis of putative interactors (e.g. SAINT and similar tools [30, 33, 38, 54–58]) allows for efficient discrimination between contaminants (e.g. proteins that bind to the solid phase support or antibody) and bona fide interactors.
While more sensitive MS instruments, an increase in the speed of bait isolation, fewer wash steps, and smarter software have dramatically improved our ability to identify interacting partners, this pipeline will probably not be sufficient to maintain all interactions; alternative strategies, most often making use of crosslinking reagents that can be applied directly to cells prior to lysis, can also be exploited (see, e.g. [35]). It must be stated that each of the approaches described above has advantages and caveats, but – performed under well-controlled conditions – have the potential to greatly expand the detection of protein-protein interactions by AP-MS.
Mapping topologies
All of the approaches highlighted above are aimed at defining protein complexes in the biochemical sense: i.e. providing a “parts list” of complex composition. Understanding how these parts are assembled into a functional unit is also clearly important. In a best-case scenario, information from binary approaches (e.g. Y2H) may already be available, and used to model protein complex topology. Crosslinking followed by mass spectrometric identification of the crosslinked residues in protein partners is also increasingly used (for reviews, see [90–94]). Since this approach also identifies likely direct interactions, crosslinking data could easily be integrated within the networks generated by AP-MS to identify some of the topological elements (Fig. 1F).
It is also possible to use “binary” approaches to systematically test for direct interactions between proteins detected by AP-MS. To determine the viability of such an approach, we have tested several different methods. Using Y2H, we performed a pilot re-scoring of ~ 1000 high-confidence AP-MS interactions (P Braun, pers. comm.). This assay yielded a fairly low (<10%) validation rate, likely due to a combination of false negatives in Y2H (where assay sensitivity is ~25% [53]), indirect interactions identified by AP-MS, and perhaps false positives in AP-MS. Combined with the tedious cherry-picking required for assembling the large number of individual protein pairs for such an analysis, this method may not be the most efficient way to identify direct interactions in an AP-MS dataset, especially since genome-wide screens by Y2H are underway and should in theory test all possible pairs. In another study, we used LUMIER [95] to test ~50 baits against a total of 600 interacting proteins, in an attempt to identify direct interactions in a single high-confidence interaction network (M Taipale, pers. comm.). LUMIER monitors the recovery of a luciferase-tagged bait protein with a FLAG-tagged prey, following immunoprecipitation. LUMIER validation was more successful than Y2H, although the percentage of interactions that are truly direct in the LUMIER assay is unclear (in this method, two proteins are co-expressed in a human cell line, and could therefore be bridged by one or more additional endogenous proteins). Finally, in a much smaller test case, we successfully identified direct protein-protein interactions by programming reticulocyte lysates to express nuclear proteins, which are normally not expressed in red blood cells. Here, we demonstrated that the catalytic subunit of PP4 interacts directly with PP4R2, and that this dimer was necessary for the recruitment of a third member of the complex, PP4R3 [49]. In this case, all interactions were also recapitulated by Y2H [49].
It may also be possible to retest AP-MS interactions to look for direct interactors by employing assays with a strong bias for close proximity, using methods such as protein fragment complementation (PCA [96, 97]); the use of fluorescent proteins for PCA has the added advantage of providing information regarding the subcellular location in which the interaction takes place.
Ideally, retesting could also be done using purified proteins from a phylogenetically distant host (e.g. a bacterial expression system for eukaryotic proteins); to date, this is widely considered to be the gold standard for the identification of direct protein-protein interactions. With the availability of cDNA and ORFeome collections, and the ongoing construction of protein collections [98–101], systematic retesting of proteins by expression in bacteria (or other hosts) may be scaled-up. While this type of testing can certainly be done using standard pull-down experiments and SDS-PAGE, protein array technologies [102, 103] could afford higher throughput. However, some difficulties remain with testing interactions using bacterially expressed recombinant proteins: e.g. many classes of proteins are not easily expressed (especially as full length polypeptides), and interactions which require, for example, a post-translational modification may be missed using this method. In summary, while it is not yet clear which of the approaches mentioned above (or others) may be the most efficient for providing information about direct interactions in AP-MS data to better understand the architecture of protein complexes, there are a number of possibilities that are becoming increasingly available. Furthermore, as high throughput mapping efforts using many different approaches continue, merging of datasets may eventually provide much of this information.
As an alternative to the use of external data sources, it is possible in some cases to map the organization of protein complexes using quantitative MS data as a proxy. For example, if a bait protein retrieves only a single high abundance interactor and many lower abundance interactors, it is unlikely that the high abundance interaction partner is bridged by another protein. In a similar way, if an interactor remains associated with the bait under conditions where most of the other interactors are displaced (e.g. by increasing the stringency of the washes), it is more likely to be a direct binding partner than an indirect interactor. An alternative is to progressively dissociate protein interactions in the mass spectrometer; this has been done for several large complexes, including the multisubunit translation initiation factor eIF3 [104] (for recent reviews of MS of intact complexes, see [92, 105]).
To better understand protein complex topology, it can be informative to place additional focus on putative scaffolds in a given dataset. For example, based on quantitative MS data we postulated that the striatin molecule could act to bridge the phosphatase (PP2A) and kinase (a family of Sterile 20 kinases known as GCKIII) components of the STRIPAK complex. To explore this hypothesis, we performed AP-MS on a series of epitope-tagged striatin truncation mutants [106]. This and subsequent studies indeed revealed that striatin is a scaffold, but that the kinase is likely recruited to the phosphatase via the CCM3 protein (mutated in Cerebral Cavernous Malformations) [107]. To confirm this model, we immunoprecipitated the kinase and analyzed by quantitative mass spectrometry the recovery of interaction partners, following the depletion of CCM3 and striatin by RNAi. A similar approach – using genetic deletion in S. cerevisiae – was employed by the Washburn group to define the network architecture of both the SAGA and ADA chromatin remodeling complexes [108], and the Rpd3 histone deacetylase complex [109]. Despite potential complicating issues (e.g. the expression level of a given protein may be influenced by the absence of interacting partners), this type of approach – especially in the context of modern quantification methods – offers great promise for the systematic analysis of complex topologies. In the case of S. cerevisiae, the approach consists of simply transforming a plasmid coding for the protein of interest into a relevant strain, or crossing strains in which endogenous proteins have been epitope tagged to strains in which a single complex component has been deleted (such crosses are now routinely used, and can even be conducted in a large-scale, automated fashion). In human cell systems, the limiting factor (at least in our hands) is the establishment of stable cell lines expressing tagged bait proteins: though still relatively expensive, transient knock down of suspected direct interactors is now robust, and enables the global analysis of protein complex organization.
We also note that – while not directly performed in the experiments described above – the inclusion of absolute peptide or protein standards within this type of framework may be extremely useful for elucidating the stoichiometry of components of a given complex. In this respect, the concatenated peptide strategy (qConCAT) mentioned above is particularly appealing, as each of the peptides in the qConCAT are present at identical molarities, thereby enabling determination of the molecular stoichiometry for multiple proteins in a complex. Using such approaches, we were able to determine that striatin is likely present as a trimer within STRIPAK (Kean et al., unpublished).
One interactome to many
While we have discussed some approaches to enable the integration of quantitative information into large-scale AP-MS interaction maps, we have not discussed what these maps actually mean. The majority of protein-protein interaction maps have been generated under a single physiological condition, and usually in only one organism or cell line, resulting in a steady-state (or static) interactome. For example, the bulk of the data currently available from medium or high-throughput human interactomes have been generated from derivatives of the HEK293 cell line (a smaller number of experiments have employed other immortalized cell lines). This concerted focus on a single cell line does have advantages in terms of benchmarking interactomes from different research groups, and in establishing a baseline for a draft map of a complete interactome in a human cell. However, there are clearly many interactions which may not be detected under these conditions, e.g. because certain proteins are not expressed in these cells (e.g. we have never detected CIP2A, a PP2A inhibitor upregulated in certain cancer cells, since it is not expressed in HEK293 cells [110]). We may also miss interesting protein-protein interactions that occur only after exposure to certain hormones, growth factors or stresses, only during apoptosis, only in highly confluent cells, or only during a given developmental stage. Standard AP-MS methods can also miss interactions that occur with; (i) membrane proteins, because buffer conditions that liberate proteins from membranes are often not compatible with maintaining protein-protein interactions in solution, and (ii) amongst chromatin-associated proteins that can be pelleted with the DNA during lysate preparation. As such, it is unclear what fraction of physiologically-relevant interactions will ultimately be identified by current efforts to systematically map protein-protein interactions in one cell type, and under one condition.
There has been an increase in efforts to produce more “dynamic” views of interactomes using AP-MS. (The LUMIER approach mentioned above was also designed with this type of analysis in mind, and can be used to monitor changes imparted by signaling events [95]). Systematic methods to map dynamic changes include the use of isotopic labeling approaches, and increasingly, quantification based on spectral counts [31, 35] or ion intensities of precursor peptide (MS1) or fragment ions (MS2). As quantitative methods and the accompanying software become more robust, there will be a major increase in interaction maps comparing cell- or tissue-specific interactions, or attempts to address changes in subsets of a network. It is not realistic to expect that every possible protein product will be monitored across all cell types (or tissues), or following treatment with every stimulus. However, as data become increasingly available regarding the function of each of the proteins encoded in a genome (e.g. via systematic RNA interference screens), and as the transcriptomes and proteomes of various cell types and tissues become known, cell types and screening conditions can be specifically selected based on the biological process of interest. For example, Glatter et al. were interested in defining the interaction network surrounding the insulin receptor / target of rapamycin pathway in Drosophila, and therefore profiled interactions in Kc167 cells following insulin stimulation using a spectral count based label-free approach [35]. Their study, in addition to identifying new components of the pathway, revealed that 22% of the detected interactions were regulated by insulin. A spectral count-based approach was also utilized by Li et al., to map interaction network dynamics regulating interferon production, centered on 58 known innate immunity regulators. This work revealed ~20% regulated interactions (following treatment with mimics of infection), and enabled them to establish the role of Mind Bomb proteins in the anti-RNA viral innate immune response [111]. Baker et al., employed a SILAC approach to reveal light-modulated interactions with the circadian clock protein FRQ in Neurospora [112]. To begin identifying cell fate decisions specified by the ERK kinase, and its dynamically-regulated interactors, von Kriegsheim et al. employed a SILAC approach to quantify interactors in rat PC12 cells stimulated for different times with nerve growth factor (NGF) or epidermal growth factor (EGF). This work revealed key differences between protein-protein associations modulated by the two different growth factors [113].
Lastly, while the quantitative approaches described above use spectral counting or SILAC for quantification, another quantification method that is gaining in popularity in the proteomics community exploits quantification of the product ions in MS2 spectra. In the standard approach known as selected reaction monitoring (SRM) [114–116], a prerequisite for quantification is to establish a robust list of peptides and product ions (these pairs are called transitions) to be recorded and quantified. Although the set-up phase of an SRM assay is time-consuming, once in place, the assay is rapid and extremely sensitive. Recently, Bisson et al. combined affinity purification with SRM (in a modified approach they call AP-SRM), and used it to better understand membrane-proximal phosphotyrosine signaling events by performing quantitative proteomics in HEK293T cells stimulated with EGF and other growth factors. Due to the combination of low cost per sample, sensitivity and accuracy, AP-SRM has great potential to enable the generation of time-resolved interactomes (e.g. Bisson et al. looked at six times points after EGF treatment) and to screen condition-specific interactions (in this case, six different growth factors). However, AP-SRM also has drawbacks, first in the need to optimize the quantification method (i.e. select the transitions to follow), but more importantly, the fact that one can only quantify what they expect to be present in the sample. These drawbacks may be eliminated in a variation on the theme of quantification in the MS2 spectra recently implemented as a pipeline on fast scanning, high resolution mass spectrometers. This approach, referred to as SWATH MS, enables sensitivity and precision similar to that of SRM [117], but because it analyzes the entire contents of a sample it can be re-interrogated at a later stage for any protein or peptide of interest. We have recently shown that SWATH can be used (similar to AP-SRM) to characterize changes in interactomes, with the added advantage of rapidity in method building, and the possibility to retrospectively analyze the data (Lambert et al., in prep.). In summary, methods harnessing the quantitative power of mass spectrometry to study interaction dynamics are becoming more robust and sensitive, and will undoubtedly lead to an increase in the number of studies producing such data. While this is exciting, how these types of data are recorded in public repositories, and how they are displayed, will remain issues that the field must deal with.
Perspective
In this review, we have attempted to raise awareness for; (i) the need to promote a better understanding of what AP-MS data can provide, and how this type of data differs from that generated by “binary” detection methods, (ii) to advocate for recording quantitative MS information in public repositories, and (iii) to take advantage of this data to better understand protein-protein interactions. While at the moment there is no single “winning” genome-scale technique that enables structural and dynamic analyses of all types of interactomes, many encouraging results which in principle should be scalable are coming to the fore. One remaining challenge will be determining how to visually and computationally represent the multiple layers of data that will be generated by future experiments focused on dynamic changes in protein-protein interactions. Lastly, while computational biologists have learned to deal with noise in interaction data (especially for making general conclusions regarding the behavior of a system), the systems biology community faces a daunting task in convincing other biologists that datasets acquired in high- or medium-throughput studies are both of high quality and biologically meaningful. This is necessary to engage the global scientific community in finally bridging the gap between the hairball and the atomic level understanding of protein-protein interactions.
Highlights.
Data generated by AP-MS is different from “binary” data and need to be better understood.
Quantitative data in AP-MS can be used to understand complex organization.
Quantitative AP-MS is poised to help understand interactome dynamics.
Acknowledgments
We thank Drs Leonard Foster, Jack Greenblatt, Pascal Braun and Mikko Taipale for communicating unpublished data, and Hyungwon Choi and Pascal Braun for critical review of the manuscript. Work in the Gingras and Raught labs is funded by the CIHR (MOP-84314 to ACG; MOP-81268 and MOP-119289 to BR) and the NIH (R01RR024031). BR holds the Canada Research Chair (CRC) in Proteomics and Molecular Medicine. ACG holds the CRC in Functional Proteomics and the Lea Reichmann Chair in Cancer Proteomics.
Abbreviations
- AP-MS
Affinity purification coupled to mass spectrometry
- Y2H
Yeast two hybrid
- TAP
Tandem Affinity Purification
- QconCAT
Quantification concatemer
- STRIPAK
Striatin Interacting Phosphatase And Kinase
- LUMIER
Luminescence-based Mammalian Interactome Mapping
- cDNA
complementary DNA
- ORF
Open Reading Frame
- MS1
Precursor ion mass spectrum
- MS2
Also called MS/MS or tandem mass spectrum; product ion mass spectrum
- SRM
Single Reaction Monitoring
- SILAC
Stable isotope labeling with amino acids in cell culture
- EGF
Epidermal growth factor
- NGF
Nerve growth factor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Temple G, Gerhard DS, Rasooly R, Feingold EA, Good PJ, Robinson C, Mandich A, Derge JG, Lewis J, Shoaf D, et al. The completion of the Mammalian Gene Collection (MGC) Genome Res. 2009;19:2324–2333. doi: 10.1101/gr.095976.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechtel S, Rosenfelder H, Duda A, Schmidt CP, Ernst U, Wellenreuther R, Mehrle A, Schuster C, Bahr A, Blocker H, et al. The full-ORF clone resource of the German cDNA Consortium. BMC Genomics. 2007;8:399. doi: 10.1186/1471-2164-8-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, Hu Z, Venkatesan K, Bethel G, Martin P, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rolfs A, Hu Y, Ebert L, Hoffmann D, Zuo D, Ramachandran N, Raphael J, Kelley F, McCarron S, Jepson DA, et al. A biomedically enriched collection of 7000 human ORF clones. PLoS One. 2008;3:e1528. doi: 10.1371/journal.pone.0001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X, Boehm JS, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, Green TM, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuyama A, Yoshida M. Systematic cloning of an ORFeome using the Gateway system. Methods Mol Biol. 2009;577:11–24. doi: 10.1007/978-1-60761-232-2_2. [DOI] [PubMed] [Google Scholar]
- 8.Howson R, Huh WK, Ghaemmaghami S, Falvo JV, Bower K, Belle A, Dephoure N, Wykoff DD, Weissman JS, O’Shea EK. Construction, verification and experimental use of two epitope-tagged collections of budding yeast strains. Comp Funct Genomics. 2005;6:2–16. doi: 10.1002/cfg.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 17.Formstecher E, Aresta S, Collura V, Hamburger A, Meil A, Trehin A, Reverdy C, Betin V, Maire S, Brun C, et al. Protein interaction mapping: a Drosophila case study. Genome Res. 2005;15:376–384. doi: 10.1101/gr.2659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanyon CA, Liu G, Mangiola BA, Patel N, Giot L, Kuang B, Zhang H, Zhong J, Finley RL., Jr A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 2004;5:R96. doi: 10.1186/gb-2004-5-12-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Huo K, Ma L, Tang L, Li D, Huang X, Yuan Y, Li C, Wang W, Guan W, et al. Toward an understanding of the protein interaction network of the human liver. Mol Syst Biol. 2011;7:536. doi: 10.1038/msb.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 21.Bandyopadhyay S, Chiang CY, Srivastava J, Gersten M, White S, Bell R, Kurschner C, Martin CH, Smoot M, Sahasrabudhe S, et al. A human MAP kinase interactome. Nat Methods. 2010;7:801–805. doi: 10.1038/nmeth.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhtar MS, Carvunis AR, Dreze M, Epple P, Steinbrenner J, Moore J, Tasan M, Galli M, Hao T, Nishimura MT, et al. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science. 2011;333:596–601. doi: 10.1126/science.1203659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lam M, Stagljar I. Strategies for membrane interaction proteomics: No mass spectrometry required. Proteomics. 2012 doi: 10.1002/pmic.201100471. in press. [DOI] [PubMed] [Google Scholar]
- 25.Braun P. Interactome mapping for analysis of complex phenotypes: insights from benchmarking binary interaction assays. Proteomics. 2012 doi: 10.1002/pmic.201100598. in press. [DOI] [PubMed] [Google Scholar]
- 26.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 27.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 28.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 29.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 30.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2012;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, et al. Global landscape of HIV-human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goudreault M, D’Ambrosio LM, Kean MJ, Mullin MJ, Larsen BG, Sanchez A, Chaudhry S, Chen GI, Sicheri F, Nesvizhskii AI, et al. A PP2A phosphatase high density interaction network identifies a novel striatin-interacting phosphatase and kinase complex linked to the cerebral cavernous malformation 3 (CCM3) protein. Mol Cell Proteomics. 2009;8:157–171. doi: 10.1074/mcp.M800266-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Glatter T, Schittenhelm RB, Rinner O, Roguska K, Wepf A, Junger MA, Kohler K, Jevtov I, Choi H, Schmidt A, et al. Modularity and hormone sensitivity of the Drosophila melanogaster insulin receptor/target of rapamycin interaction proteome. Mol Syst Biol. 2011;7:547. doi: 10.1038/msb.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glatter T, Wepf A, Aebersold R, Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol Syst Biol. 2009;5:237. doi: 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sardiu ME, Cai Y, Jin J, Swanson SK, Conaway RC, Conaway JW, Florens L, Washburn MP. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci U S A. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, et al. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 40.Hutchins JR, Toyoda Y, Hegemann B, Poser I, Heriche JK, Sykora MM, Augsburg M, Hudecz O, Buschhorn BA, Bulkescher J, et al. Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science. 2010;328:593–599. doi: 10.1126/science.1181348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedman AA, Tucker G, Singh R, Yan D, Vinayagam A, Hu Y, Binari R, Hong P, Sun X, Porto M, et al. Proteomic and functional genomic landscape of receptor tyrosine kinase and ras to extracellular signal-regulated kinase signaling. Sci Signal. 2011;4:rs10. doi: 10.1126/scisignal.2002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O’Connor L, Li M, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuhner S, van Noort V, Betts MJ, Leo-Macias A, Batisse C, Rode M, Yamada T, Maier T, Bader S, Beltran-Alvarez P, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. [DOI] [PubMed] [Google Scholar]
- 46.Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol. 2010;6:397. doi: 10.1038/msb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerrien S, Aranda B, Breuza L, Bridge A, Broackes-Carter F, Chen C, Duesbury M, Dumousseau M, Feuermann M, Hinz U, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–846. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen GI, Tisayakorn S, Jorgensen C, D’Ambrosio LM, Goudreault M, Gingras AC. PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J Biol Chem. 2008;283:29273–29284. doi: 10.1074/jbc.M803443200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gingras AC, Caballero M, Zarske M, Sanchez A, Hazbun TR, Fields S, Sonenberg N, Hafen E, Raught B, Aebersold R. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol Cell Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Turinsky AL, Razick S, Turner B, Donaldson IM, Wodak SJ. Literature curation of protein interactions: measuring agreement across major public databases. Database (Oxford) 2010;2010:baq026. doi: 10.1093/database/baq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braun P, Tasan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, de Smet AS, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skarra DV, Goudreault M, Choi H, Mullin M, Nesvizhskii AI, Gingras AC, Honkanen RE. Label-free quantitative proteomics and SAINT analysis enable interactome mapping for the human Ser/Thr protein phosphatase 5. Proteomics. 2011;11:1508–1516. doi: 10.1002/pmic.201000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi H, Larsen B, Lin ZY, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras AC, Nesvizhskii AI. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Breitkreutz A, Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, Lin ZY, Breitkreutz BJ, Stark C, Liu G, et al. A global protein kinase and phosphatase interaction network in yeast. Science. 2010;328:1043–1046. doi: 10.1126/science.1176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lavallee-Adam M, Cloutier P, Coulombe B, Blanchette M. Modeling contaminants in AP-MS/MS experiments. J Proteome Res. 2011;10:886–895. doi: 10.1021/pr100795z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi H, Glatter T, Gstaiger M, Nesvizhskii AI. SAINT-MS1: Protein-Protein Interaction Scoring Using Label-free Intensity Data in Affinity Purification-Mass Spectrometry Experiments. J Proteome Res. 2012 doi: 10.1021/pr201185r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaake RM, Wang X, Huang L. Profiling of protein interaction networks of protein complexes using affinity purification and quantitative mass spectrometry. Mol Cell Proteomics. 2010;9:1650–1665. doi: 10.1074/mcp.R110.000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Oeljeklaus S, Meyer HE, Warscheid B. New dimensions in the study of protein complexes using quantitative mass spectrometry. FEBS Lett. 2009;583:1674–1683. doi: 10.1016/j.febslet.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 61.Trinkle-Mulcahy L. Resolving protein interactions and complexes by affinity purification followed by label-based quantitative mass spectrometry. Proteomics. 2012 doi: 10.1002/pmic.201100438. in press. [DOI] [PubMed] [Google Scholar]
- 62.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci U S A. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brun V, Dupuis A, Adrait A, Marcellin M, Thomas D, Court M, Vandenesch F, Garin J. Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol Cell Proteomics. 2007;6:2139–2149. doi: 10.1074/mcp.M700163-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nat Methods. 2005;2:587–589. doi: 10.1038/nmeth774. [DOI] [PubMed] [Google Scholar]
- 65.Rivers J, Simpson DM, Robertson DH, Gaskell SJ, Beynon RJ. Absolute multiplexed quantitative analysis of protein expression during muscle development using QconCAT. Mol Cell Proteomics. 2007;6:1416–1427. doi: 10.1074/mcp.M600456-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5:2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 67.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Griffin NM, Yu J, Long F, Oh P, Shore S, Li Y, Koziol JA, Schnitzer JE. Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat Biotechnol. 2010;28:83–89. doi: 10.1038/nbt.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol. 2010;189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rinner O, Mueller LN, Hubalek M, Muller M, Gstaiger M, Aebersold R. An integrated mass spectrometric and computational framework for the analysis of protein interaction networks. Nat Biotechnol. 2007;25:345–352. doi: 10.1038/nbt1289. [DOI] [PubMed] [Google Scholar]
- 71.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 72.Wepf A, Glatter T, Schmidt A, Aebersold R, Gstaiger M. Quantitative interaction proteomics using mass spectrometry. Nat Methods. 2009;6:203–205. doi: 10.1038/nmeth.1302. [DOI] [PubMed] [Google Scholar]
- 73.Chang CY, Picotti P, Huettenhain R, Heinzelmann-Schwarz V, Jovanovic M, Aebersold R, Vitek O. Protein significance analysis in selected reaction monitoring (SRM) measurements. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M111.014662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- 75.Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci U S A. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwenk J, Harmel N, Zolles G, Bildl W, Kulik A, Heimrich B, Chisaka O, Jonas P, Schulte U, Fakler B, et al. Functional proteomics identify cornichon proteins as auxiliary subunits of AMPA receptors. Science. 2009;323:1313–1319. doi: 10.1126/science.1167852. [DOI] [PubMed] [Google Scholar]
- 77.Schwenk J, Metz M, Zolles G, Turecek R, Fritzius T, Bildl W, Tarusawa E, Kulik A, Unger A, Ivankova K, et al. Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature. 2010;465:231–235. doi: 10.1038/nature08964. [DOI] [PubMed] [Google Scholar]
- 78.Lambert JP, Pawson T, Gingras AC. Mapping physical interactions within chromatin by proteomic approaches. Proteomics. 2012 doi: 10.1002/pmic.201100547. in press. [DOI] [PubMed] [Google Scholar]
- 79.Lambert JP, Fillingham J, Siahbazi M, Greenblatt J, Baetz K, Figeys D. Defining the budding yeast chromatin-associated interactome. Mol Syst Biol. 2010;6:448. doi: 10.1038/msb.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lambert JP, Mitchell L, Rudner A, Baetz K, Figeys D. A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol Cell Proteomics. 2009;8:870–882. doi: 10.1074/mcp.M800447-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muller CS, Haupt A, Bildl W, Schindler J, Knaus HG, Meissner M, Rammner B, Striessnig J, Flockerzi V, Fakler B, et al. Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc Natl Acad Sci U S A. 2010;107:14950–14957. doi: 10.1073/pnas.1005940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gebert N, Gebert M, Oeljeklaus S, von der Malsburg K, Stroud DA, Kulawiak B, Wirth C, Zahedi RP, Dolezal P, Wiese S, et al. Dual function of Sdh3 in the respiratory chain and TIM22 protein translocase of the mitochondrial inner membrane. Mol Cell. 2011;44:811–818. doi: 10.1016/j.molcel.2011.09.025. [DOI] [PubMed] [Google Scholar]
- 83.Stroud DA, Oeljeklaus S, Wiese S, Bohnert M, Lewandrowski U, Sickmann A, Guiard B, van der Laan M, Warscheid B, Wiedemann N. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J Mol Biol. 2011;413:743–750. doi: 10.1016/j.jmb.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 84.von der Malsburg K, Muller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 85.Alkhaja AK, Jans DC, Nikolov M, Vukotic M, Lytovchenko O, Ludewig F, Schliebs W, Riedel D, Urlaub H, Jakobs S, et al. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol Biol Cell. 2012;23:247–257. doi: 10.1091/mbc.E11-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang X, Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol Cell Proteomics. 2008;7:46–57. doi: 10.1074/mcp.M700261-MCP200. [DOI] [PubMed] [Google Scholar]
- 87.Mousson F, Kolkman A, Pijnappel WW, Timmers HT, Heck AJ. Quantitative proteomics reveals regulation of dynamic components within TATA-binding protein (TBP) transcription complexes. Mol Cell Proteomics. 2008;7:845–852. doi: 10.1074/mcp.M700306-MCP200. [DOI] [PubMed] [Google Scholar]
- 88.Oeljeklaus S, Reinartz BS, Wolf J, Wiese S, Tonillo J, Podwojski K, Kuhlmann K, Stephan C, Meyer HE, Schliebs W, et al. Identification of Core Components and Transient Interactors of the Peroxisomal Importomer by Dual-Track Stable Isotope Labeling with Amino Acids in Cell Culture Analysis. J Proteome Res. 2012 doi: 10.1021/pr3000333. [DOI] [PubMed] [Google Scholar]
- 89.Chen GI, Gingras AC. Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods. 2007;42:298–305. doi: 10.1016/j.ymeth.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 90.Sinz A. Investigation of protein-protein interactions in living cells by chemical crosslinking and mass spectrometry. Anal Bioanal Chem. 2010;397:3433–3440. doi: 10.1007/s00216-009-3405-5. [DOI] [PubMed] [Google Scholar]
- 91.Petrotchenko EV, Borchers CH. Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom Rev. 2010;29:862–876. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- 92.Stengel F, Aebersold R, Robinson CV. Joining Forces: Integrating Proteomics and Cross-linking with the Mass Spectrometry of Intact Complexes. Mol Cell Proteomics. 2012;11:R111 014027. doi: 10.1074/mcp.R111.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rappsilber J. The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J Struct Biol. 2011;173:530–540. doi: 10.1016/j.jsb.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 96.Michnick SW, Ear PH, Landry C, Malleshaiah MK, Messier V. Protein-fragment complementation assays for large-scale analysis, functional dissection and dynamic studies of protein-protein interactions in living cells. Methods Mol Biol. 2011;756:395–425. doi: 10.1007/978-1-61779-160-4_25. [DOI] [PubMed] [Google Scholar]
- 97.Michnick SW, Ear PH, Landry C, Malleshaiah MK, Messier V. A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein-protein interactions in living cells. Methods Enzymol. 2010;470:335–368. doi: 10.1016/S0076-6879(10)70014-8. [DOI] [PubMed] [Google Scholar]
- 98.Colwill K, Graslund S. A roadmap to generate renewable protein binders to the human proteome. Nat Methods. 2011;8:551–558. doi: 10.1038/nmeth.1607. [DOI] [PubMed] [Google Scholar]
- 99.Nilsson P, Paavilainen L, Larsson K, Odling J, Sundberg M, Andersson AC, Kampf C, Persson A, Al-Khalili Szigyarto C, Ottosson J, et al. Towards a human proteome atlas: high-throughput generation of mono- specific antibodies for tissue profiling. Proteomics. 2005;5:4327–4337. doi: 10.1002/pmic.200500072. [DOI] [PubMed] [Google Scholar]
- 100.Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, et al. Protein production and purification. Nat Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kohl T, Schmidt C, Wiemann S, Poustka A, Korf U. Automated production of recombinant human proteins as resource for proteome research. Proteome Sci. 2008;6:4. doi: 10.1186/1477-5956-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu S, Xie Z, Qian J, Blackshaw S, Zhu H. Functional protein microarray technology. Wiley Interdiscip Rev Syst Biol Med. 2011;3:255–268. doi: 10.1002/wsbm.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mattoon DR, Schweitzer B. Profiling protein interaction networks with functional protein microarrays. Methods Mol Biol. 2009;563:63–74. doi: 10.1007/978-1-60761-175-2_4. [DOI] [PubMed] [Google Scholar]
- 104.Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci U S A. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Suk-Joon H, Ruotolo B. Integrating mass spectrometry of intact protein complexes into structural proteomics. Proteomics. 2012 doi: 10.1002/pmic.201100520. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kean MJ, Ceccarelli DF, Goudreault M, Sanches M, Tate S, Larsen B, Gibson LC, Derry WB, Scott IC, Pelletier L, et al. Structure-function analysis of core STRIPAK Proteins: a signaling complex implicated in Golgi polarization. J Biol Chem. 2011;286:25065–25075. doi: 10.1074/jbc.M110.214486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ceccarelli DF, Laister RC, Mulligan VK, Kean MJ, Goudreault M, Scott IC, Derry WB, Chakrabartty A, Gingras AC, Sicheri F. CCM3/PDCD10 heterodimerizes with germinal center kinase III (GCKIII) proteins using a mechanism analogous to CCM3 homodimerization. J Biol Chem. 2011;286:25056–25064. doi: 10.1074/jbc.M110.213777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, Florens L, Workman JL, Washburn MP. Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol Syst Biol. 2011;7:503. doi: 10.1038/msb.2011.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sardiu ME, Gilmore JM, Carrozza MJ, Li B, Workman JL, Florens L, Washburn MP. Determining protein complex connectivity using a probabilistic deletion network derived from quantitative proteomics. PLoS One. 2009;4:e7310. doi: 10.1371/journal.pone.0007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Junttila MR, Puustinen P, Niemela M, Ahola R, Arnold H, Bottzauw T, Ala-aho R, Nielsen C, Ivaska J, Taya Y, et al. CIP2A inhibits PP2A in human malignancies. Cell. 2007;130:51–62. doi: 10.1016/j.cell.2007.04.044. [DOI] [PubMed] [Google Scholar]
- 111.Li S, Wang L, Berman M, Kong YY, Dorf ME. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol Cell. 2009;34:354–363. doi: 10.1016/j.molcel.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.von Kriegsheim A, Baiocchi D, Birtwistle M, Sumpton D, Bienvenut W, Morrice N, Yamada K, Lamond A, Kalna G, Orton R, et al. Cell fate decisions are specified by the dynamic ERK interactome. Nat Cell Biol. 2009;11:1458–1464. doi: 10.1038/ncb1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Elschenbroich S, Kislinger T. Targeted proteomics by selected reaction monitoring mass spectrometry: applications to systems biology and biomarker discovery. Mol Biosyst. 2011;7:292–303. doi: 10.1039/c0mb00159g. [DOI] [PubMed] [Google Scholar]
- 115.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gillet LC, Navarro P, Tate S, Roest H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012 doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]