Abstract
Despite improvements in treatment results for pediatric T-cell acute lymphoblastic leukemia, approximately 20% of patients relapse with dismal prognosis. PTEN inactivation and NOTCH1 activation are known frequent leukemogenic events but their effect on outcome is still controversial. We analyzed the effect of PTEN inactivation and its interaction with NOTCH1 activation on treatment response and long-term outcome in 301 ALL-BFM treated children with T-cell acute lymphoblastic leukemia. We identified PTEN mutations in 52 of 301 (17.3%) of patients. In univariate analyses this was significantly associated with increased resistance to induction chemotherapy and a trend towards poor long-term outcome. By contrast, patients with inactivating PTEN and activating NOTCH1 mutations showed marked sensitivity to induction treatment and excellent long-term outcome, which was similar to patients with NOTCH1 mutations only, and more favorable than in patients with PTEN mutations only. Notably, in the subgroup of patients with a prednisone- and minimal residual disease (MRD)-response based medium risk profile, PTEN-mutations without co-existing NOTCH1-mutations represented an MRD-independent highly significant high-risk biomarker. Mutations of PTEN highly significantly indicate a poor prognosis in T-ALL patients who have been stratified to the medium risk group of the BFM-protocol. This effect is clinically neutralized by NOTCH1 mutations. Although these results have not yet been explained by an obvious molecular mechanism, they contribute to the development of new molecularly defined stratification algorithms. Furthermore, these data have unexpected potential implications for the development of NOTCH1 inhibitors in the treatment of T-cell acute lymphoblastic leukemia in general, and in those with a combination of PTEN and NOTCH1 mutations in particular.
Introduction
Despite recent advances in the treatment of pediatric precursor T-cell acute lymphoblastic leukemia (T-ALL),1 this entity still remains a challenge because relapses carry a particularly poor prognosis.2,3 Conceptually, it would, therefore, be helpful to develop a molecular risk profile, which would enable stratification of patients early after diagnosis.4 It is known that the differentiation stage of the T-ALL clone5–7 or the activation of defined leukemogenic pathways8 may play a role in prognosis. Furthermore, in patients treated on the ALL-BFM 2000 protocol, we have shown that the activation of the NOTCH1 receptor pathway signifies a favorable prognosis,9,10 although in the context of other protocols this effect was not seen.11,12
In addition, inactivating mutations of the tumor suppressor phosphatase and tensin homolog (PTEN) are known to occur in a variety of tumors and to disturb signaling networks including the PI3K-AKT pathway. Activation of the PI3K-AKT pathway is known to play a particular role in T-ALL.8,13 The loss of PTEN function thus represents a candidate mechanism to modulate the aggressiveness of T-ALL. Indeed, deletions and point mutations but also posttranslational mechanisms of PTEN inactivation, have previously been associated with poor treatment response in some studies with a small number of patients, although this has not been found in other studies.13–17 The clinical effect of oncogene activation and tumor suppressor gene inactivation can depend on the treatment strategy.11,12,18,19 Therefore, we analyzed the incidence of PTEN point mutations and the effect of PTEN inactivation on clinical outcome in what is the largest cohort so far of 301 children with T-ALL who were treated on the ALL-BFM 2000 protocol.
Studies in cell lines have revealed that NOTCH1 can inhibit PTEN function via the transcriptional inhibitor HES1.20,21 We have, therefore, analyzed the clinical interaction of PTEN inactivation and NOTCH1 pathway activation. We show in univariate analyses that PTEN inactivated leukemias tend to be resistant to induction treatment and that affected patients show an unfavorable long-term outcome. When combined with NOTCH1-mutation status, PTEN mutated and NOTCH1 non-mutated patients show a significantly inferior outcome than the remaining cohort. Interestingly, this unfavorable effect of PTEN inactivation is counterbalanced clinically by the simultaneous presence of activating NOTCH1 mutations. These data have unexpected and potentially profound implications for the development of future treatment and stratification strategies in general, and for the use of NOTCH1 inhibitors in particular.
Design and Methods
Patients’ clinical characteristics
From August 1999 through February 2008, a total of 545 patients with T-ALL were eligible for treatment in the multicenter ALL-BFM 2000 trial; no non-Hodgkin’s lymphoma patients were included. This study was approved by the institutional review board of the Hannover Medical School and other participating institutions. Informed consent was obtained in accordance with the Declaration of Helsinki. This trial enrolled pediatric patients up to 18 years of age from 70 different treatment centers in Germany, Austria and Switzerland. The subjects were selected on the basis of availability of sufficient amounts of DNA for molecular analysis. There was no significant difference in clinical parameters (age, gender, white blood cell count at diagnosis, prednisone response, MRD at Day 78) between this subgroup of patients and the entire ALL-BFM 2000 cohort.1
Mononuclear cells were isolated from bone marrow (BM) samples and stored in liquid nitrogen or at −80°C until DNA extraction. All BM samples contained a blast percentage of 80% or more. Immunophenotyping was carried out as previously described,22 and the subclassification of T-ALL was performed according to the guidelines of the European Group for Immunological Characterization of Leukemias (EGIL).23
Early in vivo response to prednisone, defined as the cytoreduction to a 7-day prednisone treatment prophase and a single dose of intrathecal methotrexate on Day 1, served to assess the effect of early treatment.24 According to prednisone response, patients were classified into good responders (PGR: <1000 blasts/microliter at Day 8) or poor responders (PPR: ≥1000 blasts/microliter at Day 8). Treatment response was further defined by determination of minimal residual disease (MRD) kinetics that were assessed at 2 different time points: at Days 33 and 78 of treatment, respectively.25–28 Allele-specific oligonucleotide-polymerase chain reaction (PCR) protocols were used for quantitative detection of leukemic clone-specific immunoglobulin and T-cell receptor gene rearrangements on a LightCycler instrument (Roche Diagnostics, Mannheim, Germany).29 An unfavorable MRD status (≥10−4) was defined by the presence of at least one leukemic cell in 104 cells, whereas a favorable MRD status (< 10−4) was defined as the absence of detectable leukemic cells in 104 cells.9 For treatment stratification, the BFM-ALL protocol distinguishes the standard risk group (negative MRD on Days 33 and 78), the high-risk group (MRD at least one leukemic cell in 103 cells on Day 78) and the intermediate-risk group (all others). Complete remission (CR) was defined as less than 5% blasts in the regenerating BM, the absence of leukemic blasts in the peripheral blood and cerebrospinal fluid, and no evidence of localized disease. Relapse was defined as recurrence of lymphoblasts or localized leukemic infiltrates at any site.
Mutational analysis of diagnostic samples for PTEN and NOTCH1 mutations
PCR amplification of exon 7 of PTEN genes was performed with primary genomic DNA. The analysis of all other exons was performed on genomic DNA after whole genome amplification (primer sequences are shown in Online Supplementary Table S1). The mutations that have been identified were confirmed in primary DNA. Sequencing of NOTCH1 has been performed as described previously.10 PCR-amplified fragments were sequenced by GATC biotech (Konstanz, Germany) and analyzed by mutation surveyor software for identification of mutations and cross checked manually.
Statistical analysis
Event-free survival (EFS) was defined as the time from diagnosis to the date of last follow up in complete remission or first event. Events were resistance to therapy (non-response), relapse, secondary neoplasm (SN), or death from any cause. Failure to achieve remission due to early death or non-response was considered as events at time zero. Survival was defined as the time of diagnosis to death from any cause or last follow up. The Kaplan-Meier method was used to estimate survival rates, differences were compared with the two-sided log rank test. Cox’s proportional hazards model was used for uni- and multivariate analyses. Cumulative incidence (CI) functions for competing events were constructed by the method of Kalbfleisch and Prentice, and were compared with the Gray’s test. Results are presented as estimated probability of 5-year EFS (pEFS) and estimated cumulative incidence of relapse (pCIR) with standard error (± SE). Differences in the distribution of individual parameters among patient subsets were analyzed using Fisher’s exact test for categorized variables and the Mann-Whitney-U test for continuous variables. Logistical regression was used to analyze the effect of mutations on response variables (prednisone response, MRD). All statistical analyses were conducted using the SAS program (SAS-PC, v. 9.1, SAS Institute Inc., Cary, NC, USA).
Results
The clinical and immunological characteristics of the patient cohort analyzed here is comparable to that of the entire cohort of T-ALL patients included in the BFM-ALL 2000 study (Table 1).1PTEN mutations have previously been identified in approximately 20% of children with TALL and reported to signify a poor treatment response in studies with small patient numbers.13–16 In the cohort of 301 patients studied here, we found a total of 58 mutations in 52 patients (17.3%). The majority (52 of 58) of the mutations were detected in the mutational hotspot in exon 7, which is consistent with previously reported findings.14 The other mutations were detected in exons 1, 4 and 5 (Online Supplementary Table S2). Four patients had 2 mutations either both in exon 7 or in exons 4 and 7 or 5 and 7, respectively. One patient had 3 mutations: 2 in exon 7 and one in exon 1. In these patients, the diagnostic strategy we used did not allow the distinction between bialleleic compound or monoallelic double mutations to be made. In a previous study, we had also found large deletion mutations in 4 of 72 patients for whom sufficient DNA for array-CGH analyses were available (including 44 from the cohort analyzed here). Three of these patients (one with a homozygous and 2 with a heterozygous deletion) showed a poor early treatment response, whereas one patient with a heterozygous deletion showed a favorable early treatment response.8 Because the array-CGH analyses could only be performed in a subset of the entire cohort, we did not include these previously reported data in the statistical analyses of this report. However, the observed poor treatment response in 3 of these 4 patients is consistent with the clinical effect of PTEN point mutations in the entire cohort (see below).
Table 1.
Clinical and immunological characteristics of the study cohort of 301 children with T-ALL.
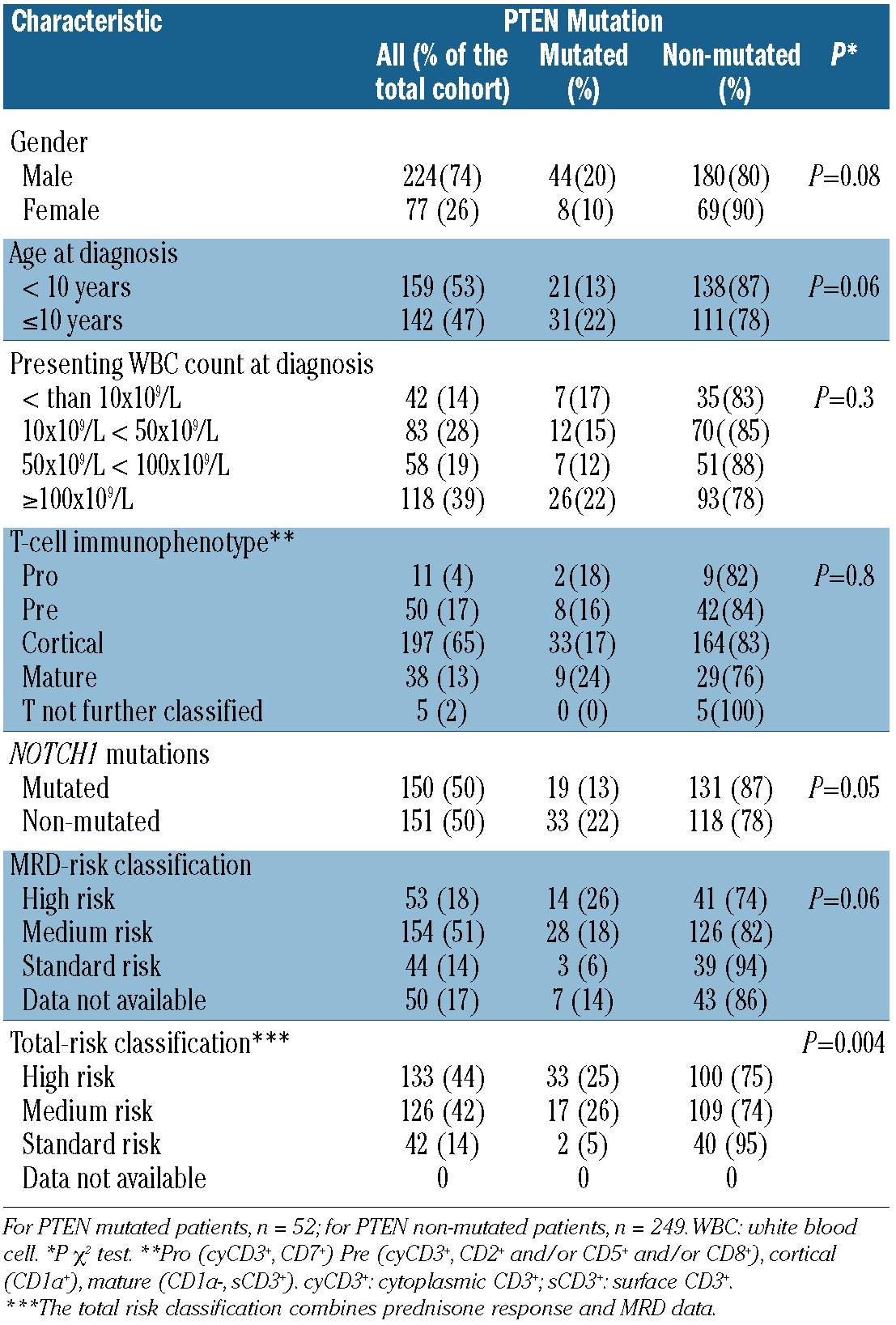
All mutations identified in the current study were small deletions or insertions which resulted either in nonsense mutations or frameshift mutations with downstream premature stop codons (48 insertions of up to 13 nucleotides, 8 deletions of up to 17 nucleotides) (Online Supplementary Table S2). The affected mRNAs may thus be targets of nonsense mediated decay quality control thus limiting the total expression of the encoded proteins30,31 or may code for inactive C-terminally truncated proteins.15 The presence of these mutations was significantly associated with the absence of activating NOTCH1 mutations. There was no significant correlation with patient age, gender, white blood cell count at the time of diagnosis, or T-cell immunophenotype (Table 1).
We next analyzed the influence of PTEN mutations on early treatment response and long-term outcome. Prednisone response was available for all the 52 patients with PTEN mutations and for 242 of 249 patients without PTEN mutations. Patients with PTEN mutations showed a poor prednisone response significantly more frequently than those patients without PTEN mutations (P=0.007), with an odds ratio in the univariate analysis of 2.4 (95%CI: 1.3–4.4, P=0.006; Table 2). Furthermore, in a multivariate analysis including variables known to be associated with prednisone response (gender, age at diagnosis, presenting WBC count at diagnosis and T-cell immunophenotype), the negative effect of PTEN mutations retained its significant effect (odds ratio 2.6, 95%CI: 1.3–5.2, P=0.005; Table 2).
Table 2.
Effect of inactivating PTEN mutations on early treatment response in ALL-BF M 2000 treated children with T-ALL.
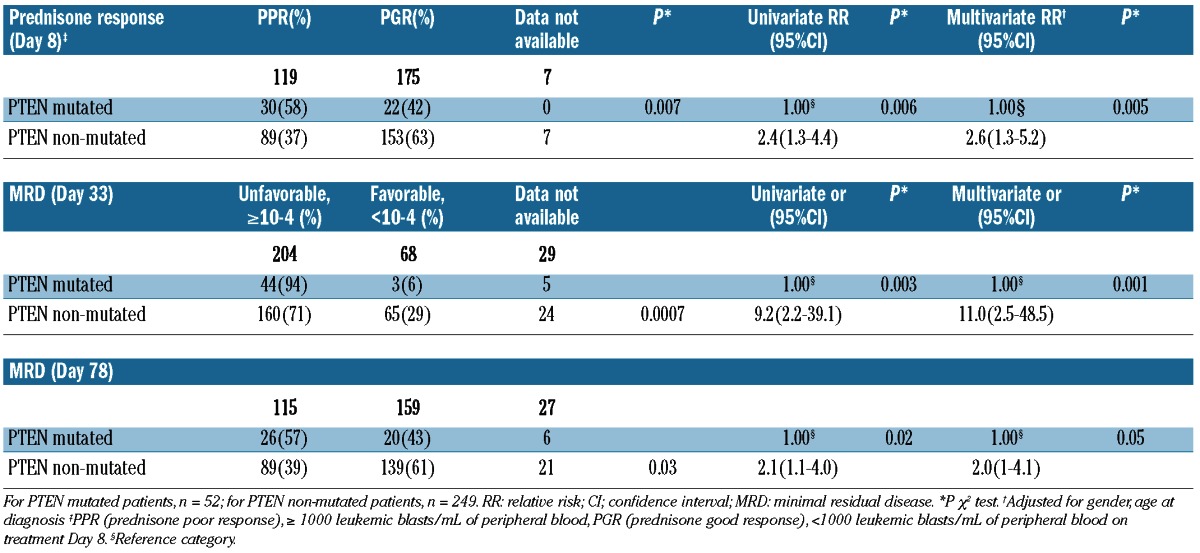
MRD data on Day 33 were available for 272 patients (47 PTEN mutated, 225 PTEN non-mutated) and in 274 patients at the end of the induction phase on Day 78 (46 PTEN mutated and 228 PTEN non-mutated). On Day 33, only 6% of the patients with PTEN mutations showed a favorable MRD response as compared to 29% of PTEN non-mutated patients (P=0.0007). On Day 78, 43% of the patients with a PTEN mutation achieved a favorable MRD response compared to 61% in the PTEN non-mutated group (P=0.03; Table 2). Logistical regression analysis showed that patients with PTEN mutations carried a 9.2-fold higher of not achieving a favorable MRD level on Day 33 (95%CI: 2.2–39.1; P=0.003) and a 2.1-fold higher risk on Day 78 (95%CI: 1.1–4.0; P=0.02), respectively. In a multivariate analysis with variables known to be associated with treatment response (gender, age at diagnosis, presenting WBC count at diagnosis and T-cell immunophenotype), the negative effect of PTEN mutation retained its significant effect on Day 33 (odds ratio 11.0, 95%CI: 2.5–48.5; P=0.001) and on Day 78 (odds ratio 2.0, 95%CI: 1.0–4.1; P=0.05) (Table 2). These effects resulted in PTEN mutated patients to be stratified into the high risk group significantly more frequently. Four out of the 5 patients with 2 or 3 PTEN mutations showed an unfavorable early treatment response.
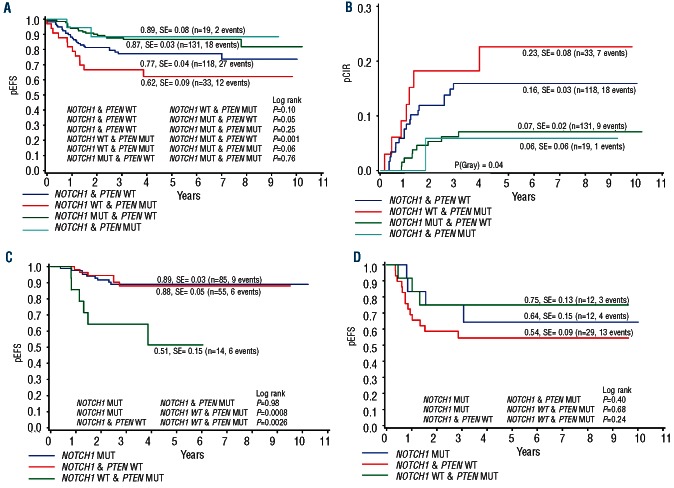
These differences between PTEN mutated and non-mutated patients in early treatment response were maintained as a trend towards an inferior pEFS of 0.72 vs. 0.82 (P=0.11, Figure 1A). There was also a slight trend towards a higher pCIR in PTEN mutated patients of 0.17 when compared to the pCIR of 0.11 observed in non-mutated patients (P=0.34, Figure 1B).
Figure 1.
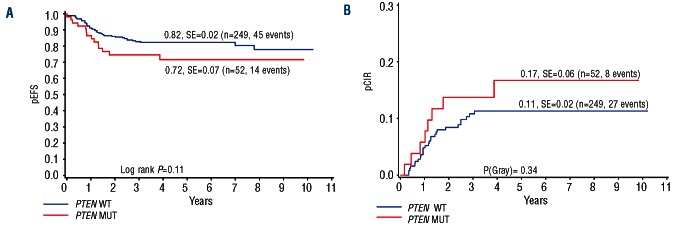
Univariate analysis shows a trend towards poor long-term outcome in children with T-ALL. Kaplan-Meier estimate of pEFS (A) and pCIR (B) in PTEN mutated and PTEN-non-mutated patients treated on the ALL-BFM 2000 protocol
The unfavorable effect of inactivating PTEN mutations is clinically neutralized by activating NOTCH1 receptor mutations
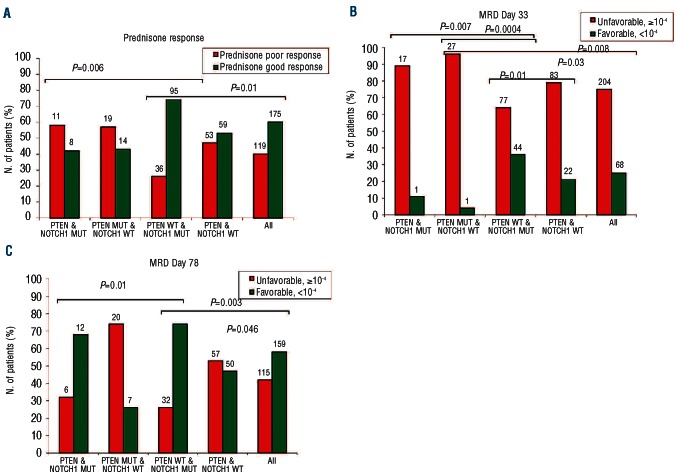
We have previously shown that activating NOTCH1 mutations are associated with a favorable early treatment response and long-term outcome in the same cohort of ALL-BFM 2000 treated patients who were analyzed here.9,10 The NOTCH1-downstream target HES1 is a negative transcriptional regulator of PTEN and thus indirectly stimulates the PI3K-AKT pathway,13 which may ultimately lead to a clinical synergism between activating NOTCH1 and inactivating PTEN mutations. We tested this hypothesis, by analyzing the clinical interaction of PTEN and NOTCH1 mutations. We thus grouped the patients according to their PTEN and NOTCH1 mutational status and compared their early treatment response and long-term outcome. In 19 of the 52 leukemias with PTEN mutations, concomitant activating NOTCH1 mutations, either in the heterodimerization or in the PEST domains, were identified. The PTEN and NOTCH1 mutation status was available in 294 patients with known prednisone response, in 272 patients with known MRD level on Day 33, and 274 patients with known MRD levels on Day 78 (Figure 2). For prednisone response and for Day 33 MRD, the unfavorable effect of PTEN mutations was maintained regardless of the presence of NOTCH1 mutations. Furthermore, the presence of PTEN mutations neutralized the known favorable effect of NOTCH1 mutations9,10 because patients with both mutations show a similar poor early treatment response to those patients with a PTEN mutation only (Figure 2A and B). These data on early treatment response are thus consistent with a dominant clinical effect of PTEN inactivation over NOTCH1 activation but do not support the notion of a clinically relevant synergism. By contrast, at the end of induction on Day 78, favorable MRD responses were observed most commonly in the NOTCH1-mutated groups regardless of the presence of PTEN mutations, whereas the group with PTEN mutations only was the least favorable, and the group with neither mutation was intermediate (Figure 2C). Therefore, at the end of induction the favorable effect of NOTCH1 receptor activation was clinically dominant over the unfavorable effect of PTEN inactivation.
Figure 2.
Effect of PTEN inactivation and NOTCH1 activation on early treatment response in children with T-ALL. (A) Prednisone response: prednisone good response (PGR; <1000 blasts/μL of peripheral blood at Day 8), Prednisone poor response (PPR; ≥1000 blasts/μL of peripheral blood at Day 8). (B) MRD response on Day 33: an unfavorable MRD status (≥10−4) was defined by the presence of at least one leukemic cell in 104 cells, whereas a favorable MRD status (<10−4) was defined as the absence of detectable leukemic cells in 104 cells (C) MRD response on Day 78: an unfavorable MRD status (≥10−4) was defined by the presence of at least one leukemic cell in 104 cells, whereas a favorable MRD status (<10−4) was defined as the absence of detectable leukemic cells in 104 cells. The number of patients is indicated on top of the columns.
We next performed a subgroup analysis of long-term outcome of the four possible PTEN/NOTCH1 combinations. The subgroup with PTEN mutations but no NOTCH1 mutations showed a significantly lower pEFS (0.62±0.09) than the rest of the cohort (pEFS 0.83±0.02; P=0.005) (Figure 3A) and a strong trend for a higher pCIR (0.23 vs. 0.11; P=0.07) (Figure 3B). Subgroups with NOTCH1 mutations, with or without PTEN mutations, were the most favorable (pEFS 0.87 and 0.89; pCIR 0.06 and 0.07) (Figure 4A), whereas the group with neither mutation was intermediate (pEFS 0.77, pCIR 0.16) (Figure 4B). Notably, subgroup analyses showed that the unfavorable PTEN-effect was restricted to the medium risk group with a good prednisone response and an intermediate MRD-response. The 14 patients with PTEN-mutations in this group (total n=154 patients), showed a highly significantly worse outcome (pEFS 0.51) than those with a NOTCH1 but no PTEN-mutation (n=85; pEFS 0.89; P=0.0008) or with neither a NOTCH nor a PTEN mutation (n=55; pEFS 0.88; P=0.0026) (Figure 4C). By contrast, there were no significant differences between the subgroups in the conventionally defined high-risk group (total n=53 patients) including 12 patients with a PTEN mutation only (Figure 4D). Given the small size of the subgroups, the direct comparison of the difference in outcome between the PTEN-only mutated patients in either the HR- or the MR-group was not statistically significant. In the standard risk group (total n=43 patients), there was only one of the 6 patients with a PTEN-mutation and no NOTCH1-mutation with a good prednisone response and negative MRD findings on Days 33 and 78 of induction.
Figure 3.
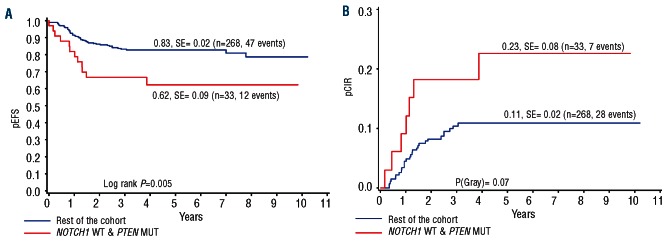
NOTCH1 activation neutralizes the unfavorable effect of PTEN inactivation on long-term outcome in children with T-ALL. Kaplan-Meier estimate of pEFS (A) and pCIR (B) in PTEN mutated and NOTCH1 non-mutated patients compared to the rest of the cohort.
Figure 4.
PTEN only mutated patients in the BFM-2000 medium-risk group show the worst long-term outcome. (A) Kaplan-Meier estimate of pEFS in the four different combinations of PTEN, NOTCH1 genotypes (total cohort n=301). (B) pCIR in the four different combinations of PTEN, NOTCH1 genotypes (total cohort n=301) (C) Kaplan-Meier estimate of pEFS in NOTCH1 and PTEN mutated, NOTCH1 and PTEN non-mutated, and NOTCH1 non-mutated and PTEN mutated patients stratified into the medium risk group (n=154) (D) Kaplan-Meier estimate of pEFS in NOTCH1 andPTEN mutated, NOTCH1 and PTEN non-mutated, and NOTCH1 non-mutated and PTEN mutated patients stratified into the high-risk group (n=53).
Taken together, these results define a subgroup of approximately 10% of the large and important medium-risk group with a poor prognosis and reveal a clinical activity of NOTCH1, which neutralizes the negative effect of PTEN inactivating mutations in this subgroup of children with T-ALL treated on the ALL-BFM 2000 protocol.
Discussion
Signaling networks including the PI3K/AKT pathway control a variety of cellular functions including differentiation, cell growth, metabolism and differentiation.32 As one of its functions, the phosphatase PTEN counterbalances the stimulatory effect of PI3K by dephosphorylating PIP3 and thus functions as a key negative regulator of this pathway.33 In addition to the other pathways that are involved in leukemogenesis, dysregulation of this pathway represents one of the most common events in tumorigenesis in general,34 and has also been identified to play an important part in T-ALL in particular.8,35–37 In TALL, both large deletions and inactivating small deletions and small insertions have been identified in a subset of approximately 15%–20% of patients.8,13–16 In addition, posttranslational modifications such as phosphorylation and oxidation have been described to also limit PTEN function in T-ALL.16 The effect of PTEN inactivation on clinical outcome has been reported in studies with smaller numbers of patients and shown to be variable and possibly dependent on the type of mutation.8,14,15,17 It is one of the important results of this study that in a large and clinically well-defined cohort of patients, inactivation of PTEN is not only associated with early treatment resistance but also with a poor long-term prognosis. This differs from previously reported results showing no prognostic effect of PTEN mutations.14,17 This difference may be explained by the modulating effect of NOTCH1 activation reported here and also by differences in treatment protocols. Differences in treatment have previously been suggested to play an important role in defining the effect of molecular risk factors in T-ALL.10 More specifically, the differences between the outcome of patients with PTEN-mutations but no NOTCH1 mutations, who have either been treated in the high-risk or in the medium-risk arm of the BFM protocol suggest (with the limitation of small sizes of the subgroups) that the unfavorable effect of PTEN inactivation may potentially be neutralized by more intensive treatment (Figure 4). The NOTCH1 pathway is a key regulator of blood cell differentiation. While the importance of NOTCH1 activity in T-cell development has been known for some time,38–40NOTCH has recently emerged to also function in myeloid, megakaryocyte and erythroid development.41–43 Gain of NOTCH1 function is a hallmark of 50%–60% of children with TALL,9,44 and is thought to function as an initiating event and a progression factor in T-ALL.45–47 Therefore, one might have predicted that activating NOTCH1 mutations should have an unfavorable effect on treatment response and long-term outcome. Interestingly, however, the clinical association of NOTCH1 activating mutations with treatment outcome is dependent on the protocol used. In ALL-BFM 2000 type protocols that are used in central Europe, Scandinavia and Japan, NOTCH1 gain of function is associated with a favorable effect,48–50 whereas in other protocols, either no effect or the expected unfavorable effect was observed.11,12 On the level of cell biology, the NOTCH and the PI3K/AKT pathways are known to interact in more complex signaling networks, which is highlighted by one of the major downstream targets of NOTCH1, the T-cell lymphomagenesis potentiating protein, HES1.51HES1 is known to down-regulate PTEN activity and thus to activate the PI3K/AKT pathway.13 This poses the interesting question of how the two signaling pathways interact in clinical terms. The second important finding in the large cohort of patients analyzed here is that early during induction the unfavorable association of PTEN loss of function with response dominates over that of NOTCH1 gain of function and that this effect is reversed later. The MRD response at the end of induction and the long-term outcome of patients with both, NOTCH1 activating and PTEN inactivating mutations are indistinguishably favorable when compared to patients with NOTCH1 mutations only. By contrast, patients with PTEN mutations but without NOTCH1 mutations in the conventionally defined medium-risk group represent a particularly unfavorable subgroup. Taken together, these data demonstrate that NOTCH1 activation and PTEN inactivation do not synergize on a clinical level in ALL-BFM 2000 treated patients. In contrast, when considering long-term outcome, NOTCH1 activation is associated with a neutralizing activity of the unfavorable effect of PTEN inactivation. The clinical association between NOTCH1 activation, PTEN inactivation and clinical outcome is surprising in the light of experimental data obtained from an analysis of cultured NOTCH1 activated cell lines and an analysis of the cell lines in xenotransplanted mice, which indicated a molecular synergism between NOTCH1 activation and PTEN inactivation.52 The discrepancy between the clinical association and molecular findings can be explained either by an unknown molecular factor that is associated with the presence of NOTCH1 mutations in primary patient samples but not in cell lines, or by a complex influence of the treatment given on the clinical effect of activated oncogenic networks. Further experimental studies will have to address this important enigma, for example in xenotransplanted primary T-ALL, to shed light on the mechanistically unexplained variable effects of NOTCH1 activation in the context of different treatment protocols.10–12
On a more practical level, the data presented here identify PTEN-mutated leukemias without activating NOTCH1 mutations as a subgroup with a particularly unfavorable prognosis with a pEFS of only 0.62. By contrast, T-ALLs with activating NOTCH1 mutations, with or without PTEN mutations, show a favorable prognosis with a pEFS of 0.87 and 0.89, respectively. Interestingly, subgroup analyses showed that the effect of PTEN mutations in patients without NOTCH1 mutations is restricted to patients who have been stratified to the medium-risk group (pEFS 0.51) and have not received intensified high-risk treatment. This combination of biomarkers thus defines a subgroup of patients with an unfavorable prognosis who may benefit from a new molecular stratification algorithm in future T-ALL protocols and from treatment intensification.
Interestingly, the effect of PTEN mutations is particularly evident on Day 8 when prednisone response is assessed and on Day 33 MRD. By contrast, the neutralizing effect of NOTCH1 mutations becomes apparent on Day 78 MRD and in long-term survival. These observations suggest that drugs introduced between Days 33 and 78 of the protocol (cyclophosphamide, 6-mercaptopurine, cytarabin) may be particularly important in mediating the differences of treatment response between early and later time points. These data also suggest that it may be beneficial to introduce these drugs earlier during induction.
Finally, the data presented here have unexpected and potentially profound implications for the development of NOTCH1 inhibitors for clinical use in T-ALL. Based on the role of NOTCH1 activity in T-ALL leukemogenesis53,54 and the discovery of the common occurrence of activating NOTCH1 mutations in T-ALL,44 the therapeutic inhibition of the NOTCH1 pathway has been a compelling perspective.55 The data of this study suggest that some patients, and notably those with a combination of PTEN inactivation and NOTCH1 activation, may not benefit from the use of NOTCH1 inhibitors in multimodal treatment regimens, although the mechanism of the unexpected clinical interactions between NOTCH1 activation and PTEN inactivation remains to be explained.
Acknowledgments
The authors acknowledge all participants of the ALL-BFM 2000 study.
Footnotes
The online version of this article has a Supplementary Appendix.
Funding
This study was supported by a grant from the German Ministry of Education and Research (BMBF, NGFN Plus), the “Tour der Hoffnung” and the Manfred Lautenschläger Stiftung.
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Schrappe M, Valsecchi MG, Bartram CR, Schrauder A, Panzer-Grumayer R, Moricke A, et al. Late MRD response determines relapse risk overall and in subsets of childhood T-cell ALL: results of the AIEOP-BFM-ALL 2000 study. Blood. 2011; 118(8):2077–84 [DOI] [PubMed] [Google Scholar]
- 2.Herold R, von Stackelberg A, Hartmann R, Eisenreich B, Henze G. Acute lymphoblastic leukemia-relapse study of the Berlin-Frankfurt-Munster Group (ALL-REZ BFM) experience: early treatment intensity makes the difference. J Clin Oncol. 2004; 22(3):569–70 [DOI] [PubMed] [Google Scholar]
- 3.Tallen G, Ratei R, Mann G, Kaspers G, Niggli F, Karachunsky A, et al. Long-term outcome in children with relapsed acute lymphoblastic leukemia after time-point and site-of-relapse stratification and intensified short-course multidrug chemotherapy: results of trial ALL-REZ BFM 90. J Clin Oncol. 2010;28(14):2339–47 [DOI] [PubMed] [Google Scholar]
- 4.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: an update. J Clin Oncol. 2011;29(5):551–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crist WM, Shuster JJ, Falletta J, Pullen DJ, Berard CW, Vietti TJ, et al. Clinical features and outcome in childhood T-cell leukemia-lymphoma according to stage of thymocyte differentiation: a Pediatric Oncology Group Study. Blood. 1988;72(6):1891–7 [PubMed] [Google Scholar]
- 6.Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10(2):147–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wuchter C, Ruppert V, Schrappe M, Dorken B, Ludwig WD, Karawajew L. In vitro susceptibility to dexamethasone- and doxorubicin-induced apoptotic cell death in context of maturation stage, responsiveness to interleukin 7, and early cytoreduction in vivo in childhood T-cell acute lymphoblastic leukemia. Blood. 2002;99(11): 4109–15 [DOI] [PubMed] [Google Scholar]
- 8.Remke M, Pfister S, Kox C, Toedt G, Becker N, Benner A, et al. High-resolution genomic profiling of childhood T-ALL reveals frequent copy-number alterations affecting the TGF-beta and PI3K-AKT pathways and deletions at 6q15-16.1 as a genomic marker for unfavorable early treatment response. Blood. 2009. 114: (5)1053–62 [DOI] [PubMed] [Google Scholar]
- 9.Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig WD, Tolle G, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108(4): 1151–7 [DOI] [PubMed] [Google Scholar]
- 10.Kox C, Zimmermann M, Stanulla M, Leible S, Schrappe M, Ludwig WD, et al. The favorable effect of activating NOTCH1 receptor mutations on long-term outcome in T-ALL patients treated on the ALL-BFM 2000 protocol can be separated from FBXW7 loss of function. Leukemia. 2010; 24(12):2005–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clappier E, Collette S, Grardel N, Girard S, Suarez L, Brunie G, et al. NOTCH1 and FBXW7 mutations have a favorable impact on early response to treatment, but not on outcome, in children with T-cell acute lymphoblastic leukemia (T-ALL) treated on EORTC trials 58881 and 58951. Leukemia. 2010;24(12):2023–31 [DOI] [PubMed] [Google Scholar]
- 12.Zuurbier L, Homminga I, Calvert V, te Winkel ML, Buijs-Gladdines JG, Kooi C, et al. NOTCH1 and/or FBXW7 mutations predict for initial good prednisone response but not for improved outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on DCOG or COALL protocols. Leukemia. 2010;24(12):2014–22 [DOI] [PubMed] [Google Scholar]
- 13.Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114(3):647–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jotta PY, Ganazza MA, Silva A, Viana MB, da Silva MJ, Zambaldi LJ, et al. Negative prognostic impact of PTEN mutation in pediatric T-cell acute lymphoblastic leukemia. Leukemia. 2010;24(1):239–42 [DOI] [PubMed] [Google Scholar]
- 16.Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest. 2008;118(11):3762–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuurbier L, Petricoin EF, Vuerhard MJ, Calvert V, Kooi C, Buijs-Gladdines J, et al. The significance of PTEN and AKT aberrations in pediatric T-cell acute lymphoblastic leukemia. Haematologica. 2012;97(9): 1405–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kox C, Zimmermann M, Stanulla M, Leible S, Schrappe M, Ludwig WD, et al. The favorable effect of activating NOTCH1 receptor mutations on long-term outcome in T-ALL patients treated on the ALL-BFM 2000 protocol can be separated from FBXW7 loss of function. Leukemia. 2010;24(12):2005–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulozik AE. Taking childhood leukemia personally. Blood. 2010;116(23):4737–8 [DOI] [PubMed] [Google Scholar]
- 20.Demarest RM, Ratti F, Capobianco AJ. It’s T-ALL about Notch. Oncogene. 2008;27 (38):5082–91 [DOI] [PubMed] [Google Scholar]
- 21.Silva A, Jotta PY, Silveira AB, Ribeiro D, Brandalise SR, Yunes JA, et al. Regulation of PTEN by CK2 and Notch1 in primary T-cell acute lymphoblastic leukemia: rationale for combined use of CK2- and gamma-secretase inhibitors. Haematologica. 2010; 95(4):674–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig WD, Rieder H, Bartram CR, Heinze B, Schwartz S, Gassmann W, et al. Immunophenotypic and genotypic features, clinical characteristics, and treatment outcome of adult pro-B acute lymphoblastic leukemia: results of the German multicenter trials GMALL 03/87 and 04/89. Blood. 1998;92(6):1898–909 [PubMed] [Google Scholar]
- 23.Bene MC, Castoldi G, Knapp W, Ludwig WD, Matutes E, Orfao A, et al. Proposals for the immunological classification of acute leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia. 1995;9(10): 1783–6 [PubMed] [Google Scholar]
- 24.Riehm H, Reiter A, Schrappe M, Berthold F, Dopfer R, Gerein V, et al. [Corticosteroid-dependent reduction of leukocyte count in blood as a prognostic factor in acute lymphoblastic leukemia in childhood (therapy study ALL-BFM 83)]. Klin Padiatr. 1987; 199(3):151–60 [DOI] [PubMed] [Google Scholar]
- 25.Flohr T, Schrauder A, Cazzaniga G, Panzer-Grumayer R, van der Velden V, Fischer S, et al. Minimal residual disease-directed risk stratification using real-time quantitative PCR analysis of immunoglobulin and T-cell receptor gene rearrangements in the international multicenter trial AIEOP-BFM ALL 2000 for childhood acute lymphoblastic leukemia. Leukemia. 2008;22(4):771–82 [DOI] [PubMed] [Google Scholar]
- 26.Hansen-Hagge TE, Yokota S, Bartram CR. Detection of minimal residual disease in acute lymphoblastic leukemia by in vitro amplification of rearranged T-cell receptor delta chain sequences. Blood. 1989;74(5): 1762–7 [PubMed] [Google Scholar]
- 27.van Dongen JJ, Seriu T, Panzer-Grumayer ER, Biondi A, Pongers-Willemse MJ, Corral L, et al. Prognostic value of minimal residual disease in acute lymphoblastic leukaemia in childhood. Lancet. 1998;352 (9142):1731–8 [DOI] [PubMed] [Google Scholar]
- 28.Willemse MJ, Seriu T, Hettinger K, d’Aniello E, Hop WC, Panzer-Grumayer ER, et al. Detection of minimal residual disease identifies differences in treatment response between T-ALL and precursor BALL. Blood. 2002;99(12):4386–93 [DOI] [PubMed] [Google Scholar]
- 29.van der Velden VH, Hochhaus A, Cazzaniga G, Szczepanski T, Gabert J, van Dongen JJ. Detection of minimal residual disease in hematologic malignancies by real-time quantitative PCR: principles, approaches, and laboratory aspects. Leukemia. 2003;17(6):1013–34 [DOI] [PubMed] [Google Scholar]
- 30.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36 (8):801–8 [DOI] [PubMed] [Google Scholar]
- 31.Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem J. 2010; 430(3):365–77 [DOI] [PubMed] [Google Scholar]
- 32.Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6 (3):184–92 [DOI] [PubMed] [Google Scholar]
- 33.Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325(5945):1261–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat Rev Cancer. 2011;11(4):289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G, et al. Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med. 2011;208 (5):901–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Subramaniam PS, Whye DW, Efimenko E, Chen J, Tosello V, De Keersmaecker K, et al. Targeting nonclassical oncogenes for therapy in T-ALL. Cancer Cell. 2012;21 (4):459–72 [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012; 481(7380):157–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268 (5208):225–32 [DOI] [PubMed] [Google Scholar]
- 39.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999; 284(5415):770–6 [DOI] [PubMed] [Google Scholar]
- 40.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7(9):678–89 [DOI] [PubMed] [Google Scholar]
- 41.Cornejo MG, Mabialah V, Sykes SM, Khandan T, Lo Celso C, Lopez CK, et al. Crosstalk between NOTCH and AKT signaling during murine megakaryocyte lineage specification. Blood. 2011;118(5):1264–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473(7346):230–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mercher T, Cornejo MG, Sears C, Kindler T, Moore SA, Maillard I, et al. Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell. 2008;3(3):314–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–71 [DOI] [PubMed] [Google Scholar]
- 45.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996; 183(5):2283–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eguchi-Ishimae M, Eguchi M, Kempski H, Greaves M. NOTCH1 mutation can be an early, prenatal genetic event in T-ALL. Blood. 2008;111(1):376–8 [DOI] [PubMed] [Google Scholar]
- 47.Joshi I, Minter LM, Telfer J, Demarest RM, Capobianco AJ, Aster JC, et al. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood. 2009;113(8):1689–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asnafi V, Buzyn A, Le Noir S, Baleydier F, Simon A, Beldjord K, et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) study. Blood. 2009;113(17):3918–24 [DOI] [PubMed] [Google Scholar]
- 49.Malyukova A, Dohda T, von der Lehr N, Akhoondi S, Corcoran M, Heyman M, et al. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 2007;67(12):5611–6 [DOI] [PubMed] [Google Scholar]
- 50.Park MJ, Taki T, Oda M, Watanabe T, Yumura-Yagi K, Kobayashi R, et al. FBXW7 and NOTCH1 mutations in childhood T cell acute lymphoblastic leukaemia and T cell non-Hodgkin lymphoma. Br J Haematol. 2009;145(2):198–206 [DOI] [PubMed] [Google Scholar]
- 51.Dudley DD, Wang HC, Sun XH. Hes1 potentiates T cell lymphomagenesis by up-regulating a subset of notch target genes. PLoS One. 2009;4(8):e6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Real PJ, Tosello V, Palomero T, Castillo M, Hernando E, de Stanchina E, et al. Gammasecretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat Med. 2009;15(1):50–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gothert JR, Brake RL, Smeets M, Duhrsen U, Begley CG, Izon DJ. NOTCH1 pathway activation is an early hallmark of SCL T leukemogenesis. Blood. 2007;110(10):3753–62 [DOI] [PubMed] [Google Scholar]
- 54.Armstrong F, Brunet de la Grange P, Gerby B, Rouyez MC, Calvo J, Fontenay M, et al. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood. 2009;113(8):1730–40 [DOI] [PubMed] [Google Scholar]
- 55.Cullion K, Draheim KM, Hermance N, Tammam J, Sharma VM, Ware C, et al. Targeting the Notch1 and mTOR pathways in a mouse T-ALL model. Blood. 2009;113 (24):6172–81 [DOI] [PMC free article] [PubMed] [Google Scholar]