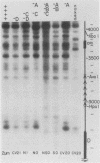
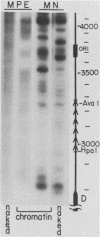
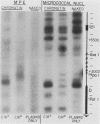
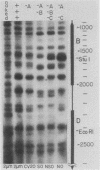
Abstract
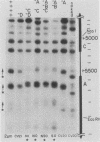
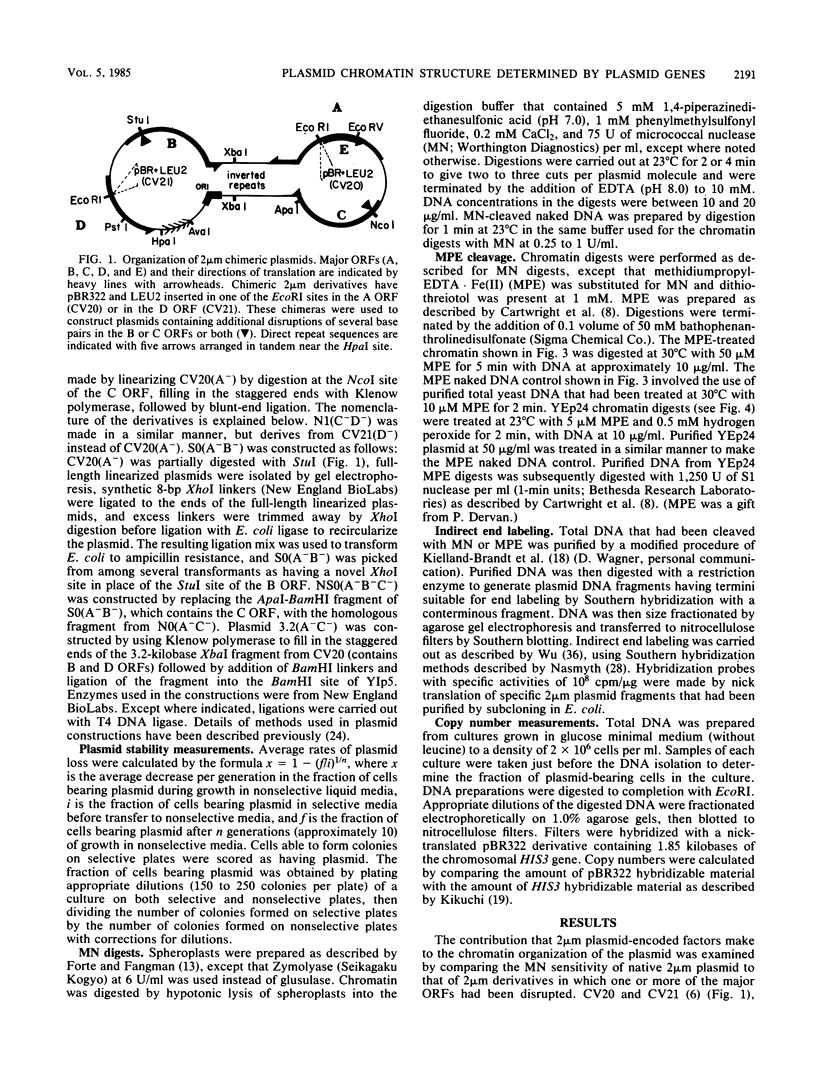
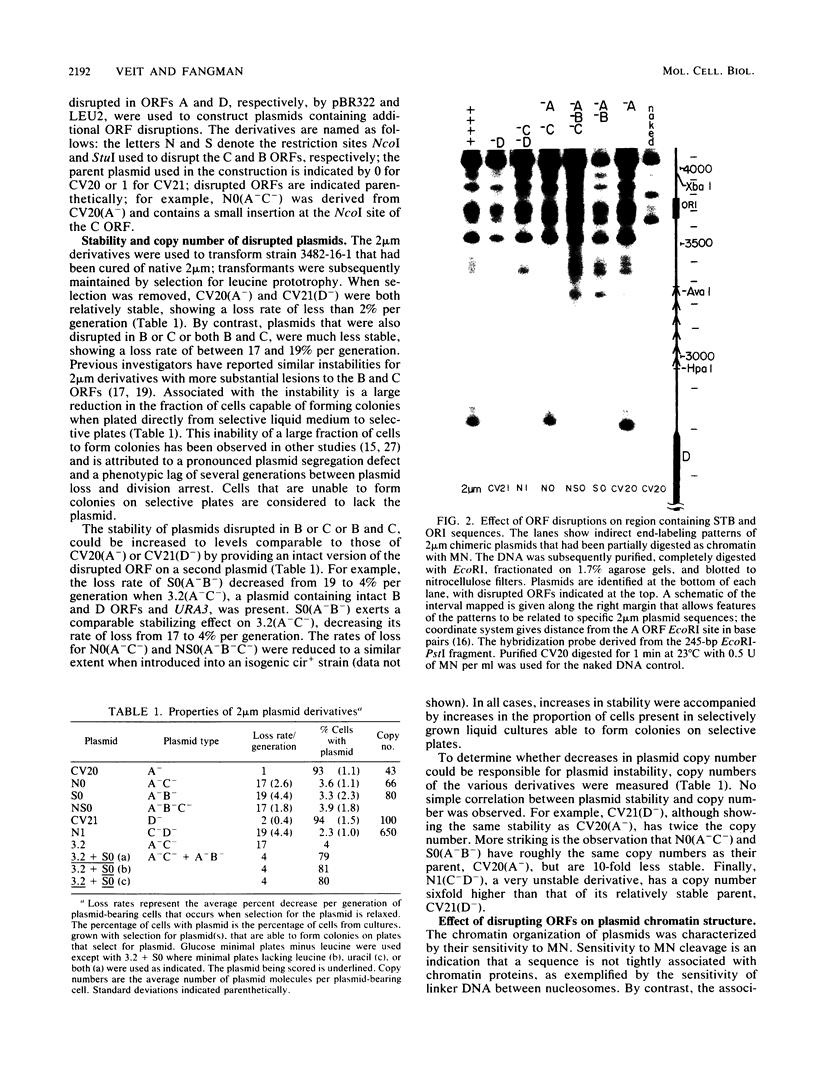
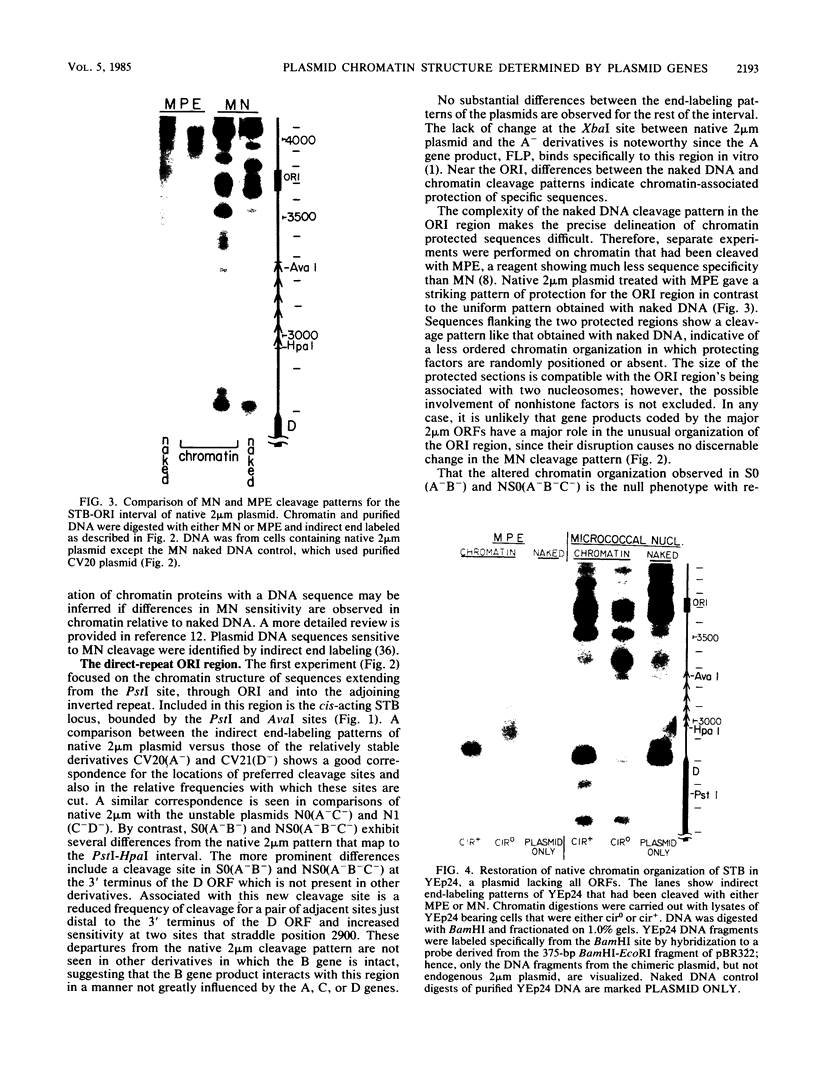
We have used gene disruptions and nuclease probes to assess the roles of yeast 2 micron plasmid genes in plasmid chromatin organization. The chromatin structure at the replication origin is not dependent on any of the four major open reading frames, A, B, C, or D. While stable plasmid maintenance is known to depend on a cis-acting locus STB and genes B and C, we find that only gene B influences STB chromatin. Other interactions between plasmid gene products and sequences may reflect gene regulation: the chromatin organization at the 5' end of gene A, which codes for a site-specific recombinase, depends on both gene B and gene C. Since disruption of gene C results in an increase in plasmid copy number that is dependent on gene A, we propose that gene C (and probably gene B) control copy number by regulating the level of the gene A recombinase.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews B. J., Proteau G. A., Beatty L. G., Sadowski P. D. The FLP recombinase of the 2 micron circle DNA of yeast: interaction with its target sequences. Cell. 1985 Apr;40(4):795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- Botstein D., Falco S. C., Stewart S. E., Brennan M., Scherer S., Stinchcomb D. T., Struhl K., Davis R. W. Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene. 1979 Dec;8(1):17–24. doi: 10.1016/0378-1119(79)90004-0. [DOI] [PubMed] [Google Scholar]
- Brewer B. J., Fangman W. L. Preferential inclusion of extrachromosomal genetic elements in yeast meiotic spores. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5380–5384. doi: 10.1073/pnas.77.9.5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach J. R., Guarascio V. R., Jayaram M. Recombination within the yeast plasmid 2mu circle is site-specific. Cell. 1982 May;29(1):227–234. doi: 10.1016/0092-8674(82)90107-6. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Li Y. Y., Feldman J., Jayaram M., Abraham J., Nasmyth K. A., Hicks J. B. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- Broach J. R. The yeast plasmid 2 mu circle. Cell. 1982 Feb;28(2):203–204. doi: 10.1016/0092-8674(82)90337-3. [DOI] [PubMed] [Google Scholar]
- Cartwright I. L., Hertzberg R. P., Dervan P. B., Elgin S. C. Cleavage of chromatin with methidiumpropyl-EDTA . iron(II). Proc Natl Acad Sci U S A. 1983 Jun;80(11):3213–3217. doi: 10.1073/pnas.80.11.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhart E., Hollenberg C. P. The presence of a defective LEU2 gene on 2 mu DNA recombinant plasmids of Saccharomyces cerevisiae is responsible for curing and high copy number. J Bacteriol. 1983 Nov;156(2):625–635. doi: 10.1128/jb.156.2.625-635.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagrelius T. J., Livingston D. M. Location of DNAase I sensitive cleavage sites in the yeast 2 micron plasmid DNA chromosome. J Mol Biol. 1984 Feb 15;173(1):1–13. doi: 10.1016/0022-2836(84)90400-5. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. Chromatin. Nature. 1978 Jan 12;271(5641):115–122. doi: 10.1038/271115a0. [DOI] [PubMed] [Google Scholar]
- Forte M. A., Fangman W. L. Naturally occurring cross-links in yeast chromosomal DNA. Cell. 1976 Jul;8(3):425–431. doi: 10.1016/0092-8674(76)90155-0. [DOI] [PubMed] [Google Scholar]
- Futcher A. B., Cox B. S. Copy number and the stability of 2-micron circle-based artificial plasmids of Saccharomyces cerevisiae. J Bacteriol. 1984 Jan;157(1):283–290. doi: 10.1128/jb.157.1.283-290.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher A. B., Cox B. S. Maintenance of the 2 microns circle plasmid in populations of Saccharomyces cerevisiae. J Bacteriol. 1983 May;154(2):612–622. doi: 10.1128/jb.154.2.612-622.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Jayaram M., Li Y. Y., Broach J. R. The yeast plasmid 2mu circle encodes components required for its high copy propagation. Cell. 1983 Aug;34(1):95–104. doi: 10.1016/0092-8674(83)90139-3. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y. Yeast plasmid requires a cis-acting locus and two plasmid proteins for its stable maintenance. Cell. 1983 Dec;35(2 Pt 1):487–493. doi: 10.1016/0092-8674(83)90182-4. [DOI] [PubMed] [Google Scholar]
- Livingston D. M. A sequence of the yeast 2 micron DNA plasmid chromosome near the origin of replication is exposed to restriction endonuclease digestion. J Mol Biol. 1982 Sep 25;160(3):397–410. doi: 10.1016/0022-2836(82)90304-7. [DOI] [PubMed] [Google Scholar]
- Livingston D. M., Hahne S. Isolation of a condensed, intracellular form of the 2-micrometer DNA plasmid of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3727–3731. doi: 10.1073/pnas.76.8.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston D. M., Kupfer D. M. Control of Saccharomyces cerevisiae 2microN DNA replication by cell division cycle genes that control nuclear DNA replication. J Mol Biol. 1977 Oct 25;116(2):249–260. doi: 10.1016/0022-2836(77)90215-7. [DOI] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil J. B., Friesen J. D. Expression of the Herpes simplex virus thymidine kinase gene in Saccharomyces cerevisiae. Mol Gen Genet. 1981;184(3):386–393. doi: 10.1007/BF00352510. [DOI] [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Szostak J. W. Pedigree analysis of plasmid segregation in yeast. Cell. 1983 Oct;34(3):961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Nelson R. G., Fangman W. L. Nucleosome organization of the yeast 2-micrometer DNA plasmid: a eukaryotic minichromosome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6515–6519. doi: 10.1073/pnas.76.12.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligy V. L., Thomas D. Y., Miki B. L. Saccharomyces cerevisiae plasmid, Scp or 2 mum: intracellular distribution, stability and nucleosomal-like packaging. Nucleic Acids Res. 1980 Aug 11;8(15):3371–3391. doi: 10.1093/nar/8.15.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdson D. C., Gaarder M. E., Livingston D. M. Characterization of the transmission during cytoductant formation of the 2 micrometers DNA plasmid from Saccharomyces. Mol Gen Genet. 1981;183(1):59–65. doi: 10.1007/BF00270139. [DOI] [PubMed] [Google Scholar]
- Thoma F., Bergman L. W., Simpson R. T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984 Aug 25;177(4):715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Vetter D., Andrews B. J., Roberts-Beatty L., Sadowski P. D. Site-specific recombination of yeast 2-micron DNA in vitro. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7284–7288. doi: 10.1073/pnas.80.23.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. S. Genetic effects of methyl benzimidazole-2-yl-carbamate on Saccharomyces cerevisiae. Mol Cell Biol. 1982 Sep;2(9):1064–1079. doi: 10.1128/mcb.2.9.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Zakian V. A., Brewer B. J., Fangman W. L. Replication of each copy of the yeast 2 micron DNA plasmid occurs during the S phase. Cell. 1979 Aug;17(4):923–934. doi: 10.1016/0092-8674(79)90332-5. [DOI] [PubMed] [Google Scholar]