Abstract
Trithorax and polycomb group proteins antagonistically regulate the transcription of many genes, and cancer can result from the disruption of this regulation. Deregulation of trithorax function occurs through chromosomal translocations involving the trithorax gene MLL, leading to the expression of MLL fusion proteins and acute leukemia. It is poorly understood how MLL fusion proteins block differentiation, a hallmark of leukemogenesis. We analyzed the effect of acute depletion of menin, a close partner of MLL that is critical for MLL and MLL-AF9 recruitment to target genes, on MLL-AF9 leukemia cell differentiation using an in vivo model. We performed cDNA microarray analysis of menin-regulated genes from primary leukemia cells to determine menin-regulated pathways involved in suppressing MLL-AF9 leukemia cell differentiation. We found that menin binds the promoter of the polycomb gene Ezh2, and promotes its expression. EZH2 interacts with the differentiation-promoting transcription factor C/EBPα and represses C/EBPα target genes. Menin depletion reduces MLL binding to the Ezh2 locus, EZH2 expression, and EZH2 binding and repressive H3K27 methylation at C/EBPα target genes, thereby inducing the expression of pro-differentiation C/EBPα targets. In conclusion, our results show that in contrast to its classical role antagonizing trithorax function, the polycomb group protein EZH2 collaborates with trithorax-associated menin to block MLL-AF9 leukemia cell differentiation, uncovering a novel mechanism for suppression of C/EBPα and leukemia cell differentiation, through menin-mediated upregulation of EZH2.
Introduction
Trithorax (Trx) and polycomb (PcG) group proteins have crucial yet opposing roles in the regulation of key genes involved in development and stem cell maintenance.1 The expression of these genes is tightly regulated during cell differentiation, with PcG complexes repressing and Trx promoting transcription.2 PcG and Trx proteins regulate transcription by influencing chromatin structure, in part through covalent modification of histones. One group of genes regulated in this manner is HOX genes.3HOX gene expression is maintained at high levels in hematopoietic progenitors, and is coordinately decreased as these cells differentiate.4
The Trx group protein MLL maintains HOX gene expression in hematopoietic progenitors, at least in part through trimethylation of histone H3 at lysine 4 (H3K4m3) via its C-terminal SET domain.5 Chromosomal translocations involving the MLL gene result in the formation of MLL fusion proteins, which disrupt normal MLL function, causing acute leukemia. MLL translocated leukemias represent approximately 10% of adult acute leukemias and the majority of cases of infant leukemia; these patients have a poor prognosis.6,7
MLL fusion proteins promote the expression of a subset of wild-type (WT) MLL target genes, including HOX genes, through recruitment of the histone H3K79 methyltransferase Dot1L and the pTEFb complex, enhancing transcriptional elongation.8–12 MLL fusion proteins lack a large C-terminal portion of the WT MLL protein, including the histone H3 lysine 4 (H3K4)-methylating SET domain. This functional deficiency is remedied by expression of WT MLL from the non-translocated MLL allele. WT MLL works in concert with MLL fusion proteins to upregulate HOX genes and promote leukemia, highlighting a critical role for WT MLL in this disease.13
WT MLL and MLL fusion proteins are recruited to target genes through binding to menin, which is encoded by the Men1 gene.13–16 X-ray crystallographic studies have recently shown that menin interacts with the identical N-terminal sequences of both WT MLL and MLL fusion proteins via a deep central pocket, demonstrating that menin is a close partner of these proteins.17,18 This interaction is required for leukemic transformation, highlighting a central role for menin in MLL fusion protein leukemia.19
A major hallmark of leukemia and a consequence of MLL fusion protein expression is a block in hematopoietic differentiation.20 MLL-AF9 (MA9) leukemia cells have a block in the myeloid lineage at the granulocyte-macrophage progenitor stage, with cells expressing high levels of the cell surface receptor c-kit being enriched for leukemia-initiating cells.21–24 While HOX genes are at least partially responsible for this suppression of differentiation, it remains unclear how MLL-fusion protein leukemia cells are blocked at the progenitor stage.25,26 In addition, global analysis has identified over 200 direct MLL fusion protein target genes, some of which could also have a role in blocking differentiation.12
C/EBPα is a leucine zipper transcription factor that promotes myeloid differentiation in part through the activation of differentiation-associated genes. Many leukemogenic oncogenes and pathways inhibit the expression/function of C/EBPα as a means of blocking differentiation, including Bcr-Abl, AML-ETO, and Notch/Trib2.27 However, little is known as to whether repression of C/EBPα is involved in blocking differentiation in MLL fusion protein leukemias.
Polycomb repressive complex 2 (PRC2) consists of Suz12, EED, RbAp46/48, and the catalytic component EZH2, which methylates H3K27, leading to transcriptional repression. EZH2 point mutations are found in about 10% of cases of myelodysplastic syndrome, suggesting that PRC2 acts as a tumor suppressor in the myeloid lineage.28,29 However, recent work has demonstrated that ectopic expression of EZH2 causes a block in myeloid differentiation, leading to the development of myeloproliferative disease.30 Thus, the role of EZH2 in hematopoietic development and leukemia is not well understood. Utilizing murine models for MA9 leukemia, we set out to examine the role of the Trx protein MLL and its partner, menin, in regulating MA9 cell differentiation in vivo, and found that the polycomb protein EZH2 is a collaborating factor in suppressing C/EBPα and differentiation in MA9-induced leukemia.
Design and Methods
Cell culture
AT-1 cells were generated from bone marrow cells of Men1f/f;Cre-ER mice by transduction with retrovirus expressing MA9 and cultured in medium with 15% fetal bovine serum for long-term myeloid culture (Cat #06500, Stem Cell Technologies) and 10 ng/mL interleukin-3.16 In AT-1 cells, Men1f/f was excised by treating the cells with 4-OHT (200 nM). THP-1 cells were maintained in RPMI-1640 containing 10% fetal bovine serum and 1% Pen/Strep supplemented with 0.05 mM 2-mercaptoethanol. HEK293T and RAW264.7 cells were cultured in DMEM supplemented with 10% fetal bovine serum and 1% Pen/Strep.
Murine transplantation experiments with Men1 or WT Mll excision
Men1f/f; Cre-ER or Mllf/f; Cre-ER mice (6–8 weeks old) were injected with 5-FU and sacrificed 5 days later for bone marrow isolation. Bone marrow was pre-stimulated with a mixture of cytokines as previously described.16 Cells were then transduced with MLL-AF9-ires-GFP and 106 cells were transplanted retro-orbitally into lethally irradiated (900 rads) recipient mice (BL6xSJLF1, Taconic) with 2.5×105 normal bone marrow cells. Mice with a high percentage of GFP+ in the peripheral blood (21–24 days post-transplant) were gavaged with corn oil control or corn oil with tamoxifen (200 mg/kg body wt) (MP Biomedicals) daily over a 5-day period, with no gavage on the third day. Organs from mice were fixed, processed and stained with hematoxylin and eosin for microscopic analysis.
Chromatin immunoprecipitation assay, immunoprecipitation, and antibodies
Chromatin immunoprecipitation (ChIP) assays were performed as previously described using the Imgenex kit.16 Briefly, 106-107 formaldehyde cross-linked cells were lysed and sonicated to obtain sheared DNA. This lysate was then incubated with control IgG or an antigen-specific antibody overnight, then bound to beads and washed. Samples were eluted from the beads and incubated at 65°C overnight to reverse crosslinking. Eluted DNA was quantified using the 7500 fast real-time polymerase chain reaction (PCR) system (Applied Biosystems) with the Quantitect Sybr Green kit (Qiagen), and normalized to input DNA, as well as total H3 for histone modifications. Oligos used for ChIP real-time PCR are provided in the Online Supplementary Material.
RNA isolation and quantitative real-time polymerase chain reaction
RNA was isolated using Trizol (Ambion) and the RNeasy kit (Qiagen). One microgram of total RNA was used to make cDNA with the SS III RT system (Invitrogen), and real-time PCR was performed using the 7500 fast real-time PCR system (Applied Biosystems) and Quantitect Sybr Green kit (Qiagen). Transcript levels were normalized to GAPDH between samples, and relative quantity was calculated using the ΔΔCt method.
Results
Acute menin depletion causes MA9 cell differentiation in culture
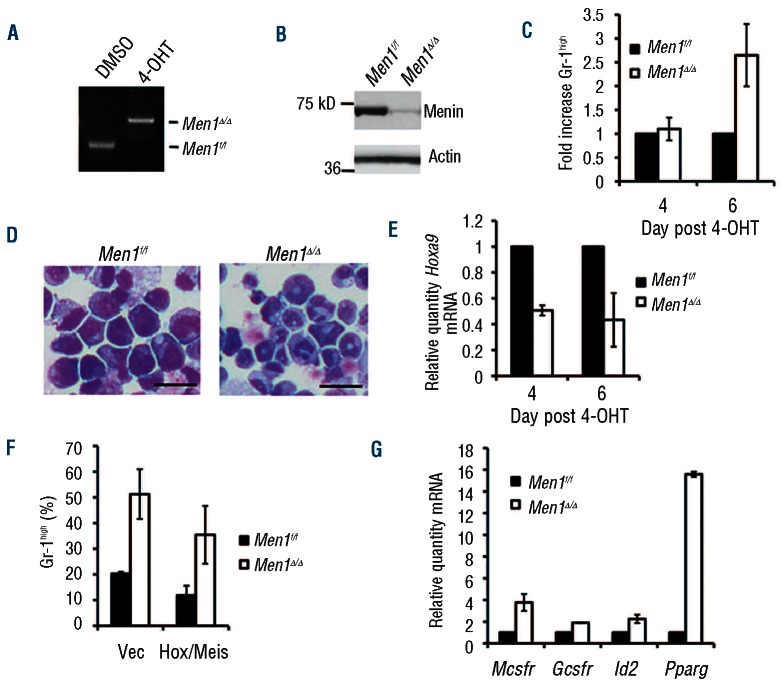
To determine the effect of acute menin depletion on MA9 cell differentiation, we utilized the murine MA9 cell line AT-1, which contains floxed Men1 alleles and the Cre-ER transgene, allowing Men1 excision by addition of 4-hydroxytamoxifen (4-OHT) to the culture medium.16 4-OHT treatment effectively excised the floxed Men1 alleles within 2 days (Figure 1A), and markedly reduced menin protein expression by day 4 (Figure 1B). We measured cell differentiation by flow cytometry using the terminal myeloid differentiation marker Gr-1, and found little effect on Gr-1 cell surface expression 4 days after Men1 excision, but a marked increase in the percentage of cells expressing high levels of Gr-1 by day 6 (Figure 1C, Online Supplementary Figure S1A), indicating that enhanced terminal myeloid differentiation follows reduced menin expression. This increase in Gr-1high cells also corresponded with a change in cell morphology consistent with myeloid differentiation (Figure 1D, right).
Figure 1.
Acute menin depletion leads to MA9 cell differentiation in culture. (A) Genotype for Men1 excision in AT-1 cells 2 days after 4-OHT treatment. (B) Western blot for menin in control and Men1-excised AT-1 cells 4 days after 4-OHT treatment. (C) Flow cytometry for Gr-1 cell surface expression in control and Men1-excised AT-1 cells. (D) Wright-Giemsa staining of control and Men1-excised AT-1 cells 6 days after 4-OHT treatment. (E) Real-time PCR examining Hoxa9 transcript levels in control and Men1-excised AT-1 cells. (F) Flow cytometry for Gr-1 cell surface expression in control and Men1-excised AT-1 cells with or without Hoxa9/Meis1 overexpression day 6 after 4-OHT treatment. (G) Real-time PCR examining transcript levels of genes associated with differentiation in control and Men1-excised AT-1 cells.
The ability to observe cell differentiation in this setting then allowed us to explore how menin depletion affects genes that regulate this process over time. The direct menin/MA9 target gene Hoxa9, which is at least partially responsible for the MA9-mediated differentiation block,25,26 was decreased as early as day 4 after 4-OHT treatment, prior to cell differentiation (Figure 1E). However, overexpression of Hoxa9 in combination with its cofactor Meis1 (Online Supplementary Figure S1B-C) did not completely rescue differentiation caused by menin depletion (Figure 1F), suggesting an additional function for menin in suppressing MA9 leukemia cell differentiation. Mcsfr, Gcsfr, Id2 and Pparg, genes associated with myeloid differentiation, were upregulated by day 6 after 4-OHT treatment (Figure 1G). To determine whether menin is also important for blocking the differentiation of human MLL fusion protein-expressing leukemia cells, we used shRNA targeting MEN1 in the human MA9 cell line THP-1. Menin knockdown increased cell surface expression of CD11b, a prominent myeloid differentiation marker in human leukemia cells, and also increased MCSFR transcript levels (Online Supplementary Figure S1D-E). Together, these results indicate that menin depletion upregulates genes associated with myeloid differentiation and causes MA9 cell differentiation.
Acute menin depletion causes MA9 cell differentiation in vivo
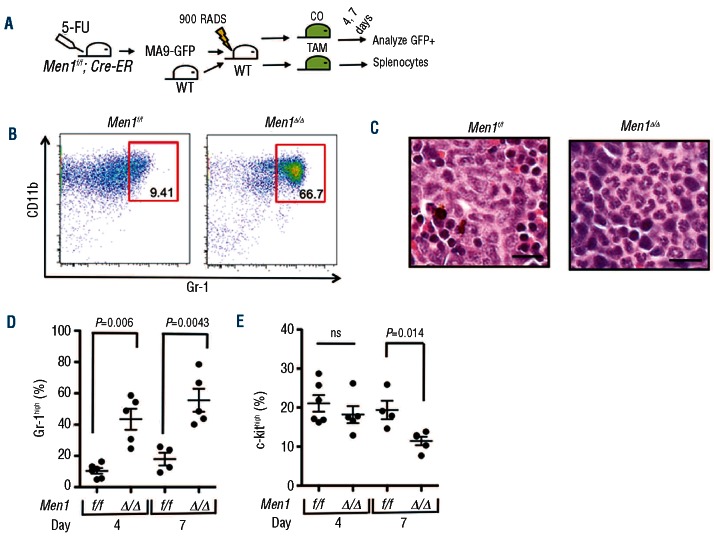
To determine whether menin depletion is pathologically relevant to MA9-induced leukemia, we transduced Men1f/f; Cre-ER bone marrow with MA9-ires-GFP retrovirus and transplanted these cells into lethally irradiated recipient mice (Figure 2A). When we observed substantial GFP+ cells in the peripheral blood of recipient mice, we treated the mice with either corn oil as the control or corn oil with tamoxifen by oral gavage to excise Men1 in the transplanted cells. At days 4 and 7 after the initial gavage we isolated splenocytes from each treatment group for analysis by flow cytometry. Flow cytometry analysis revealed a striking increase in the percentage of Gr-1high MA9 (GFP+) cells in Men1-excised splenocytes as early as 4 days after the initial gavage, and this effect became even more pronounced at day 7 (Figure 2B,D). Consistent with this increased Gr-1high population, we observed mature differentiated myeloid cells in hematoxylin and eosin-stained spleen sections from mice treated with tamoxifen to deplete menin (Figure 2C). In contrast to the immediate increase in the percentage of Gr-1high cells, the c-kithigh population, which is enriched for leukemia-initiating cells, was unchanged at day 4 post-treatment, but decreased significantly by day 7 (Figure 2E). Further analysis of the relationship between c-kit and Gr-1 revealed an increase in the c-kithigh/Gr-1high population at day 4 (Online Supplementary Figure S2A), followed by a loss of this population at day 7 (Online Supplementary Figure S2B). We also observed increased staining for the apoptotic marker annexin V at day 7 (Online Supplementary Figure S2C). These results suggest that as a consequence of menin depletion, c-kithigh cells first gain expression of Gr-1, then either lose c-kit expression during the process of differentiation or undergo cell death, leading to the depletion of c-kithigh MA9 leukemia-initiating cells.
Figure 2.
Menin depletion results in MA9 cell differentiation in vivo. (A) A schematic for examining the acute effect of menin depletion on MA9 cells in vivo. Men1f/f; Cre-ER bone marrow was transduced with MA9-ires-GFP and transplanted into lethally irradiated recipients. Recipient mice were treated with corn oil (CO) as a control or with CO plus tamoxifen (TAM) to excise the floxed Men1 allele. GFP+ cells were analyzed for MA9 cell immunophenotype. (B) FACS plot showing increased Gr-1high percentage in Men1-excised MA9 primary cells 7 days after-initial TAM treatment. (C) Hematoxylin and eosin staining of spleen sections from control (left) and TAM-treated (right) Men1f/f; Cre-ER MA9 primary recipients 7 days after-initial TAM treatment. (D–E) A summary of the Gr-1high (D) and c-kithigh (E) MA9 cell population in response to menin depletion 4 and 7 days after-initial TAM treatment.
WT MLL depletion causes MA9 cell differentiation in vivo
Due to the observed effect of menin depletion on MA9 cells, we hypothesized that acute depletion of WT MLL, which interacts with menin and is required to maintain MA9-mediated transformation,13 would also cause MA9 cell differentiation in vivo. To test this hypothesis, we isolated control and WT Mll-excised MA9 (GFP+) splenocytes for plating in methylcellulose and performed flow cytometry to evaluate leukemia cell differentiation status. WT MLL-depleted MA9 splenocytes were deficient in colony formation in methylcellulose as compared to their WT MLL-containing counterparts, suggesting that MA9 cells lacking WT MLL are deficient in leukemia-initiating cells when derived from an in vivo setting (Online Supplementary Figure S3A). Consistent with the effect of menin depletion, flow cytometry analysis of MA9 splenocytes lacking WT MLL demonstrated a significant increase in the percentage of Gr-1high cells, and a significant decrease in the c-kithigh population compared to that in controls, suggesting that MA9 cells also undergo differentiation in response to WT MLL depletion (Online Supplementary Figure S3B-E). These findings demonstrate that a key Trx group gene, Mll, is critical for blocking MA9 leukemia cell differentiation.
Given that WT MLL depletion caused a less pronounced increase in Gr-1high MA9 cells than that of menin, we wondered whether Gr-1low cells lacking WT MLL were defective in propagating leukemic disease. To address this question, we isolated control and WT Mll-excised MA9 splenocytes (Online Supplementary Figure S4A-B) from primary recipients, sorted into either Gr-1low or Gr-1high MA9 cell populations, and transplanted these cells into lethally irradiated secondary recipient mice. WT MLL-depleted Gr-1low recipients had a significantly longer survival rate than the WT MLL-containing Gr-1low cohort (Online Supplementary Figure S3F), likely due to the decreased population of c-kithigh cells in this group (Online Supplementary Figure S3E). This survival effect may have been less pronounced due to the eventual outgrowth of MA9 cells resistant to Mll excision in about half of the tamoxifen-treated Gr-1low recipients (Online Supplementary Figure S4C). Consistent with previously published results, Gr-1high recipient mice from each treatment group had significantly longer survival than their Gr-1low counterparts (Online Supplementary Figure S3F).24 Together, these data demonstrate a role for WT MLL in maintaining a population of Gr-1low/c-kithigh MA9 cells, which possess the ability to propagate leukemic disease.
Menin depletion leads to the upregulation of C/EBPα target genes in MA9 cells in vivo
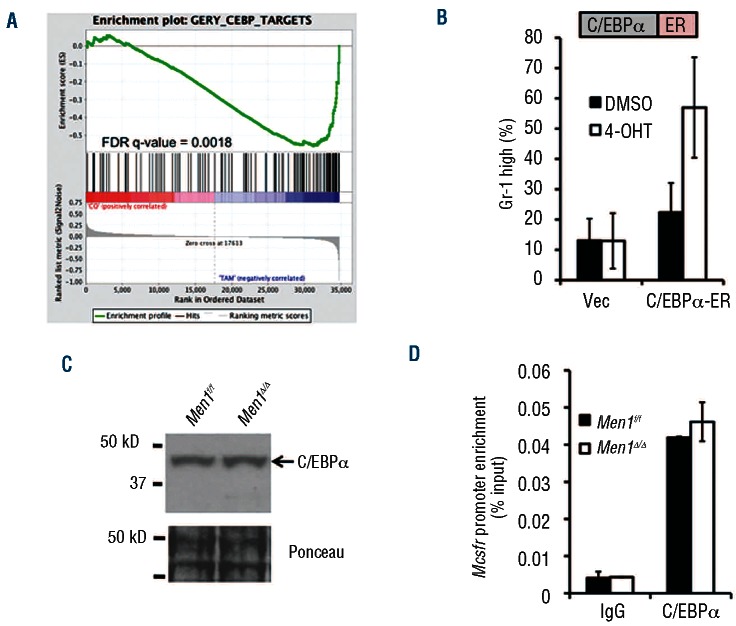
To investigate a potential mechanism for the menin/WT MLL-mediated block in myeloid differentiation, we performed a cDNA microarray using primary GFP+ MA9 splenocytes from either control or Men1-excised recipient mice. We isolated cells 4 days after initial gavage to determine genes affected by menin depletion, and performed gene set enrichment analysis of the microarray data to identify groups of genes regulated by menin that are associated with differentiation. This analysis revealed a significant overlap of menin-regulated genes with C/EBP transcription factor target genes (Figure 3A, Online Supplementary Figure S5), consistent with up-regulation of C/EBPα target genes Mcsfr, Gcsfr, Id2 and Pparg in AT-1 cells, leading us to investigate a role for menin in repressing C/EBPα function.
Figure 3.
Men1 excision leads to C/EBPα target gene upregulation but does not affect C/EBPα expression. (A) Gene set enrichment analysis analysis comparing Men1f/f; Cre-ER MA9 primary cells treated with tamoxifen in vivo to the C/EBP target data set. (B) Flow cytometry analysis of Gr-1 cell surface expression in vector or C/EBPα-ER transduced AT-1 cells 2 days after 4-OHT treatment. (C) Western blot for C/EBPα expression in control or Men1-excised AT-1 cells 6 days after 4-OHT treatment. (D) ChIP for C/EBPα enrichment at the Mcsfr promoter in control or Men1-excised AT-1 cells 6 days after 4-OHT treatment.
As multiple leukemogenic pathways affect the expression level of C/EBPα p42 or increase the ratio of the dominant negative p30 isoform over p42,27 we first explored whether menin depletion affected C/EBPα expression or target gene binding in AT-1 cells. Although overexpression of C/EBPα-ER followed by activation via addition of 4-OHT to the culture medium was able to drive differentiation (Figure 3B), we failed to observe a menin-dependent effect on C/EBPα protein levels (Figure 3C). In addition, there was no detectable expression of C/EBPα p30, which has been reported to promote leukemia development (Figure 3C).32 ChIP assay indicated that C/EBPα was able to bind the promoter of its target gene Mcsfr regardless of menin expression (Figure 3D). The lack of a direct effect on C/EBPα protein led us to investigate a potential role for menin in actively repressing C/EBPα target genes
Menin promotes EZH2 expression in MA9 cells
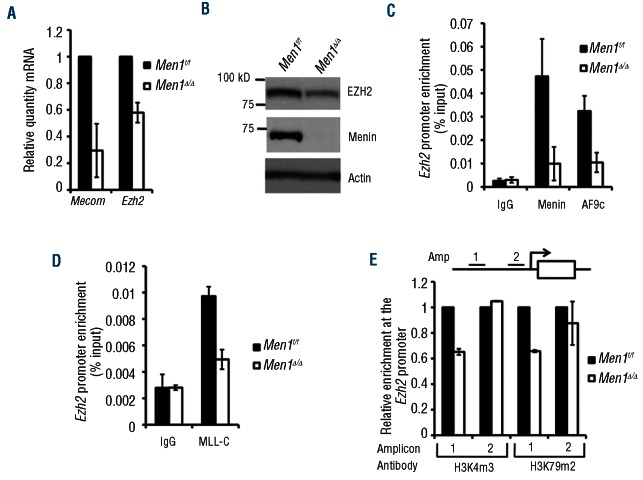
In exploring potential repressors of C/EBPα function that may be regulated by menin/WT MLL, we observed that Mecom, which encodes the transcription factor Evi-1, was the most down-regulated gene in response to acute menin depletion in vivo and was also decreased in Men1-excised AT-1 cells (Figure 4A). Evi-1 is directly activated by MA9, and is required for the maintenance of MLL-ENL transformed cells.33 An Evi-1 oncogenic fusion protein has been shown to repress C/EBPα at target genes, and Evi-1 has been reported to mediate transcriptional repression of the tumor suppressor PTEN through its interaction with PcG proteins, including EZH2.34–36
Figure 4.
Menin promotes EZH2 expression in MA9 cells. (A) Real-time PCR analysis of Mecom and Ezh2 transcript levels in control or Men1-excised AT-1 cells 6 days after 4-OHT treatment. (B) Western blot for EZH2 in control or Men1-excised AT-1 cells 6 days after 4-OHT treatment. (C–D) ChIP assays for menin, AF9c (C), and MLL-C (D) binding at the Ezh2 promoter in control or Men1-excised AT-1 cells 6 days after 4-OHT treatment. (E) ChIP assays for H3K4m3 and H3K79m2 at the Ezh2 promoter 6 days after 4-OHT treatment.
Surprisingly, we found that expression of EZH2, the catalytic component of polycomb repressive complex 2 (PRC2), was also decreased upon menin depletion in MA9-expressing cells (Figure 4A-B), while expression of the closely related EZH1 was unaffected (Online Supplementary Figure S6B). This effect on EZH2 occurs independently of Hoxa9/Meis1, as overexpression of these genes is unable to rescue the decrease in Ezh2 transcript levels due to menin depletion (Online Supplementary Figure S6A). These findings suggest that in addition to Mecom, Ezh2 is a direct target of menin/MA9. Along these lines, ChIP assay showed enrichment for menin and the portion of AF9 found in MA9 (AF9c) at the Ezh2 and Mecom promoters. As expected, this enrichment was diminished by menin depletion (Figure 4C). ChIP using an MLL-C antibody, which specifically detects WT MLL, found that WT MLL also bound the Ezh2 promoter in a menin-dependent fashion (Figure 4D). In addition, H3K4m3 and H3K79m2, which are associated with upregulation of gene transcription,1,37 were reduced at a certain part of the Ezh2 locus (Figure 4E). Together, these results demonstrate that menin promotes EZH2 expression, suggesting that menin/WT MLL/MA9 promote EZH2-mediated repression of C/EBPα target genes, leading to a block in MA9 leukemia cell differentiation.
EZH2 interacts with C/EBPα and represses its transcriptional activity
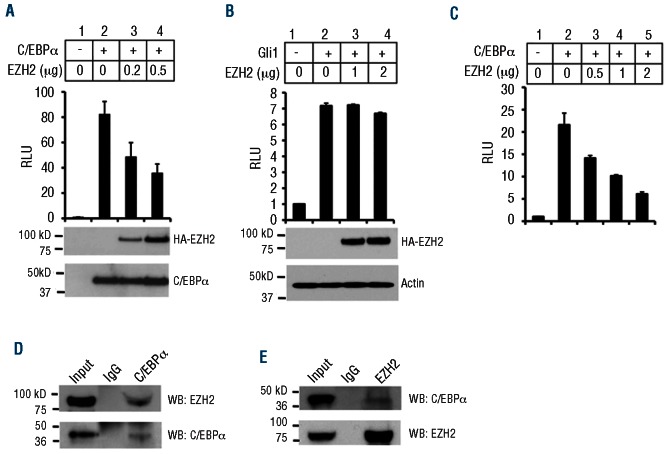
To test whether EZH2 inhibits C/EBPα function, we first determined whether EZH2 could repress C/EBPα-mediated transcriptional activation. To this end, we transfected 293T cells with a C/EBPα binding site-containing promoter-driven luciferase reporter,31 C/EBPα, and increasing amounts of EZH2 expression plasmids. C/EBPα robustly activated the luciferase reporter, and EZH2 repressed C/EBPα-mediated activation in a dose-dependent manner (Figure 5A). As a control, EZH2 was unable to repress Gli1-activated luciferase (Figure 5B). EZH2 also repressed C/EBPα-activated luciferase in myeloid (RAW264.7) cells (Figure 5C). As EZH2 could repress C/EBPα-mediated activation, we decided to test the possibility that EZH2 physically interacts with C/EBPα in MA9 cells by performing immunoprecipitation experiments using endogenously expressed proteins in THP-1 cells. While it is unclear whether C/EBPα directly or indirectly binds EZH2, immunoprecipitation of C/EBPα was able to bring down EZH2 (Figure 5D), and immunoprecipitation of EZH2 pulled down C/EBPα (Figure 5E), showing that these proteins physically interact at endogenous levels in MA9 leukemia cells.
Figure 5.
EZH2 interacts with C/EBPα in MA9 cells and represses C/EBPα target genes. (A) Luciferase assay in HEK 293T cells with a C/EBPα binding site-containing promoter-driven luciferase plasmid, C/EBPα and increasing amounts of EZH2. (B) Luciferase assay in HEK 293T cells with a Gli-1 binding site-containing promoter-driven luciferase plasmid, Gli-1 and increasing amounts of EZH2. (C) Luciferase assay in RAW264.7 cells with a C/EBPα binding site-containing promoter-driven luciferase plasmid, C/EBPα and increasing amounts of EZH2. (D) Immunoprecipitation (IP) for C/EBPα followed by western blotting for EZH2 (top) or C/EBPα (bottom). (E) IP for EZH2 followed by western blotting for C/EBPα (top) and EZH2 (bottom) in THP-1 cells.
EZH2 binds C/EBPα target genes and suppresses MA9 cell differentiation
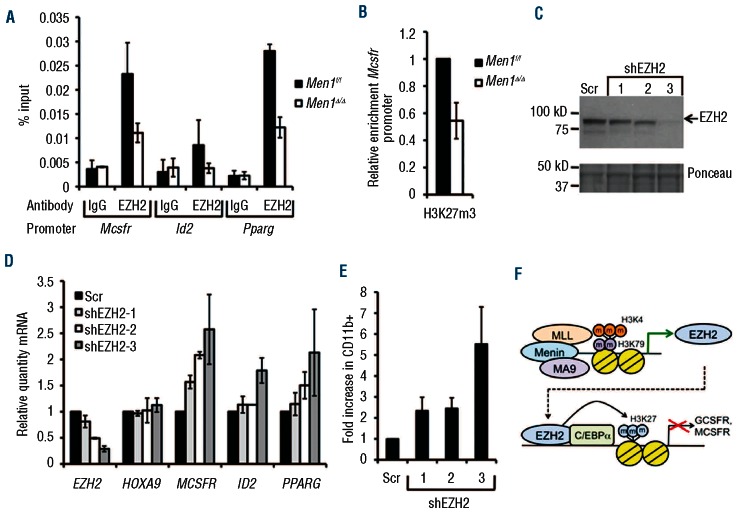
Since EZH2 interacts with C/EBPα in MA9 cells and represses its transcriptional activity, we reasoned that EZH2 could bind C/EBPα target genes to mediate direct repression. To explore this possibility, we performed ChIP to determine whether EZH2 bound to the promoter of C/EBPα target genes in AT-1 cells. ChIP assay showed that EZH2 binding was enriched at the Mcsfr, Id2, and Pparg loci, and menin depletion by addition of 4-OHT led to decreased promoter occupancy of EZH2 (Figure 6A). As a control, menin did not bind the Mcsfr promoter (Online Supplementary Figure S6D). As EZH2 mediates transcriptional repression through catalysis of histone H3 lysine 27 trimethylation (H3K27m3), we also tested whether H3K27m3 is reduced at the Mcsfr promoter in response to Men1 excision. Consistent with EZH2 ChIP results, H3K27m3 was decreased at the Mcsfr promoter, while H3K4m3 was increased in menin-depleted cells (Figure 6B, Online Supplementary Figure S6C), supporting a model in which EZH2 interacts with C/EBPα at target gene promoters, leading to increased H3K27m3 and transcriptional repression of C/EBPα target genes in MA9 cells.
Figure 6.
EZH2 knockdown induces MA9 cell differentiation. (A) ChIP assay for EZH2 enrichment at the Mcsfr, Id2, and Pparg loci in control or Men1-excised AT-1 cells 6 days after 4-OHT treatment. (B) ChIP assay for H3K27m3 enrichment at the Mcsfr promoter in control or Men1-excised AT-1 cells 6 days after 4-OHT treatment. (C) Western blot for EZH2 expression in Scr control and EZH2 knockdown THP-1 cells. (D) Real-time PCR analysis of Scr control and EZH2 KD THP-1 cells for C/EBPα target genes. (E) Flow cytometry analysis of CD11b cell surface expression in Scr and EZH2 knockdown THP-1 cells. (F) A model for the role of EZH2 in MA9 leukemia.
Since EZH2 occupies the Mcsfr promoter in MA9 cells and inhibits C/EBPα-mediated transcriptional activation, we tested whether loss of EZH2 causes upregulation of C/EBPα target genes and MA9 cell differentiation using shRNA to knock down EZH2 in THP-1 cells. Transduction of THP-1 cells with each of three different shRNA resulted in a reduction of EZH2 expression compared to the scrambled (Scr) control (Figure 6C-D). EZH2 knockdown led to a dose-dependent increase in MCSFR, ID2, and PPARG transcript levels, but did not affect HOXA9 expression (Figure 6D). EZH2 knockdown also caused a dose-dependent increase in the percentage of CD11b positive cells and decrease in cell growth (Figure 6E, Online Supplementary Figure S7). Taken together, these results highlight EZH2 as a necessary component for the MA9-mediated block in myeloid differentiation, revealing a novel mechanism by which myeloid differentiation is inhibited via EZH2-mediated repressive histone methylation.
Discussion
Depletion of the Trx protein MLL or its partner menin triggers MA9 cell differentiation
One of the major mechanisms for MA9-mediated leukemogenesis is causing a block in mature myeloid differentiation,38 but it is poorly understood how MA9 cells are blocked in their mature differentiation. Trx and PcG proteins are well known for their antagonizing function in regulating gene expression, but there is some evidence that they cooperate to promote leukemia.39,40 However, little is known as to how Trx/PcG proteins might work together to regulate differentiation in acute myeloid leukemia.
We found that excision of either Men1 or Mll in MA9-expressing leukemia cells in vivo significantly increased the mature myeloid differentiation of MA9 leukemia cells and decreased the population of c-kithigh cells, which are enriched for leukemia-initiating cells. In addition to c-kit, Gr-1 expression is a functional indicator, as Gr-1high cells are deficient in the ability to cause transplantable leukemic disease (Online Supplementary Figure S3F).
Interestingly, menin depletion increased the Gr-1high population as early as 4 days after the initial tamoxifen treatment, before the c-kithigh population was diminished (Figure 2D-E). Closer examination of flow cytometry data revealed an increase in c-kithigh cells expressing Gr-1 at the day 4 time point, and this population was lost at day 7, either due to decreased c-kit expression or apoptotic cell death, as annexin V staining was also increased as a result of menin loss at day 7 (Online Supplementary Figure S2C). These results suggest that increased Gr-1 expression is an early indicator of MA9 cell differentiation, followed by c-kit loss and/or cell death following Men1 deletion. While Hoxa9 and Meis1 are well known for their crucial role in MLL- fusion protein-induced leukemia,25,26,41 we found that menin blocks the mature differentiation of MA9 leukemia cells independently of Hoxa9/Meis1 (Figure 1F). However, Hoxa9/Meis1 overexpression is able to compensate for defects in colony formation in methylcellulose due to menin depletion.16 It is possible that in more stressed conditions, such as liquid culture, Hoxa9/Meis1 alone are unable to compensate for menin loss.
The acute effect of menin depletion in MA9 cells had a more drastic effect than that of WT MLL on differentiation and apoptosis. WT MLL knockdown in human MA9 cells causes decreased H3K79m2 at target genes, suggesting that WT MLL is required for MA9 recruitment.13 Additionally, in MLL-null MEF cells, MA9 is unable to bind the Hoxa9 promoter.42 However, there is no physical interaction between WT MLL and MA9, suggesting that MA9 recruitment resulting from WT MLL function at target gene promoters is an indirect process. In contrast, menin directly binds both WT MLL and MA9 via their common N-terminal domains and is required for their recruitment to target genes. The acute effect of menin loss could be more drastic than that of WT MLL loss alone, because menin is directly responsible for the recruitment of both WT MLL and MA9 to target genes to enhance their transcription.
Menin promotes EZH2 transcription in MA9 leukemia cells
Little is known about the role of Trx complex components, menin and MLL in regulating the expression of their rival polycomb genes. We observed a substantial reduction in EZH2 expression following Men1 excision (Figure 4A-B). Menin, AF9c, and WT MLL bind the Ezh2 promoter, and their binding is decreased in response to menin loss (Figure 4C-D), suggesting that menin recruits MA9 and WT MLL to the Ezh2 promoter to upregulate EZH2 expression. It is not yet clear whether menin regulates the expression of EZH2 in other leukemias or normal hematopoietic cells, or why menin might promote the expression of protein complexes that oppose its function at common target genes. One possibility is that during development or in stem/progenitor cells, menin/MLL promote the expression of PRC components to preserve “bivalent” histone methylation, with both H3K4m3 and H3K27m3 at relevant promoters, leaving these genes repressed, but poised for activation.2 The maintenance of bivalency is critical for the regulated expression of these genes, and may be the rationale underlying menin/MLL-mediated activation of PRC protein expression in a developmental context.
EZH2 represses C/EBPα target genes in MA9 leukemia cells
C/EBPα is a critical transcription factor that controls myeloid differentiation,43 and its normal function is inhibited through mutation, aberrant expression, or oncogene-mediated suppression, contributing to a block in myeloid differentiation in various leukemias.43,44 Little is known about whether and/or how C/EBPα is regulated in MA9 leukemia cells. Forced expression of C/EBPα drives MLL-fusion protein cell differentiation (Figure 3B),45 but C/EBPα protein levels and target gene promoter binding are unchanged in response to menin depletion (Figure 3C-D), suggesting an alternative method for menin-mediated repression of C/EBPα target genes in MA9 leukemia cells.
We have found that EZH2 physically associates with C/EBPα, binds to the promoter of C/EBPα target genes in MA9 cells, and represses C/EBPα target genes (Figures 5A-E, 6A). Menin does not affect C/EBPα expression. Rather, menin induces expression of EZH2, which then suppresses the expression of C/EBPα targets and blocks the differentiation of MA9 cells. These findings reveal a previously unappreciated mechanism for suppressing C/EBPα and MA9 leukemia cell differentiation (Figure 6F).
Consistent with our findings, recent reports describe that EZH2 depletion causes primary leukemia cell differentiation.46–48 However, EZH2 knockout seems to have a less severe effect on MA9 cells in vivo.46 One potential reason for this milder effect is partial compensation for loss of EZH2 function by the closely related EZH1 protein, which can also be found in the PRC2 complex and catalyzes H3K27 methylation. In fact, depletion of the core PRC2 component EED, which is essential for both EZH1 and EZH2 function, more effectively extends the life span of MA9 leukemic mice than does EZH2 knockout.46 In addition, combined knockdown of EZH1 and EZH2 more effectively reduces MA9 cell growth than EZH2 knockdown alone,47 providing further evidence that EZH1 can at least partially compensate for EZH2 loss under certain conditions.
The role of EZH2 in regulating C/EBPα function during normal hematopoiesis and in other leukemia types remains to be examined. Although EZH2 is required for stem/progenitor cell expansion in developmental hematopoiesis, EZH2 depletion in adult bone marrow is less severe, with no effect on LSK cells, but impaired differentiation of lymphoid cells, and increased myelo-erythroid progenitors, suggesting that EZH2 does regulate at least some stages of myeloid differentiation.49 Future studies will clarify whether EZH2 specifically regulates C/EBPα function in MA9 cells or in a broader spectrum of cell types.
A context-dependent role for EZH2 in hematopoietic malignancies
The role of EZH2 in hematopoietic malignancies is complex, and seems to be context-dependent. Mono- or bi-allelic mutations were found in 12% of patients with myelodysplastic/myeloproliferative disorders, suggesting that EZH2 may be a tumor suppressor in the myeloid lineage.28,29 However, in lymphomas of germinal center origin, a specific recurring point mutation in the SET domain of EZH2, converting tyrosine 646 to cytosine has been observed.50,51 This mutation causes increased H3K27 methyltransferase activity, suggesting that EZH2 function promotes tumorigenesis in this disease.52,53 EZH2 is involved in suppressing PTEN expression and promoting AKT signaling in Evi-1-induced leukemia cells,36 yet we found that EZH2 knockdown did not affect PTEN expression in MA9 leukemia cells, suggesting an alternative mechanism for EZH2 in regulating MA9 cell differentiation.
Consistent with the role of EZH2 in regulating differentiation, EZH2 has been reported to promote MA9 leukemias.46,47 It has been unclear as to how mature differentiation of MA9-induced leukemia is blocked. We found for the first time that in MA9-induced leukemia, menin plays a key role in promoting the expression of the polycomb protein EZH2. EZH2 cooperates with menin to epigenetically suppress the expression of pro-differentiation C/EBPα targets and block the mature differentiation of MA9 leukemia cells (Figure 6F). These findings unravel a novel mechanism for EZH2 in blocking MLL-AF9 leukemia cell differentiation.
Acknowledgments
This work was supported in part by the National Institutes of Health (grant R01 CA113962 to XH), the Leukemia and Lymphoma Society (TRP grant to XH), and a T32 training grant (CA 9140-36 to AT). We thank Dr. Patricia Ernst for providing Mllf/f mice. We appreciate stimulating discussions with Drs. Warren Pear and Martin Carroll, and members of our laboratory. We thank Ms. Hong Wei at the Histology Core Facility for histological studies.
Footnotes
The online version of this article has a Supplementary Appendix.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12(12):799–814 [DOI] [PubMed] [Google Scholar]
- 2.Mills AA. Throwing the cancer switch: reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10(10):669–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanson RD, Hess JL, Yu BD, Ernst P, van Lohuizen M, Berns A, et al. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci USA. 1999;96(25):14372–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pineault N, Helgason CD, Lawrence HJ, Humphries RK. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp Hematol. 2002;30(1):49–57 [DOI] [PubMed] [Google Scholar]
- 5.Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, et al. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10(5):1107–17 [DOI] [PubMed] [Google Scholar]
- 6.Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, Kazemier KM, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351(6):533–42 [DOI] [PubMed] [Google Scholar]
- 7.Daser A, Rabbitts TH. Extending the repertoire of the mixed-lineage leukemia gene MLL in leukemogenesis. Genes Dev. 2004;18(9):965–74 [DOI] [PubMed] [Google Scholar]
- 8.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121(2):167–78 [DOI] [PubMed] [Google Scholar]
- 10.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117(25):6912–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller D, Garcia-Cuellar MP, Bach C, Buhl S, Maethner E, Slany RK. Misguided transcriptional elongation causes mixed lineage leukemia. PLoS Biol. 2009;7(11):e1000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang QF, Wu G, Mi S, He F, Wu J, Dong J, et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood. 2011;117(25):6895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiel AT, Blessington P, Zou T, Feather D, Wu X, Yan J, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17(2):148–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, et al. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24(13):5639–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13(4):587–97 [DOI] [PubMed] [Google Scholar]
- 16.Chen YX, Yan J, Keeshan K, Tubbs AT, Wang H, Silva A, et al. The tumor suppressor menin regulates hematopoiesis and myeloid transformation by influencing Hox gene expression. Proc Natl Acad Sci USA. 2006;103(4):1018–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Gurung B, Wan B, Matkar S, Veniaminova NA, Wan K, et al. The same pocket in menin binds both MLL and JUND but has opposite effects on transcription. Nature. 2012;482(7386):542–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murai MJ, Chruszcz M, Reddy G, Grembecka J, Cierpicki T. Crystal structure of menin reveals binding site for mixed lineage leukemia (MLL) protein. J Biol Chem. 2011;286(36):31742–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caslini C, Yang Z, El-Osta M, Milne TA, Slany RK, Hess JL. Interaction of MLL amino terminal sequences with menin is required for transformation. Cancer Res. 2007;67(15):7275–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5(4):311–21 [DOI] [PubMed] [Google Scholar]
- 21.Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10(4):257–68 [DOI] [PubMed] [Google Scholar]
- 22.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–22 [DOI] [PubMed] [Google Scholar]
- 23.Chen W, Kumar AR, Hudson WA, Li Q, Wu B, Staggs RA, et al. Malignant transformation initiated by Mll-AF9: gene dosage and critical target cells. Cancer Cell. 2008;13(5):432–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faber J, Krivtsov AV, Stubbs MC, Wright R, Davis TN, van den Heuvel-Eibrink M, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113(11):2375–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17(13):3714–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPalpha differentiation pathway in human cancer. J Clin Oncol. 2009;27(4):619–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42(8):665–7 [DOI] [PubMed] [Google Scholar]
- 29.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42(8):722–6 [DOI] [PubMed] [Google Scholar]
- 30.Herrera-Merchan A, Arranz L, Ligos JM, de Molina A, Dominguez O, Gonzalez S. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun. 2012;3:623. [DOI] [PubMed] [Google Scholar]
- 31.Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, et al. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer Cell. 2006;10(5):401–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirstetter P, Schuster MB, Bereshchenko O, Moore S, Dvinge H, Kurz E, et al. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 2008;13(4):299–310 [DOI] [PubMed] [Google Scholar]
- 33.Goyama S, Yamamoto G, Shimabe M, Sato T, Ichikawa M, Ogawa S, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008;3(2):207–20 [DOI] [PubMed] [Google Scholar]
- 34.Tokita K, Maki K, Mitani K. RUNX1/EVI1, which blocks myeloid differentiation, inhibits CCAAT-enhancer binding protein alpha function. Cancer Sci. 2007;98(11): 1752–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arai S, Yoshimi A, Shimabe M, Ichikawa M, Nakagawa M, Imai Y, et al. Evi-1 is a transcriptional target of mixed-lineage leukemia oncoproteins in hematopoietic stem cells. Blood. 2011;117(23):6304–14 [DOI] [PubMed] [Google Scholar]
- 36.Yoshimi A, Goyama S, Watanabe-Okochi N, Yoshiki Y, Nannya Y, Nitta E, et al. Evi1 represses PTEN expression and activates PI3K/AKT/mTOR via interactions with polycomb proteins. Blood. 2011;117(13): 3617–28 [DOI] [PubMed] [Google Scholar]
- 37.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28(8):2825–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiner S, Birke M, Garcia-Cuellar MP, Zilles O, Greil J, Slany RK. MLL-ENL causes a reversible and myc-dependent block of myelomonocytic cell differentiation. Cancer Res. 2001;61(17):6480–6 [PubMed] [Google Scholar]
- 39.Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, et al. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20(5):563–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith LL, Yeung J, Zeisig BB, Popov N, Huijbers I, Barnes J, et al. Functional crosstalk between Bmi1 and MLL/Hoxa9 axis in establishment of normal hematopoietic and leukemic stem cells. Cell Stem Cell. 2011;8(6):649–62 [DOI] [PubMed] [Google Scholar]
- 41.Zeisig BB, Milne T, Garcia-Cuellar MP, Schreiner S, Martin ME, Fuchs U, et al. Hoxa9 and Meis1 are key targets for MLL-ENL-mediated cellular immortalization. Mol Cell Biol. 2004;24(2):617–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milne TA, Kim J, Wang GG, Stadler SC, Basrur V, Whitcomb SJ, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38(6):853–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nerlov C. C/EBPalpha mutations in acute myeloid leukaemias. Nat Rev Cancer. 2004;4(5):394–400 [DOI] [PubMed] [Google Scholar]
- 44.Reckzeh K, Cammenga J. Molecular mechanisms underlying deregulation of C/EBPalpha in acute myeloid leukemia. Int J Hematol. 2010;91(4):557–68 [DOI] [PubMed] [Google Scholar]
- 45.Matsushita H, Nakajima H, Nakamura Y, Tsukamoto H, Tanaka Y, Jin G, et al. C/EBPalpha and C/EBPvarepsilon induce the monocytic differentiation of myelomonocytic cells with the MLL-chimeric fusion gene. Oncogene. 2008;27(53):6749–60 [DOI] [PubMed] [Google Scholar]
- 46.Neff T, Sinha AU, Kluk MJ, Zhu N, Khattab MH, Stein L, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci USA. 2012;109(13):5028–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi J, Wang E, Zuber J, Rappaport A, Taylor M, Johns C, et al. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene. 2013; 32(7):930–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka S, Miyagi S, Sashida G, Chiba T, Yuan J, Mochizuki-Kashio M, et al. Ezh2 augments leukemogenecity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 2012;120(5):1107–17 [DOI] [PubMed] [Google Scholar]
- 49.Mochizuki-Kashio M, Mishima Y, Miyagi S, Negishi M, Saraya A, Konuma T, et al. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 2011;118(25):6553–61 [DOI] [PubMed] [Google Scholar]
- 50.Ryan RJ, Nitta M, Borger D, Zukerberg LR, Ferry JA, Harris NL, et al. EZH2 codon 641 mutations are common in BCL2-rearranged germinal center B cell lymphomas. PLoS One. 2011;6(12):e28585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci USA. 2010;107(49): 20980–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117(8):2451–9 [DOI] [PMC free article] [PubMed] [Google Scholar]