Abstract
Background
Anthrax and its etiologic agent remain a biological threat. Anthrax vaccine is highly effective, but vaccine-induced IgG antibody responses vary widely following required doses of vaccinations. Such variation can be related to genetic factors, especially genomic copy number variants (CNVs) that are known to be enriched among genes with immunologic function. We have tested this hypothesis in two study populations from a clinical trial of anthrax vaccination.
Methods
We performed CNV-based genome-wide association analyses separately on 794 European Americans and 200 African-Americans. Antibodies to protective antigen were measured at week 8 (early response) and week 30 (peak response) using an enzyme-linked immunosorbent assay. We used DNA microarray data (Affymetrix 6.0) and two CNV detection algorithms, hidden markov model (PennCNV) and circular binary segmentation (GeneSpring) to determine CNVs in all individuals. Multivariable regression analyses were used to identify CNV-specific associations after adjusting for relevant non-genetic covariates.
Results
Within the 22 autosomal chromosomes, 2,943 non-overlapping CNV regions were detected by both algorithms. Genomic insertions containing HLA-DRB5, DRB1 and DQA1/DRA genes in the major histocompatibility complex (MHC) region (chromosome 6p21.3) were moderately associated with elevated early antibody response (β = 0.14, p = 1.78×10−3) among European Americans, and the strongest association was observed between peak antibody response and a segmental insertion on chromosome 1, containing NBPF4, NBPF5, STXMP3, CLCC1, and GPSM2 genes (β = 1.66, p = 6.06×10−5). For African-Americans, segmental deletions spanning PRR20, PCDH17 and PCH68 genes on chromosome 13 were associated with elevated early antibody production (β = 0.18, p = 4.47×10−5). Population-specific findings aside, one genomic insertion on chromosome 17 (containing NSF, ARL17 and LRRC37A genes) was associated with elevated peak antibody response in both populations.
Conclusion
Multiple CNV regions, including the one consisting of MHC genes that is consistent with earlier research, can be important to humoral immune responses to anthrax vaccine adsorbed.
Introduction
Anthrax, caused by Bacillus anthracis, is one of the most likely biological threat candidates as infection can involve the skin, gastrointestinal tract, or lungs [1], [2], [3]. Anthrax can be and has been used in biological warfare and bioterrorism. Throughout history, anthrax infections have killed hundreds of thousands of animals and people. The development of an animal vaccine in 1881 substantially reduced the threat of anthrax to livestock and humans [4]. However, anthrax still has a fatality rate of 80% or higher [5] and the long-lived, soil-borne anthrax bacterial spore remains a public health concern with intermittent outbreaks in livestock and humans. European investigators recently confirmed 14 deaths among 119 suspected anthrax cases [6]. In the United States in 2001, 22 cases of anthrax were linked to anthrax contaminated envelopes mailed to persons working in the news media and government [2]. As recently as August 2012, the Colorado Department of Agriculture investigators confirmed anthrax in one deceased cow and suspected exposure to anthrax in about 50 other dead cattle [7]. Prevention of the disease depends upon the efficacy of the licensed anthrax vaccine adsorbed (AVA-Biothrax™, Bioport Corporation, Lansing MI) [8]. AVA-Biothrax™ is a cell-free filtrate prepared from an avirulent strain of B. anthracis, the product contains protective antigen as its major component. It was licensed for use in 1970 after a single trial demonstrated a vaccine efficacy of 93% [9]. Like others, anthrax vaccine confers immunity by simulating a natural infection, but responses to this and other vaccines vary in human populations.
Genetic differences in vaccine response have been explained by the polymorphic nature of gene families involved in various pathways, specifically immune response. Genetic variations such as single nucleotide polymorphisms (SNPs), insertion/deletion, gene duplication and copy number variants (CNVs) are common in the human genome. Many polymorphic genetic variations have been implicated as independent cofactors in infection and immunity [10], [11], [12]. Studies among monozygotic and dizygotic twin pairs (for hepatitis B, oral polio, tetanus, Haemophilus influenzae type b and diphtheria vaccine response) have suggested significant heritability (44–77%) with both HLA and non-HLA genes [13], [14], [15], [16], [17]. We have previously shown that several genes, including those in the major histocompatibility (MHC) region are associated with longitudinal variation of antibody response [18], [19].
Most genetic epidemiological studies, including our own previous work on vaccine response [18], [19], have focused on SNPs. This concentration has overshadowed the importance of structural variants, especially CNVs [20]. CNVs are the result of duplications, deletions, insertions and other complex rearrangements of DNA segments often defined to be larger than 10 kb. These structural genetic variations have been shown to be involved in pathogenesis and treatment of immune related diseases [21], [22], [23], [24]. CNVs are frequently enriched in genes associated with immunity, inflammation, and host defense; they are likely under positive selection for their contribution to the enhanced ability of humans to adapt to their environment [25], [26]. We hypothesized that common CNVs play a role in differential antibody response to the AVA-Biothrax™ vaccine.
Methods
Ethics Statement
The parent study and this sub-study conformed to the procedures for informed written consent (parental permission was obtained wherever required) approved by institutional review boards (IRB) at all sponsoring organizations and to human-experimentation guidelines set forth by the United States Department of Health and Human Services and finally reviewed and approved by the UAB IRB.
Study Population
The Anthrax Vaccine Research Program clinical trial (clinicaltrials.gov identifier NCT00119067, hereafter referred to as AVA000) was a multicenter, randomized, double blind trial of 1,563 healthy individuals aged 16 to 61 years at baseline and enrolled into the study between 2002 and 2008. The design of the study and participant characteristics have been described in detail previously [18], [19], [27]. Briefly, of the 1,563 participants, 1,303 were randomly assigned to seven arms: group 1 received the licensed regimen (8 doses, subcutaneously (SQ)), while group 2 also received 8 doses but with intramuscular (IM) administration. Groups 3 through 5 received between 4 and 7 IM doses, and groups 6a (SQ) and 6b (IM) received saline placebo. Of the 1,303 participants, 1,078 eligible individuals were genotyped using the Affymetrix SNP Array 6.0. All genetic and clinical data have been deposited to the Immunology Database and Analysis Portal (ImmPort) system. Based on previously reported principal component analysis [18], we selected a sample of 794 European Americans and 200 African Americans to be included in genome-wide CNV analyses.
Measurement of IgG Antibody to Protective Antigen
IgG antibodies to protective antigen (AbPA) were measured using a quantitative enzyme-linked immunosorbent assay (ELISA) as described by Semenova et al [28]. Following administration of 3 or 4 doses of anthrax vaccine either IM or SQ, AbPA was measured at 4, 8, 26 and 30 weeks post-baseline. The impact of route of administration and number of vaccinations on the AbPA response has been discussed previously [1]. We excluded AbPA measurements taken after missed vaccinations or at times that deviated significantly from those specified in the study protocol, as previously described [18].
Genotyping and Quality Control
Genetic data were obtained from all participating vaccinees using the Affymetrix Genome-Wide Human SNP Array 6.0. SNP genotyping methods, quality control, and association methods have been previously described in detail [19]. In the present study, we only considered interrogated CNVs. The array consisted of 744,000 probes, evenly spaced along the genome along with additional 202,000 probes targeting 5,677 CNV regions from the Toronto Database of Genomic Variants.
CNV Calling
CNV calling algorithms can often generate varying results according to array normalization, definition of the reference genome, type of calling algorithm implemented, and parameters provided to the algorithm [29]. In this study, CNVs were called using two algorithms: the hidden Markov model calling algorithm PennCNV [30]; and the change-point calling algorithm GeneSpring GX 11 [31]. To reduce false positives, we selected CNVs that were concordant between both algorithms (see below). All the samples in each batch (96 samples) were used to create a reference signal at a given SNP for CNV calls in both algorithms. All annotated CNV genomic regions were based on the hg18 (NCBI) human genome assembly.
PennCNV Software and Algorithm
The PennCNV algorithm considers the total signal intensity and allelic intensity ratio at each SNP, the distances between SNPs, and the frequency of each SNP via a Hidden Markov model (HMM) [30]. The PennCNV-Affymetrix protocol, as described in the manufacturer’s guide, (http://www.openbioinformatics.org/penncnv/penncnv_tutorial_affy_gw6.html) was utilized for transforming the intensities from the raw CEL files into log R ratios (LRR) and B allele frequencies (BAFs). The software’s default settings for the HMM were employed.
GeneSpring Software and Algorithm
GeneSpring software [31] utilizes a Circular Binary Segmentation (CBS) algorithm to smooth the outliers and define change points (using default p-value <0.002 for a t-test) to identify segment breaks [32]. Once the segment breaks are identified, CNVs and confidence scores are assigned.
Concordance of Detected CNV
PennCNV produces a discrete, integer copy number state of 0, 1, 2, 3 or 4 copies. Generally, 0 represents deletion in both chromosomes; 1, deletion in one chromosome; 3, insertion in one chromosome; and 4, two or more insertions. GeneSpring provides a continuous value that does not always clearly discriminate between single and double deletions or insertions. In order to assess concordance between the two algorithms, we classified the calls from both algorithms as either deletion, insertion, or normal without accounting for single or double insertion/deletion.
CNV and CNV Region
We determined CNV regions (CNVRs) using the method suggested in PLINK [33]: we defined CNV predictor variables at individual genomic positions when an individual had a CNV that spanned the given position. The genomic positions used were the transition points, that is, all starting and ending positions of the called CNV for any individual in the sample. We only considered CNVs with at least two consecutive CNV probes spanning at least 10 bp showing consistent deletion or duplication. We further limited the analysis to transition points that were spanned by CNVs called in at least 10 individuals. When a test of association at a transition point was of interest (statistically significant), we defined the spanning CNVR as the largest common region containing all CNVs that spanned the given transition point.
Statistical Analysis
The outcome variable analyzed for association with each CNV was base 10 logarithm (log10) AbPA at 8 weeks for early antibody response and at 30 weeks for peak antibody response. Values below detection threshold were treated as missing and excluded from the analysis. We regressed outcomes on the transition point of each confirmed CNV in our analyses. The CNV data or each transition point defined above were represented in the linear regression as either: a) any deletion versus all others (D2-); or b) any insertion versus all others (I2+). All analyses in each CNV transition point were performed separately among European American and African American populations. Adjustment for age, sex, route of administration (SQ vs. 3-IM vs. 4-IM), and the first three principal component values, from the combined population, were applied in all models [19]. For significantly associated CNV transition points, all genes were identified within the respective CNVR. As described above, administrations were performed by either 3 or 4 doses of anthrax vaccine by IM or SQ; thus we also performed separate analysis for each vaccine delivery group and performed a meta-analysis among European Americans and African Americans separately, using the DerSimonian and Laird’s linear random-effects model with the “rma” function in R package “metaphor” [34], [35]. Since CNV were relatively less frequent, when we performed the analysis in the three vaccine administration groups separately, we were under-powered to observe the associations in each group; however, we estimated the beta and p-values from the meta-analysis and confirmed the direction of the association, both in separate and overall meta-analysis to the main analysis with adjustment for the route of vaccine administration.
Results
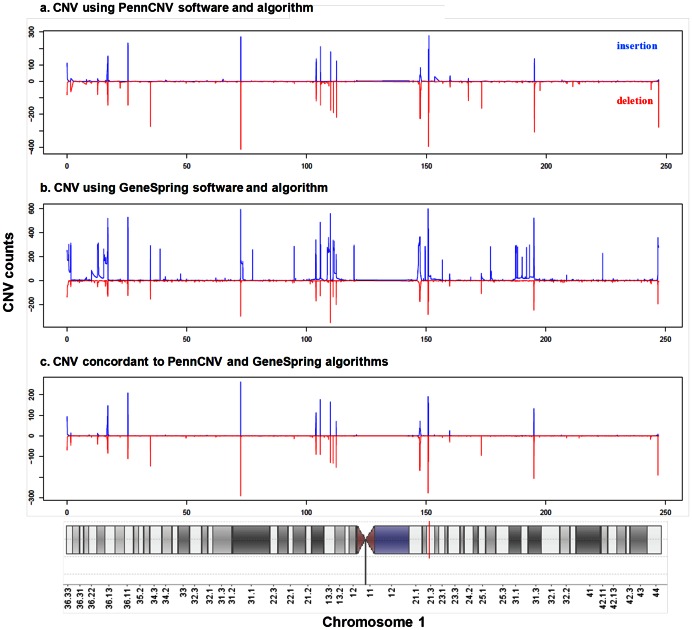
The characteristics of the 794 European American and 200 African American participants included in the analysis have been described in detail previously [18]. There were 2,943 concordant, non-overlapping, unique CNVRs spanning at least 10 bp and with a minimum count of 10 individuals in the study population that were used in the analyses (Figure 1).
Figure 1. Distribution of insertion and deletion CNVs across chromosome 1 among European Americans: a) CNV calls based on the hidden Markov model calling algorithm PennCNV, b) CNV calls based on the change-point calling algorithm GeneSpring GX 11, and c) CNV calls based on the concordance between PennCNV and GeneSpring.
On the vertical axis, the top peaks (blue) demonstrate the actual count of insertions and the bottom peaks (red) demonstrate the actual count of deletions. The X axis shows chromosome 1 location.
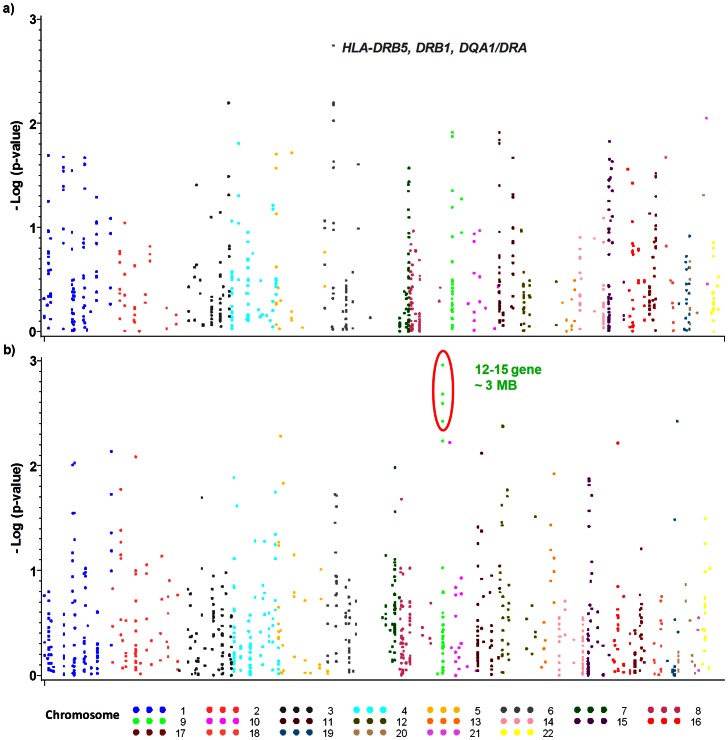
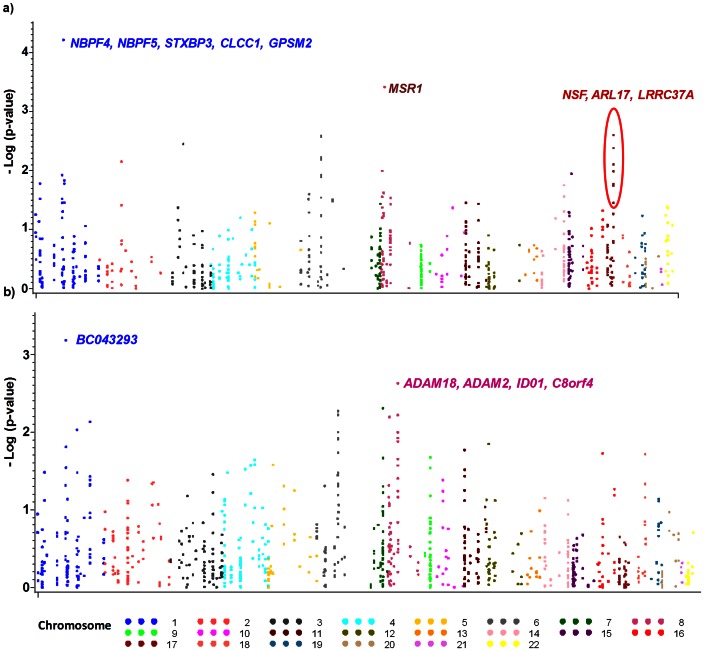
The genome-wide association signals for concordant insertion and deletion CNVs with antibody response among European Americans for early vaccine response at week 8 are shown in Figures 2a and 2b, respectively; and at week 30 for peak vaccine response in Figures 3a and 3b, respectively (all results presented in table S1). The strongest significant associations of CNVs (confirmed by both adjusted analysis and meta-analysis) with early and peak anthrax antibody responses are displayed in Table 1. Among European Americans, the genomic insertions in the positions – chr6∶32,539,530–32,701,486 (Figure 2a) are associated with early elevated antibody response (β = 0.14; p = 1.78×10−3). This CNVR contains HLA-DRB5, DRB1, DQA1/DRA genes. Similarly, insertions of genomic regions chr8∶16,251,325–16,333,182, and chr1∶108,729,983–109,312,876 (Figure 3a) have moderate effect on elevated peak antibody (β = 0.61; p = 9.29×10−3 and β = 1.66; p = 6.06×10−5 respectively). The chromosome 1 region contains multiple genes - NBPF4, NBPF5, STXBP3, CLCC1, and GPSM2. Also, deletions in chr1∶105,814,277–106,015,829, containing BC043293 gene as well as deletions in the CNVR chr8∶39,344,897–40,302,956 (Figure 3b), containing ADAM18, ADAM2, ID01, C8orf4 genes are associated with elevated peak antibody response.
Figure 2. Genome-wide CNV association for early anthrax antibody response among European Americans: a) any insertions at 8 week AbPA titer, b) any deletions at 8 weeks AbPA titer.
Association was assessed using linear regression models adjusted for age, sex, route of administration (SQ vs. IM), and the principal components analysis (PCA) of ancestry. The X axis shows chromosomal locations, and P values were plotted on the Y axis using logarithmic scale.
Figure 3. Genome-wide CNV association for peak anthrax antibody response among European Americans: a) any insertions at 30 week AbPA titer, and b) any deletions at 30 week AbPA titer.
Association was assessed using single linear regression adjusted for age, sex, route of administration (SQ vs. IM), and the principal components analysis (PCA) of ancestry. The X axis shows chromosomal locations, and P values were plotted on the Y axis using a logarithmic scale.
Table 1. CNV regions associated with week 8 and week 30 IgG antibody response to anthrax vaccine adsorbed in European Americans and African Americans.
| CNVtype | Chr | CNV Marker point* | CNV probe ID† | Common CNV genomic region* | Gene | Adjusted analysis Beta p | Meta-analysis Beta p | |||||
| European American | ||||||||||||
| Early response (8 weeks) | ||||||||||||
| I2+ | 6 | 32,562,253 | CN_1175484 | 32,539,530–32,701,485 | HLA-DRB5, DRB1, DQA1/DRA | 0.14 | 1.78E-03 | 0.15 | 0.01 | |||
| D2− | 9 | 43,542,722 | CN_1322511 | 41,682,297–44,780,914 | 12–15 loci | −0.12 | 1.10E-03 | −0.15 | 0.05 | |||
| Peak response (30 weeks) | ||||||||||||
| D2− | 1 | 105,826,321 | SNP_A-8310560 | 105,814,277 -106,015,829 | BC043293 | 1.04 | 6.52E-04 | 1.33 | 1.87E-04 | |||
| I2+ | 1 | 109,298,377 | CN_451797 | 108,729,983–109,312,876 | NBPF4, NBPF5, STXBP3, CLCC1, GPSM2 | 1.66 | 6.06E-05 | 1.70 | 5.28E-05 | |||
| D2− | 8 | 39,452,128 | CN_1283719 | 39,344,897–40,302,956 | ADAM18, ADAM2, ID01, C8orf4 | 0.12 | 2.35E-03 | 0.13 | 1.97E-03 | |||
| I2+ | 17 | 42,078,158 | CN_739340 | 41,521,621–42,150,374 | NSF, ARL17, LRRC37A | 0.12 | 2.48E-03 | 0.13 | 1.02E-03 | |||
| African American | ||||||||||||
| Early response (8 weeks) | ||||||||||||
| D2− | 13 | 56,661,148 | CN_654225 | 54,593,069–58,010,252 | PRR20, PCDH17, PCH68 | 0.18 | 4.47E-05 | 0.21 | 8.57E-04 | |||
| D2− | 13 | 68,146,300 | SNP_A-8363651 | 68,140,180–68,166,243 | PCDH9, KLHL1 | −0.82 | 1.79E-03 | −0.81 | 1.47E-03 | |||
| Peak response (30 weeks) | ||||||||||||
| I2+ | 3 | 164,106,591 | CN_991857 | 163,985,816–164,117,009 | BC073807 | 0.29 | 1.25E-03 | 0.51 | 2.49E-03 | |||
| I2+ | 5 | 886,628 | CN_1131184 | 751,371–1,048,270 | ZDHHC11, BRD9, TRIP13 | −0.27 | 2.73E-03 | −0.22 | 3.23E-03 | |||
| D2− | 11 | 55,133,210 | CN_587565 | 55,108,112–55,414,942 | OR4C11, OR4P4, SPRYD5 | −0.33 | 1.46E-03 | −0.36 | 2.76E-03 | |||
| I2+ | 17 | 42,120,174 | CN_739357 | 41,521,621–42,150,374 | NSF, ARL17, LRRC37A | 0.60 | 2.14E-04 | 0.11 | 2.89E-03 | |||
I2+ = any insertion, D2- = any deletion;
genomic position based on hg18;
CNV probe ID based on the Affymetrix 6.0 microarray.
With the smaller sample-size of African American populations in our study, all results are presented in table S2. Of special interest in African Americans, deletions in the CNVR chr13∶54,593,069–58,010,252 containing PRR20, PCDH17, PCH68 genes (Table 1 and Table S2) are significantly associated with increased antibody levels in the early period (Table 1 - β = 0.18, p = 4.47×10−5). In contrast, deletions in the CNVR chr13∶68,140,180–68,166,243 that contains PCDH9, KLHL1 genes are associated with reduced early antibody response. For the peak response, insertions in the genomic region chr3∶163,985,816–164,117.009 that contains BC073807 gene (Table 1 and Table S2) are associated with mildly increased antibody response (β = 0.24; p = 1.25×10−3). Interestingly, in both European Americans and African Americans, insertions in the genomic region chr17∶41,521,621–42,150,374 (Table 1) are associated with elevated peak antibody response. This region consists of NSF, ARL17 and LRRC37A genes.
Discussion
Vaccination against anthrax is very effective with generally high but variable antibody response. Using genome-wide association testing, we focused on CNV, an important but not yet commonly evaluated source of genetic variation, for its association with AbPA response to AVA-Biothrax™ in European American and African Americans. The strongest associations were found in regions containing NBPF4, NBPF5, STXBP3, CLCC1, GPSM2 on chromosome 1 (adjusted analysis p = 6.06×10−5, meta-analysis p = 5.28×10−5) in European Americans, and in regions containing PRR20, PCDH17, PCH68 (adjusted analysis p = 4.47×10−5, meta-analysis p = 8.57×10−4) in African-Americans. CNVs containing MHC class II genes, namely HLA–DRB5, DRB1, and DQA1/DRA on chromosome 6 were also identified on chromosome 6 in European Americans only. Interestingly, CNVs containing NSF, ARL17 and LRRC37A genes on chromosome 17 were associated with elevated antibody response in subjects of both races.
The association of MHC class II genes -HLA–DRB1, DQA1 with anthrax vaccine antibody response has been previously reported in SNP analysis [18]. In a GWAS of hepatitis B vaccine response, markers in HLA-DR and HLA-DP showed independent associations with antibody response [36]. Similarly, markers in HLA-DRB5 were found to be associated with elevated antibody response. In our own previous study in this population [37], carriage of three haplotypes in this region (DRB1*1501-DQA1*0102-DQB1*0602, DRB1*0101-DQA1*0101-DQB1*0501, and DRB1*0102-DQA1*0101-DQB1*0501) were all significantly associated with lower AbPA levels in the longitudinal analyses [18]. Recently in an animal study [37], MHC class II locus has been associated with high antibody titers to a component of anthrax toxin, lethal factor whereas non-MHC class loci were discovered to influence antibody titers to protective antigen.
Interestingly, as an internal replication, CNVs in the genomic region for chromosome 17 containing NSF, ARL17, LRRC37A genes were associated with increased antibody response in both European Americans and African Americans. The NSF gene, N-ethylmaleimide-sensitive factor, is one of the essential components of the cellular membrane fusion machinery, an important homeostatic process [38]. NSF gene has been associated with Parkinson’s disease [39]. In one study ARL17, ADP-ribosylation factor-like 17 was one of the genes reported to be differentially regulated in the human monocytes after treatment with anthrax lethal toxin (LT) [40]. ARL17 is localized to the Golgi apparatus and is thought to be involved in modulation of vesicle budding and is known to be an activator of cholera toxin catalytic subunit, an ADP-ribosyltransferase [41]. LRRC37A (leucine-rich repeat containing 37A) belongs to the complex LRRC37 family that has been mapped to 18 distinct loci in the human genome on the q arm of chromosome 17. LRRC37A protein localizes to cellular vesicles, including Golgi apparatus and to the plasma membrane [42]. Variants in LRC37A have also been associated with Parkinson disease [43]. However, to date there are no reports of involvement of any of these genes in immunity or vaccine response.
We also observed associations with other CNVRs (table 1); but none of the genes within those CNVRs have a known role in basic immunology or vaccine response mechanisms. STXBP3 and CLCC1 genes on chromosome 1 have been linked to insulin dependent diabetes mellitus [44]. STXBP3, syntaxin binding protein, is associated with glucose metabolism in adipocytes and contributes to fusion of intracellular GLUT4-containing vesicles with the cell surface in adipocytes [44]. CLCC1, chloride channel CLIC-like 1, produces protein located in the membranes of intracellular compartments of endoplasmic reticulum and Golgi apparatus [45]. GPSM2, G-protein-signaling modulator 2, belongs to a family of genes that modulate G proteins activation and is associated with cell division and cancer [46]. NBPF4 and NBPF5 are neuroblastoma breakpoint family genes that have not been associated with any disease.
Each CNV calling algorithm and analytical approach has its own limitations and strengths. In our study, Penn-CNV identified 5,812 different CNVRs, with an average of 70 CNVs in each individual; whereas GeneSpring identified 7,271 different CNVRs, with an average of 168 in each individual. Using only the CNVs with concordant calls from two different methods will miss potential CNVs from the filtering due to inherent difficulties in defining CNVs. For example, the HMM algorithm has difficulty with heterogenous CNV regions. However, the requirement for concordance provides higher confidence in the fidelity of the CNV calls. CNVs consistently called from two or more different calling algorithms are more reliable than those CNVs called from a single algorithm [47]. Even with our more stringent definition, the number of CNV calls per person seems to be higher in our study than other studies, very likely because we have considered CNVs as small as 10 bp (but occurring in at least 10 individuals), rather than the conventional size of 10 kb that is often used. However, that difference is not a major concern for our analysis because most of the significant associations were observed with large CNVs.
Of note, in this study we examined the association of the CNVs in both early and peak vaccine response. The biological mechanisms of early and peak response may well be different, and our studies suggest that CNVs associated with differential levels of antibody were different at the two phases. Our study involved two ethnic populations nested within a multi-center vaccine trial. While this cohort is the largest for an anthrax vaccine genetic study, our analysis in African Americans remains underpowered. Nevertheless, one region of the genome seems to have a constant effect in both European American and African American populations. With our conservative approach to CNV calls, we only examined 2,943 CNVs in total; thus, our associations seem to meet the stringent correction criteria for multiple tests. As with most vaccine genetic studies, another limitation of the present study is lack of a readily available replication cohort. Although the results from this study are preliminary, they show the potential for identification of gene copy number that could play a role in vaccine response. Weaponized anthrax spores remain a lethal bioterrorism threat to military personnel and civilian populations; continued research is therefore needed to understand whether and how genetic profiles can predict vaccine response.
Supporting Information
Complete association results of all CNV tranisition points with early (8 weeks) and peak (30 weeks) antibody responses to anthrax vaccine in 794 European Americans.
(XLSX)
Complete association results of all CNV tranisition points with early (8 weeks) and peak (30 weeks) antibody responses to anthrax vaccine in 200 African Americans.
(XLSX)
Acknowledgments
The authors gratefully acknowledge the other site principal investigators from the Anthrax Vaccine Research Program (AVRP) human clinical trial (clinicaltrials.gov identifier NCT00119067, also referred to as AVA000) for their efforts in data collection: Janine Babcock, M.D. (Walter Reed Army Institute of Research), Wendy Keitel, M.D. (Baylor College of Medicine), and Harry Keyserling, M.D. (Emory University School of Medicine), Gregory A Poland (Mayo Clinic), and Scott D Parker (University of Alabama at Birmingham). The authors would also like to thank Conrad Quinn, PhD for helpful information concerning the trial and clinical data; Nicholas M Pajewski, PhD and Jelai Wang for their assistance with the data management. All genetic and clinical data have been deposited in the Immunology Database and Analysis Portal (ImmPort) system, as required for data acquired with NIAID funding.
Funding Statement
This work was supported by the National Institute of Allergy and Infectious Diseases through contracts N01-AI40068 and 272201000023C-0-0-1. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Maillard JM, Fischer M, McKee KT Jr, Turner LF, Cline JS (2002) First case of bioterrorism-related inhalational anthrax, Florida, 2001: North Carolina investigation. Emerg Infect Dis 8: 1035–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jernigan DB, Raghunathan PL, Bell BP, Brechner R, Bresnitz EA, et al. (2002) Investigation of bioterrorism-related anthrax, United States, 2001: epidemiologic findings. Emerg Infect Dis 8: 1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Traeger MS, Wiersma ST, Rosenstein NE, Malecki JM, Shepard CW, et al. (2002) First case of bioterrorism-related inhalational anthrax in the United States, Palm Beach County, Florida, 2001. Emerg Infect Dis 8: 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cherkasskiy BL (1999) A national register of historic and contemporary anthrax foci. Journal of applied microbiology 87: 192–195. [DOI] [PubMed] [Google Scholar]
- 5. Meselson M, Guillemin J, Hugh-Jones M, Langmuir A, Popova I, et al. (1994) The Sverdlovsk anthrax outbreak of 1979. Science 266: 1202–1208. [DOI] [PubMed] [Google Scholar]
- 6.National Health Service An outbreak of anthrax among drug users in Scotland, December 2009 to December 2010: a report on behalf of the National Anthrax Outbreak Control Team.
- 7.Colorado Department of Agriculture (August 8, 2012) Colorado Department of Agriculture Investigates Anthrax Case. media release.
- 8. From the Centers for Disease Control and Prevention. Use of anthrax vaccine in response to terrorism: supplemental recommendations of the Advisory Committee on Immunization Practices. JAMA 288: 2681–2682. [DOI] [PubMed] [Google Scholar]
- 9. Brachman P, Gold H, PLotkin S, Fekety F, Werrin M, et al. (1962) Field evaluation of a juman anthrax vaccine. Am J Publ Health 52: 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Poland GA (2006) Human leukocyte antigen haplotypes in the genetic control of immune response to measles-mumps-rubella vaccine. J Infect Dis 193: 655–663. [DOI] [PubMed] [Google Scholar]
- 11. Ovsyannikova IG, Dhiman N, Jacobson RM, Poland GA (2006) Human leukocyte antigen polymorphisms: variable humoral immune responses to viral vaccines. Expert Rev Vaccines 5: 33–43. [DOI] [PubMed] [Google Scholar]
- 12. Wang C, Tang J, Song W, Lobashevsky E, Wilson CM, et al. (2004) HLA and cytokine gene polymorphisms are independently associated with responses to hepatitis B vaccination. Hepatology 39: 978–988. [DOI] [PubMed] [Google Scholar]
- 13. Regnstrom K, Ragnarsson E, Artursson P (2003) Gene expression after vaccination of mice with formulations of diphtheria toxoid or tetanus toxoid and different adjuvants: identification of shared and vaccine-specific genes in spleen lymphocytes. Vaccine 21: 2307–2317. [DOI] [PubMed] [Google Scholar]
- 14. Lee YC, Newport MJ, Goetghebuer T, Siegrist CA, Weiss HA, et al. (2006) Influence of genetic and environmental factors on the immunogenicity of Hib vaccine in Gambian twins. Vaccine 24: 5335–5340. [DOI] [PubMed] [Google Scholar]
- 15. Newport MJ, Goetghebuer T, Marchant A (2005) Hunting for immune response regulatory genes: vaccination studies in infant twins. Expert Rev Vaccines 4: 739–746. [DOI] [PubMed] [Google Scholar]
- 16. Newport MJ, Goetghebuer T, Weiss HA, Whittle H, Siegrist CA, et al. (2004) Genetic regulation of immune responses to vaccines in early life. Genes Immun 5: 122–129. [DOI] [PubMed] [Google Scholar]
- 17. Desombere I, Willems A, Leroux-Roels G (1998) Response to hepatitis B vaccine: multiple HLA genes are involved. Tissue Antigens 51: 593–604. [DOI] [PubMed] [Google Scholar]
- 18. Pajewski NM, Parker SD, Poland GA, Ovsyannikova IG, Song W, et al. (2011) The role of HLA-DR-DQ haplotypes in variable antibody responses to anthrax vaccine adsorbed. Genes Immun 12: 457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pajewski NM, Shrestha S, Quinn CP, Parker SD, Wiener H, et al. (2012) A genome-wide association study of host genetic determinants of the antibody response to Anthrax Vaccine Adsorbed. Vaccine 30: 4778–4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Almal SH, Padh H (2012) Implications of gene copy-number variation in health and diseases. Journal of human genetics 57: 6–13. [DOI] [PubMed] [Google Scholar]
- 21. Willcocks LC, Lyons PA, Clatworthy MR, Robinson JI, Yang W, et al. (2008) Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med 205: 1573–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, et al. (2007) FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet 39: 721–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hollox EJ, Huffmeier U, Zeeuwen PL, Palla R, Lascorz J, et al. (2008) Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet 40: 23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McKinney C, Merriman ME, Chapman PT, Gow PJ, Harrison AA, et al. (2008) Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis 67: 409–413. [DOI] [PubMed] [Google Scholar]
- 25. Nguyen DQ, Webber C, Ponting CP (2006) Bias of selection on human copy-number variants. PLoS genetics 2: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. (2006) Global variation in copy number in the human genome. Nature 444: 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marano N, Plikaytis BD, Martin SW, Rose C, Semenova VA, et al. (2008) Effects of a reduced dose schedule and intramuscular administration of anthrax vaccine adsorbed on immunogenicity and safety at 7 months: a randomized trial. JAMA 300: 1532–1543. [DOI] [PubMed] [Google Scholar]
- 28. Semenova VA, Schiffer J, Steward-Clark E, Soroka S, Schmidt DS, et al. (2012) Validation and long term performance characteristics of a quantitative enzyme linked immunosorbent assay (ELISA) for human anti-PA IgG. J Immunol Methods 376: 97–107. [DOI] [PubMed] [Google Scholar]
- 29. Wineinger NE, Kennedy RE, Erickson SW, Wojczynski MK, Bruder CE, et al. (2008) Statistical issues in the analysis of DNA Copy Number Variations. Int J Comput Biol Drug Des 1: 368–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang K, Li M, Hadley D, Liu R, Glessner J, et al. (2007) PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res 17: 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agilent Technologies Inc. GeneSpring User Manual.
- 32. Chen J, Wang YP (2009) A statistical change point model approach for the detection of DNA copy number variations in array CGH data. IEEE/ACM Trans Comput Biol Bioinform 6: 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Controlled clinical trials 7: 177–188. [DOI] [PubMed] [Google Scholar]
- 35. Viechtbauer W (2010) Conducting Meta-Analyses in R with the metafor Package. Journal of Statistical Software 36: 1–48. [Google Scholar]
- 36. Png E, Thalamuthu A, Ong RT, Snippe H, Boland GJ, et al. (2011) A genome-wide association study of hepatitis B vaccine response in an Indonesian population reveals multiple independent risk variants in the HLA region. Hum Mol Genet 20: 3893–3898. [DOI] [PubMed] [Google Scholar]
- 37. Garman L, Dumas E, Kurella S, Hunt J, Crowe S, et al. (2012) MHC Class II and Non-MHC Class II Genes Differentially Influence Humoral Immunity to Bacillus anthracis Lethal Factor and Protective Antigen. Toxins 4: 1451–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thielmann Y, Weiergraber OH, Ma P, Schwarten M, Mohrluder J, et al. (2009) Comparative modeling of human NSF reveals a possible binding mode of GABARAP and GATE-16. Proteins 77: 637–646. [DOI] [PubMed] [Google Scholar]
- 39. Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, et al. (2009) Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nature genetics 41: 1308–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chauncey KM, Lopez MC, Sidhu G, Szarowicz SE, Baker HV, et al. (2012) Bacillus anthracis’ lethal toxin induces broad transcriptional responses in human peripheral monocytes. BMC immunology 13: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pasqualato S, Renault L, Cherfils J (2002) Arf, Arl, Arp and Sar proteins: a family of GTP-binding proteins with a structural device for ‘front-back’ communication. EMBO reports 3: 1035–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giannuzzi G, Siswara P, Malig M, Marques-Bonet T, Mullikin JC, et al.. (2012) Evolutionary dynamism of the primate LRRC37 gene family. Genome research. [DOI] [PMC free article] [PubMed]
- 43. Latourelle JC, Dumitriu A, Hadzi TC, Beach TG, Myers RH (2012) Evaluation of Parkinson Disease Risk Variants as Expression-QTLs. PloS one 7: e46199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Irvin MR, Shrestha S, Chen YD, Wiener HW, Haritunians T, et al. (2011) Genes linked to energy metabolism and immunoregulatory mechanisms are associated with subcutaneous adipose tissue distribution in HIV-infected men. Pharmacogenet Genomics 21: 798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagasawa M, Kanzaki M, Iino Y, Morishita Y, Kojima I (2001) Identification of a novel chloride channel expressed in the endoplasmic reticulum, golgi apparatus, and nucleus. J Biol Chem 276: 20413–20418. [DOI] [PubMed] [Google Scholar]
- 46. Blumer JB, Chandler LJ, Lanier SM (2002) Expression analysis and subcellular distribution of the two G-protein regulators AGS3 and LGN indicate distinct functionality. Localization of LGN to the midbody during cytokinesis. J Biol Chem 277: 15897–15903. [DOI] [PubMed] [Google Scholar]
- 47. Kim S-Y, Kim J-H, Chung Y-J (2012) Effect of Combining Multiple CNV Defining Algorithms on the Reliability of CNV Calls from SNP Genotyping Data. Genomics & Informatics 10: 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete association results of all CNV tranisition points with early (8 weeks) and peak (30 weeks) antibody responses to anthrax vaccine in 794 European Americans.
(XLSX)
Complete association results of all CNV tranisition points with early (8 weeks) and peak (30 weeks) antibody responses to anthrax vaccine in 200 African Americans.
(XLSX)