We show in human immunodeficiency virus–positive persons that the coronary artery disease effect of an unfavorable genetic background is comparable to previous studies in the general population, and comparable in size to traditional risk factors and antiretroviral regimens known to increase cardiovascular risk.
Keywords: HIV infection, coronary artery disease, genetics, traditional risk factors, antiretroviral therapy
Abstract
Background Persons infected with human immunodeficiency virus (HIV) have increased rates of coronary artery disease (CAD). The relative contribution of genetic background, HIV-related factors, antiretroviral medications, and traditional risk factors to CAD has not been fully evaluated in the setting of HIV infection.
Methods In the general population, 23 common single-nucleotide polymorphisms (SNPs) were shown to be associated with CAD through genome-wide association analysis. Using the Metabochip, we genotyped 1875 HIV-positive, white individuals enrolled in 24 HIV observational studies, including 571 participants with a first CAD event during the 9-year study period and 1304 controls matched on sex and cohort.
Results A genetic risk score built from 23 CAD-associated SNPs contributed significantly to CAD (P = 2.9×10−4). In the final multivariable model, participants with an unfavorable genetic background (top genetic score quartile) had a CAD odds ratio (OR) of 1.47 (95% confidence interval [CI], 1.05–2.04). This effect was similar to hypertension (OR = 1.36; 95% CI, 1.06–1.73), hypercholesterolemia (OR = 1.51; 95% CI, 1.16–1.96), diabetes (OR = 1.66; 95% CI, 1.10–2.49), ≥1 year lopinavir exposure (OR = 1.36; 95% CI, 1.06–1.73), and current abacavir treatment (OR = 1.56; 95% CI, 1.17–2.07). The effect of the genetic risk score was additive to the effect of nongenetic CAD risk factors, and did not change after adjustment for family history of CAD.
Conclusions In the setting of HIV infection, the effect of an unfavorable genetic background was similar to traditional CAD risk factors and certain adverse antiretroviral exposures. Genetic testing may provide prognostic information complementary to family history of CAD.
A major long-term concern in HIV-positive persons includes increased rates and premature onset of coronary artery disease (CAD), stroke, and peripheral vascular disease, compared to the general population [1–6]. The pathogenesis of CAD in HIV is incompletely understood; a high prevalence of smoking, proinflammatory and procoagulant mechanisms in the context of immunosuppression [7–9], adverse viral effects on endothelial and other cells, and deleterious metabolic effects such as dyslipidemia and insulin resistance after exposure to certain antiretroviral treatments have been implicated [2, 10–12].
CAD has a strong hereditary component [13, 14]. Genome-wide association studies (GWAS) have identified common genetic variants that contribute to the risk of CAD in the general population [15, 16]. The Myocardial Infarction, Assessment of Antiretroviral and Genetic Factors in Human Immunodeficiency Virus Infection (MAGNIFICENT) Consortium was established with the aim of assessing the relative contribution of traditional risk factors, HIV-related factors, antiretroviral regimen, and genetic background to CAD in HIV-positive persons. We report here on 571 white HIV-positive persons who experienced a first CAD event and 1304 HIV-positive matched controls without CAD events in 24 HIV observational studies. This represents the most comprehensive genetics–CAD study undertaken in HIV-positive persons.
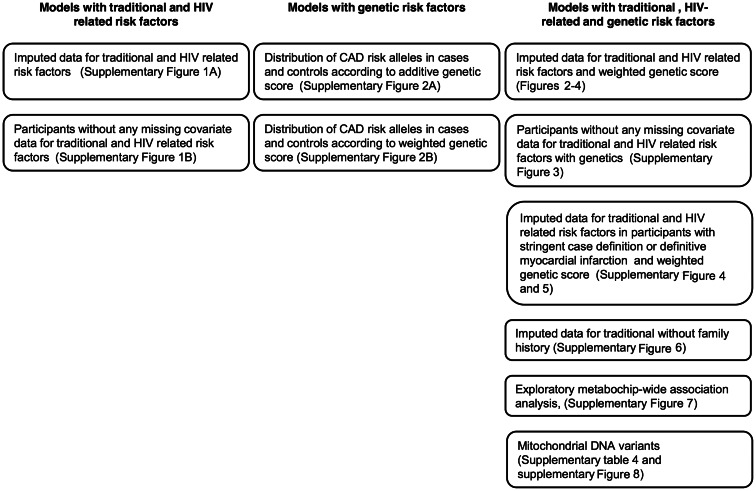
Figure 1.
Summary of the models applied and sensitivity analyses performed. Abbreviations: CAD, coronary artery disease; HIV, human immunodeficiency virus.
METHODS
Study Population, Inclusion Criteria
The MAGNIFICENT Consortium includes 24 HIV observational studies from Europe, the United States, Australia, and Argentina (Supplementary Data). Participants gave written informed consent for genetic testing. The ethics committee of each study center approved the study. Applying a case-control design, we defined cases as HIV positive, with a first CAD event during the study period (1 April 2000 through 31 March 2009). Controls were HIV positive and event free during the study period. For each case, we aimed to select 3 controls from the same cohort using risk-set sampling [17]. Controls were matched only on sex, to allow analysis of the effect of relevant nongenetic factors. Participants with cardiovascular events prior to the study period were excluded. Because most previous CAD GWAS in the general population were conducted in populations of European descent [16], the present report is restricted to participants of European descent.
CAD Events
CAD events were validated by the treating physician and defined according to the Data Collection on Adverse Events of Anti-HIV Drugs (D:A:D) study and the MONICA Project of the World Health Organization [2, 18]. CAD events included definite myocardial infarction (MI); possible MI or unstable angina; percutaneous coronary intervention including coronary angioplasty and stenting; coronary artery bypass surgery; and fatal CAD, which required evidence of CAD before death. All CAD events in participating cohorts that occurred during the study period were included.
Power Calculation, Genotyping, and Quality Control
We interrogated 23 single-nucleotide polymorphisms (SNPs) with known CAD association in GWAS meta-analysis in the general population [16]. Using the ESPRESSO-CC Power Calculator [19], with projected 600 cases and 1800 controls, the study had an 80% power to capture the effect of SNPs with minor allele frequency (MAF) ≥0.1 and CAD odds ratio (OR) ≥1.25. Genotyping was performed on the Metabochip (Illumina, Eindhoven, the Netherlands, and Broad Institute, Harvard University/Massachusetts Institute of Technology, Boston, MA), a custom array of 196 725 SNPs from gene regions associated with multiple metabolic/cardiovascular traits in GWAS [20]. The Metabochip was developed by leader groups in the field to facilitate affordable genotyping of (1) recognized SNPs and (2) the genetic regions that carried them, with the goal of discovering causal variants associated with the recognized tag SNPs. However, the study was not designed or powered for a Metabochip-wide association study, which would require a significance threshold of P < 2.5 × 10–7 (ie, P = .05 divided by the number of SNPs interrogated [196 725]).
Participants were filtered based on gender check (heterozygosity testing) and cryptic relatedness. We used a modified Eigenstrat approach to identify and exclude population outliers and to control for the possibility of spurious associations resulting from residual population stratification [21]. This method derives the principal components of the correlations among common (MAF > 5%) gene variants, which reflect population ancestry, and corrects for those correlations in the subsequent association tests by integrating the coordinates of the significant principal component axes as covariates (Eigenstrat covariates) in the models.
Nongenetic CAD Risk Factors
Covariates were selected a priori based on published CAD effect and included in the final model regardless of statistical significance: high total cholesterol (>6.2 mmol/L [22] or being on lipid-lowering medication), low high-density lipoprotein (HDL) cholesterol (<1.04 mmol/L) [22], diabetes mellitus (confirmed plasma glucose level ≥7.0 mmol/L [fasting] or ≥11.1 mmol/L [nonfasting] or taking antidiabetic medication) [23, 24], hypertension (systolic blood pressure ≥140 mm Hg or diastolic blood pressure ≥90 mm Hg or taking antihypertensive medication), smoking (never, past, or current), family history of CAD, and age (per 5-year increments [25]). HIV-related covariates were defined a priori, based on their contribution to CAD in the D:A:D study [25]: CD4+ count and HIV RNA value (closest to the event date), current antiretroviral therapy exposure, current abacavir exposure, and cumulative exposure to lopinavir and indinavir. Because few patients had ≥2 years exposure and the CAD effect of 1 year and ≥2 years of treatment was equivalent, these drug exposures were considered as binary covariates (ie, < or ≥1 year).
Missing Data
Certain covariates were unavailable or had >20% missing data (Supplementary Table 3). Mostly, these data were systematically missing in entire cohorts, and therefore assumed to be missing at random. This assumption was further checked by comparing summary statistics on nonmissing values across cohorts. There was no evidence that cohorts differed significantly in the distribution of important confounders. Therefore, single imputation using predictive mean matching was performed to replace missing data for glucose, total and HDL cholesterol, blood pressure, smoking, family history, duration of lopinavir and indinavir exposure, HIV RNA, and CD4+ count. Missing values were imputed using models with the predictors age, sex, abacavir at time of event, region, and case/control status [26]. Primary analyses utilized the imputed dataset; sensitivity analyses utilized the nonimputed dataset (Supplementary Figures 1B and 3).
Genetic Association Analyses
We built 2 a priori defined genetic risk scores using 23 SNPs (or a proxy with r2 > 0.8) with known CAD association [16] (Supplementary Table 2); (1) additive genetic score (number of CAD risk alleles; heterozygous = 1, homozygous = 2; that is, scores ranged from 0 to 46; higher scores indicate a higher CAD risk; (2) additive weighted genetic score that takes into account the effect size reported in the reference paper [16]. For a SNP with, for example, a CAD OR of 1.2: reference allele = 0, heterozygous = 0.2, homozygous risk allele = 0.4. The numbers obtained for each of the 23 SNPs were added to create an individual weighted genetic risk score.
Statistical Analysis
A summary of the models applied and sensitivity analyses performed is provided in Figure 1. First, we tested the associations of nongenetic factors using a conditional logistic regression model [27]. Then, we tested the weighted genetic score plus the 5 Eigenstrat covariates and added them to the model, by dividing study participants into 4 genetic score quartiles. We made a post hoc test for an interaction between genetic score and traditional risk factors plus factors that contributed to CAD in the D:A:D study [25]. The pseudo-r2 from each conditional logistic regression model was used as an estimate of the percentage of explained CAD variability in the study population. Analyses were done using PLINK, R, SAS version 9.2 (SAS Corporation, Cary, NC) and Stata version 12.0 (StataCorp LP, College Station, TX).
Sensitivity Analyses
To assess the robustness of results, we repeated the final model in participants with (1) complete (nonimputed) data for all covariates; (2) stringent case definition (definite MI, coronary artery bypass surgery, and fatal CAD, plus corresponding controls); (3) definite MI plus corresponding controls; (4) family history of CAD excluded from the model.
Exploratory Genetic Association Analyses
First, all 196 725 SNPs present on the Metabochip were separately tested for association with CAD by conditional logistic regression. Second, to search for additional, weaker genetic associations in the regions containing known CAD-associated genes or variants, we considered as a group all SNPs located in/near (±5 kb) the 23 CAD-associated genes [16] and as a separate group the SNPs mapping to genes associated with traits indirectly related to CAD (total, low-density lipoprotein, and HDL cholesterol; diabetes mellitus; fasting glucose level; body mass index [20]). The distribution of association P values was compared between these groups and all other SNPs genotyped on the Metabochip using the 2-sample Kolmogorov-Smirnov test. Third, we evaluated a potential association of CAD events with mitochondrial DNA (mtDNA) haplogroups.
Results
Study Population
We received DNA specimens from 702 cases and 1849 controls. Twenty-one cases and 158 controls were excluded because of registration in the cohort of the control after the event date of their matched case (n = 124), insufficient DNA quantity or quality (n = 42), sample administrative error (n = 7), nonwhite self-reported origin (n = 4), event occurred after study ended (n = 1), or missing genetic consent (n = 1). After genotyping quality control, 97 cases were excluded because they were population outliers in the Eigenstrat analysis (n = 89) or genetically related with another participant (n = 8); corresponding controls were also excluded. The final study population included 1875 participants (571 cases and 1304 controls).
Among the 571 cases, there were 273 definite MI, 48 possible MI or unstable angina, 179 percutaneous coronary interventions, 32 coronary artery bypass surgeries, and 39 fatal CAD. Characteristics of participants are shown in Table 1. The median age at first CAD event was 50 years. Cases were older than controls and more likely to be smokers, and to have elevated cholesterol and glucose levels, a family history of CAD, and current treatment with abacavir.
Table 1.
Characteristics of the Cases and Controls at the Matching Date
| Characteristic | Cases, % | Controls, % |
|---|---|---|
| Total No. | 571 | 1304 |
| Male sexa | 91.1 | 91.3 |
| Age, y, median (range) | 50.0 (22–85.5) | 45.0 (16.5–81.3) |
| Smoking | ||
| Never | 22.8 | 31.4 |
| Past | 23.3 | 21.6 |
| Current | 53.9 | 46.9 |
| Hypercholesterolemia | 45.5 | 31.8 |
| Low HDL cholesterol | 43.1 | 39.3 |
| Diabetes mellitus | 19.4 | 13.6 |
| Arterial hypertension | 43.6 | 31.1 |
| Family history of coronary artery disease | 25.7 | 15.4 |
| Receiving antiretroviral therapy | 87.7 | 79.3 |
| Currently on abacavir | 25.6 | 17.6 |
| Duration of treatment with indinavir, y, median (range) | 0 (0–8.2) | 0 (0–11.3) |
| Duration of treatment with lopinavir, y, median (range) | 0 (0–8.0) | 0 (0–8.7) |
| CD4+ T-cell count, cells/μL, median (range) | 497 (11–1688) | 500 (10–1905) |
| HIV RNA, log copies/mL, median (range) | 3.8 (0–14.6) | 3.9 (0–13.6) |
| HIV RNA | ||
| <50 copies/mL | 63.2 | 60.2 |
| <400 copies/mL | 74.1 | 68.2 |
All values are percentages unless otherwise specified.
Abbreviations: HDL, high-density lipoprotein; HIV, human immunodeficiency virus.
a Cases and controls were matched by sex and cohort.
Nongenetic Factors Contributing to CAD
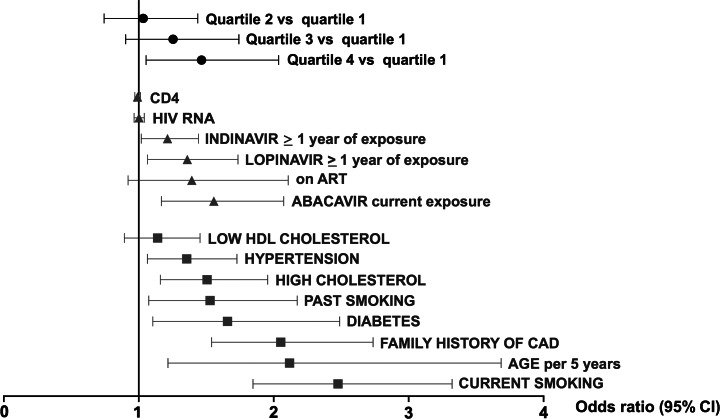
All covariates were significantly associated with the OR of a first CAD event, except low HDL cholesterol (P = .29), being on antiretroviral therapy at time of CAD event (P = .12), CD4+ count (P = .44), and HIV viremia (P = .88) (Figure 2). In the complete case analysis (participants without missing covariate data), the sample size was 720 individuals (183 cases, 537 controls). For the imputed models, the sample size was 1875 (571 cases, 1304 controls). Conditional logistic regression models for the imputed dataset were consistent with the complete case analysis as regards direction and effect size of individual covariates (Supplementary Figures 1B and 3). Therefore, the final model and the results presented hereafter are based on the imputed dataset.
Figure 2.
Contribution of traditional coronary artery disease (CAD) risk factors, HIV-related factors and weighted genetic score to CAD risk in multivariable analysis. Results are represented as the estimated effect and 95% confidence interval on the odds ratio of a first CAD event for genetic risk score quartile (black dots), HIV-related variables (gray triangles), and traditional CAD risk factors (gray squares). Results for the final, fully adjusted model (Supplementary Table 1A) and for the weighted genetic risk score (see Methods section) are shown. Abbreviations: ART, antiretroviral therapy; CAD, coronary artery disease; CI, confidence interval; HDL, high-density lipoprotein; HIV, human immunodeficiency virus.
CAD Odds Ratio According to Genetic Risk Score
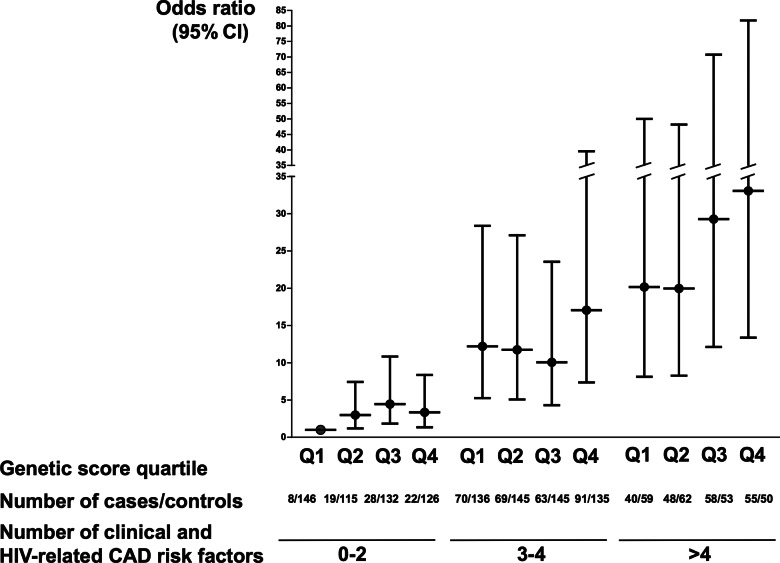
In unadjusted analysis (Table 2), participants in the third and fourth genetic risk score quartiles had an increased CAD OR, compared to the first quartile (OR = 1.34; 95% confidence interval [CI], 1–1.79; P = .05; and OR = 1.59; 95% CI, 1.19–2.13; P < .01, respectively). In the final, multivariable model, the additive and the weighted genetic score were associated with the CAD odds (P = 1.3 × 10−4 and P = 2.9 × 10−4, respectively). Cases were more likely to be in the upper 2 genetic score quartiles compared to controls (P = .01; Supplementary Figure 2). The effect of the weighted genetic score (CAD OR = 1.47 for the fourth quartile, compared to first quartile; 95% confidence interval [CI], 1.06–2.04; P = .02) was similar to the effect of established CAD risk factors and certain antiretroviral medications (Figure 2 and Supplementary Table 1A). This included family history (OR = 2.05; 95% CI, 1.54–2.74), hypertension (OR = 1.36; 95% CI, 1.06–1.73), hypercholesterolemia (OR = 1.51; 95% CI, 1.16–1.96), diabetes (OR = 1.66; 95% CI, 1.10–2.49), current smoking (OR = 2.48; 95% CI, 1.85–3.32), ≥1 year lopinavir exposure (OR = 1.36; 95% CI, 1.06–1.73) and current abacavir treatment (OR = 1.56; 95% CI, 1.17–2.07). An unfavorable genetic background had an additive effect on the CAD odds, without a significant interaction effect (P = .60; Figure 3 and Supplementary Table 1B).
Table 2.
Odds Ratio for Coronary Artery Disease According to Weighted Genetic Risk Score Quartile
| Genetic Risk Score Quartile | Genetic Risk Score and 5 Eigenstrat Covariates, Unadjusted for Nongenetic Covariates | Final Model, With Family History of CADa | Final Model, Without Family History of CADb |
|---|---|---|---|
| Quartile 2 vs quartile 1 | 1.27 (.95–1.69); P = .11 | 1.03 (.74–1.44); P = .84 | 1.04 (.75–1.44); P = .82 |
| Quartile 3 vs quartile 1 | 1.34 (1–1.79); P = .05 | 1.25 (.90–1.74); P = .18 | 1.25 (.90–1.72); P = .18 |
| Quartile 4 vs quartile 1 | 1.59 (1.19–2.13); P < .01 | 1.47 (1.06–2.04); P = .02 | 1.47 (1.06–2.03); P = .02 |
Data in parentheses are 95% confidence intervals.
Abbreviation: CAD, coronary artery disease.
a See Figure 2.
b See Supplementary Figure 7.
Figure 3.
Coronary artery disease (CAD) risk according to genetic score quartile and number of non-genetic CAD risk factors (odds ratio and 95% confidence interval). Participants are stratified into 12 groups by weighted genetic score quartile (quartile 1, 2, 3, and 4) and by the number of nongenetic risk factors (0–2, 3–4, or >4 nongenetic CAD risk factors). The first group is the reference group (odds ratio = 1), ie, participants with 0–2 nongenetic risk factors who are in genetic risk score quartile 1. The sum of all nongenetic CAD risk factors is considered (presence of risk factor = 1, absence of risk factor = 0), including traditional risk factors and additional factors that contributed significantly to CAD risk in the D:A:D study [25], ie, age, past smoking, exposure ≥1 year to lopinavir, exposure ≥1 year to indinavir, current exposure to abacavir. Abbreviations: CAD, coronary artery disease; CI, confidence interval; HIV, human immunodeficiency virus; Q, quartile.
Relative Contribution of Clinical, HIV-Related, and Genetic Factors
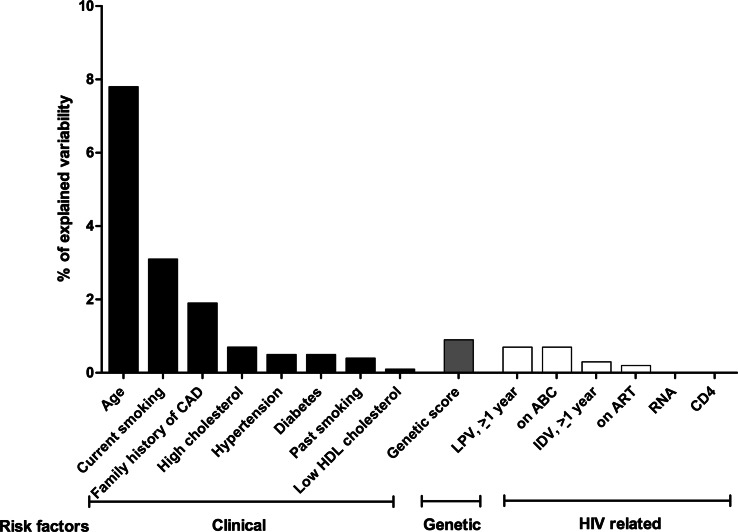
In the final model, 7.5% of the CAD odds ratio variability was explained by age, 3.1% by current smoking, 1.9% by family history, and 0.9% by genetic score. Smaller percentages were explained by traditional and HIV-related risk factors, for example, 0.7% each by hypercholesterolemia, ≥1 year lopinavir or current abacavir treatment; 0.5% each by diabetes or hypertension (Figure 4). Addition of the genetic score to the clinical model improved the fit of the model (χ2 = 6.28, P = .01).
Figure 4.
Coronary artery disease (CAD) variability explained by traditional risk factors, human immunodeficiency virus–related factors and genetic background. Variability in the CAD odds ratio explained by the final model: 21.1%. Of this, age: 7.5%, current smoking: 3.1%, past smoking: 0.4%, high total cholesterol: 0.7%, hypertension: 0.5%, diabetes: 0.5%, low high-density lipoprotein cholesterol: 0.1%, family history of CAD: 1.9%, genetic risk score: 0.9%, current antiretroviral therapy: 0.2%, current abacavir: 0.7%, lopinavir (≥1 year): 0.7%, indinavir (≥1 year): 0.3%, HIV load: 0%, CD4+ count: 0%. Abbreviations: ABC, abacavir; CAD, coronary artery disease; HDL, high-density lipoprotein; IDV, indinavir; LPV, lopinavir.
Sensitivity Analyses
Models restricted to participants with nonimputed data, stringent case definition (344 cases, 806 corresponding controls), and definite MIs (273 cases, 651 corresponding controls) showed similar results, except for a widening of confidence intervals due to reduced sample size (Supplementary Figures 3–5). Removing family history did not change the estimates for the genetic score (Table 2, Supplementary Figure 6).
Exploratory Metabochip-wide Analyses, mtDNA Variants
None of the 196 725 SNPs on the array were associated with CAD events in a metabochip-wide analysis after correction for multiple testing (Supplementary Figure 7). The global distribution of association P values was not significantly different between the 5070 SNPs within CAD-associated genes, the 36 863 SNPs within genes with a potential indirect CAD association [16], and the rest of the SNPs on the Metabochip, indicating an absence of enrichment of potentially interesting association signals in the first 2 groups. The mtDNA coverage of the Metabochip was found to be unreliable, with an excess of seemingly polymorphic (heteroplasmic) mtDNA SNPs; 80% of the participants displayed ≥1 heteroplasmic SNP out of 135 SNPs mapped to mtDNA (total, 5604 heteroplasmic calls; Supplementary Results, Supplementary Figure 8). Results were also inconsistent with the complete mtDNA genomes of 2 participants that had been previously Sanger sequenced [28], with 47% unmatched SNPs (64/135 positions, Supplementary Table 4).
DISCUSSION
This is the first large-scale analysis of clinical, HIV-related, and genetic risk factors that contribute to CAD in HIV-positive persons. Our findings suggest that the effect of an unfavorable genetic background on CAD events is comparable to well-established traditional risk factors and certain antiretroviral regimens. The genetic risk score, which was defined a priori and captures the joint effect of 23 common SNPs with known CAD association in the general population [16], remained independently associated with CAD after considering multiple nongenetic factors and in sensitivity analyses, suggesting that the effect is robust.
In this HIV-positive study population, genetic background explained a larger proportion of the CAD variability than did diabetes, hypertension, or dyslipidemia, but a smaller proportion than age or current smoking. Our exploratory analyses using the metabochip did not provide any novel insight about the genetics of CAD in HIV-positive persons. This was expected, as the study was designed to assess a panel of candidate SNPs with validated CAD association and was not powered for metabochip-wide discovery of novel gene variants.
Family history and genetic risk score contributed to CAD to a similar degree, and the effect of the genetic score did not change after adjusting for family history. This suggests that family history, which may reflect genetic background, but also environmental, social, and lifestyle factors shared among family members [29, 30], and assessment of common genetic variants capture independent, complementary effects on CAD in HIV-positive persons. This is consistent with the results by Ripatti and colleagues, a large study in the general population that assessed a similar genetic score [31].
An unfavorable genetic background had an effect on CAD comparable to certain antiretroviral agents known to increase cardiovascular risk. Because the magnitude of the CAD effect of certain drugs (eg, abacavir) is unresolved [32], we based selection of drug exposure covariates on the D:A:D study, the largest ongoing consortium of observational HIV studies [2, 25]. The increased CAD risk associated with lopinavir and indinavir is consistent with their previously recorded metabolic effects [2, 33]. Low CD4+ count or detectable HIV viremia at the time of the CAD event were not associated with CAD in our dataset, consistent with the D:A:D study [34]. Other authors have noted adverse effects of immunosuppression on CAD risk [35, 36].
Strengths of this study include the assembly of a large study population of HIV-positive persons who experienced a first CAD event during a 9-year study period; rigorous quality control of the genotyping data; exclusion of population outliers and correction for residual population stratification; physician validation of all CAD events; analysis of only SNPs and nongenetic covariates with established CAD association; and robust results in sensitivity analyses. Our study was limited by the effort required to establish the MAGNIFICENT Consortium. Even though HIV-positive populations are aging [37], the number of HIV-positive persons who have experienced CAD events is limited and not all studies include consent for genetic testing. Because demonstration of the CAD effect of 13 of the 23 SNPs required meta-analysis of >86 000 participants from multiple GWASs in the general population [16], additional CAD-associated SNPs with modest effect sizes may emerge from HIV-positive study populations larger than the MAGNIFICENT consortium. At the time of study design, GWAS-based CAD associations were essentially limited to white populations, so we restricted the present analysis to white participants; our findings may not be applicable to other populations.
Our findings suggest that genetic testing may provide prognostic information complementary to that afforded by family history, traditional risk factors, and antiretroviral regimen. Particularly in high-risk patients, knowledge of a deleterious genetic CAD predisposition might further emphasize the rationale for aggressive risk factor modification and selection of a CAD-neutral antiretroviral regimen to achieve HIV control. The clinical value of genetic testing will rely on demonstration of improved CAD risk stratification in prospective studies, as shown by Ripatti in the general population [31, 38]. This was beyond the scope of the case-control design of our consortium. Areas for future investigation include addition of genetic score to, for example, Framingham or D:A:D score in prospective HIV study settings; comparison of genetic CAD prediction in HIV-positive versus HIV-negative populations; and integration of genetic background and plasma biomarkers of inflammation, coagulation, and endothelial function to predict CAD in HIV [7, 39].
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments.
The authors acknowledge the contributions of the different research institutions, and the effort and commitment of investigators, study nurses, laboratory personnel, and participants. We thank the Cardio-Metabochip Consortium for designing the Metabochip and making it available at a competitive price, and the participants of the IdiPAZ Biobank integrated in the Spanish Hospital Biobanks Network for clinical samples.
Financial support.
This work was supported by the Swiss National Science Foundation (SNF; project 324730_127631/1), the Swiss HIV Cohort Study (SNF grant 33CS30_134277, SHCS project 599), an INFECTIGEN grant from the Universities of Geneva and Lausanne (to P. E. T.), the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, and unrestricted grants from Gilead Sciences and Merck Sharp & Dohme Switzerland to the SHCS research foundation. The IdiPAZ Biobank is supported by Instituto de Salud Carlos III, Spanish Health Ministry (RETIC RD09/0076/00073) and Farmaindustria, through the Cooperation Program in Clinical and Translational Research of the Community of Madrid Red de Investigación en SIDA grant (ISCIII-RETIC RD06/0006/1017, ISCIII-MA06/0164).
Potential conflicts of interest.
A. B. W. currently is Manager Scientific Affairs with the Crucell Vaccine Institute; the present work was initiated while she was working full-time at the Academic Medical Center (AMC) at University of Amsterdam and has been completed through her continued affiliation with the AMC. H. F. G. has been adviser and/or consultant for GlaxoSmithKline (GSK), Abbott, Gilead, Novartis, Boehringer Ingelheim, Roche, Tibotec, Pfizer, and Bristol-Myers Squibb (BMS), and his institution has received funding from Roche, Abbott, BMS, Gilead, Astra-Zeneca, GlaxoSmithKline, and MSD. C. T. has received lecture fees from Viiv and travel expenses from Viiv and Gilead. L. J. has been a consultant for BMS. E. M. has received consultancy and lecture fees from Abbott, BI, BMS, Gilead, GSK, MSD, Theratechnologies, Tibotec, and ViiV. J. R. has received advisory/lecture/consulting fees from Abbott, Bionor, BI, BMS, Gilead, GSK, Merck, Novartis, Janssen, Pfizer, Vertex, and ViiV and his institution has received funding from Abbott, MSD, and Roche. J.-C. W. has received advisory/lecture fees from Abbott, BMS, BI, Pfizer, and Tibotec and travel fees from Gilead. S. D. has received advisory fees from BMS and his institution has received consultancy fees from Viiv and travel fees from Janssen, Viiv, Gilead, Abbvie, and BMS. S. M. has received consulting/advisory fees from AbbVie, BI, BMS, Gilead, Janssen, and Roche and travel grants from the same companies plus MSD. J. G. has received consulting fees and his institution has received funding from Gilead, Abbott, BMS, MSD, and Janssen. The institution of C. B., K. P., and M. L. has received funding from BI, BMS, Gilead, GSK, Janssen, MSD, Pfizer, and Roche. M. L. has received payments for serving on data and safety monitoring boards for Sirtex Pty Ltd. A. D. has received advisory/consultant fees from Janssen, ViiV, Abbott, and Gilead, travel grants from ViiV and Abbott, and funding from ViiV and Janssen. A. D. has received advisory fees from Gilead and Viiv, consulting fees from Janssen and Abbott, and travel grants from Abbott and Viiv, and his institution has received funding from Viiv and Janssen. I. B. has received research funding and consulting/lecture fees from Abbott, Gilead, BMS, ViiV, and Janssen. J. G. G. has received consultancy fees from Abbott, Janssen, MSD, BMS, Gilead, ViiV, and Roche. J. R. A. has received advisory/speaker fees and grant support from Viiv, Tibotec, Janssen, Abbott, BMS, Gilead, and MSD. E. N. has received consultant fees from Gilead, BI, MSD, Abbott, Tibotec, GSK, and BMS. P. D. has received advisory, consultant, and/or lecture fees and has been a data safety monitoring board member for Gilead, Abbott, Janssen, BI, BMS, MSD, Theratechnologies, ViiV, and Ferrer. G. F. has received consultancy/lecture fees and funding from AbbVie, Janssen, Gilead, BMS, MSD, Pfizer, Roche, and ViiV. A. M. currently is Medical Director, HIV/Endocrinology with EMD Serono; the present work was initiated while she was working full-time at Tufts University and has been completed through her continued affiliation with Tufts. R. W.'s institution has received travel grants from Abbott, BI, BMS, Gilead, GSK, MSD, Pfizer, Roche, TRB Chemedica, and Tibotec. P. R. has been adviser for GSK, Gilead, and Janssen, and his institution has received funding and/or travel support from Gilead, ViiV, MSD, Janssen, BMS, and BI. He has served on data safety monitoring boards and endpoint adjudication committees for Janssen, and his institution has received honoraria for development of educational presentations from Gilead. H. C. B.'s institution has received travel grants, honoraria, and unrestricted research grants from GSK, BMS, Gilead, Janssen, Roche, Abbott, Tibotec, BI, and ViiV and is supported by unrestricted grants from Santésuisse and the Gottfried and Julia Bangerter-Rhyner-Foundation. P. E. T. has received advisory fees from Janssen, consultancy fees from Gilead, and honoraria from Viiv, and his institution has received advisory fees from Gilead and MSD, honoraria from Viiv, and travel expenses from MSD and Viiv. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.d'Arminio A, Sabin CA, Phillips AN, et al. Cardio- and cerebrovascular events in HIV-infected persons. AIDS (London) 2004;18:1811–7. doi: 10.1097/00002030-200409030-00010. [DOI] [PubMed] [Google Scholar]
- 2.Friis-Moller N, Reiss P, Sabin CA, et al. Class of antiretroviral drugs and the risk of myocardial infarction. N Engl J Med. 2007;356:1723–35. doi: 10.1056/NEJMoa062744. [DOI] [PubMed] [Google Scholar]
- 3.Grinspoon SK, Grunfeld C, Kotler DP, et al. State of the science conference: initiative to decrease cardiovascular risk and increase quality of care for patients living with HIV/AIDS: executive summary. Circulation. 2008;118:198–210. doi: 10.1161/CIRCULATIONAHA.107.189622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Obel N, Thomsen HF, Kronborg G, et al. Ischemic heart disease in HIV-infected and HIV-uninfected individuals: a population-based cohort study. Clin Infect Dis. 2007;44:1625–31. doi: 10.1086/518285. [DOI] [PubMed] [Google Scholar]
- 5.Periard D, Cavassini M, Taffe P, et al. High prevalence of peripheral arterial disease in HIV-infected persons. Clin Infect Dis. 2008;46:761–7. doi: 10.1086/527564. [DOI] [PubMed] [Google Scholar]
- 6.Triant VA, Lee H, Hadigan C, Grinspoon SK. Increased acute myocardial infarction rates and cardiovascular risk factors among patients with human immunodeficiency virus disease. J Clin Endocrinol Metab. 2007;92:2506–12. doi: 10.1210/jc.2006-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ford ES, Greenwald JH, Richterman AG, et al. Traditional risk factors and D-dimer predict incident cardiovascular disease events in chronic HIV infection. AIDS. 2010;24:1509–17. doi: 10.1097/QAD.0b013e32833ad914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuller LH, Tracy R, Belloso W, et al. Inflammatory and coagulation biomarkers and mortality in patients with HIV infection. PLoS Med. 2008;5:e203. doi: 10.1371/journal.pmed.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Sadr WM, Lundgren JD, et al. Strategies for Management of Antiretroviral Therapy Study G. CD4+ count-guided interruption of antiretroviral treatment. N Engl J Med. 2006;355:2283–96. doi: 10.1056/NEJMoa062360. [DOI] [PubMed] [Google Scholar]
- 10.Sabin CA, Worm SW, Weber R, et al. Use of nucleoside reverse transcriptase inhibitors and risk of myocardial infarction in HIV-infected patients enrolled in the D:A:D study: a multi-cohort collaboration. Lancet. 2008;371:1417–26. doi: 10.1016/S0140-6736(08)60423-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riddler SA, Smit E, Cole SR, et al. Impact of HIV infection and HAART on serum lipids in men. JAMA. 2003;289:2978–82. doi: 10.1001/jama.289.22.2978. [DOI] [PubMed] [Google Scholar]
- 12.Stein JH, Klein MA, Bellehumeur JL, et al. Use of human immunodeficiency virus-1 protease inhibitors is associated with atherogenic lipoprotein changes and endothelial dysfunction. Circulation. 2001;104:257–62. doi: 10.1161/01.cir.104.3.257. [DOI] [PubMed] [Google Scholar]
- 13.Lloyd-Jones DM, Nam BH, D'Agostino RB, Sr., et al. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291:2204–11. doi: 10.1001/jama.291.18.2204. [DOI] [PubMed] [Google Scholar]
- 14.Murabito JM, Pencina MJ, Nam BH, et al. Sibling cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults. JAMA. 2005;294:3117–23. doi: 10.1001/jama.294.24.3117. [DOI] [PubMed] [Google Scholar]
- 15.Clarke R, Peden JF, Hopewell JC, et al. Genetic variants associated with Lp(a) lipoprotein level and coronary disease. N Engl J Med. 2009;361:2518–28. doi: 10.1056/NEJMoa0902604. [DOI] [PubMed] [Google Scholar]
- 16.Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–8. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Essebag V, Genest J, Jr., Suissa S, Pilote L. The nested case-control study in cardiology. Am Heart J. 2003;146:581–90. doi: 10.1016/S0002-8703(03)00512-X. [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization. MONICA Manual, Part IV: Event Registration. Section 1: Coronary Event Registration Data Component. 1999. Available at: www.ktl.fi/publications/monica/manual/part4/iv-1.htm. Accessed 17 December 2012. [Google Scholar]
- 19.Burton PR, Hansell AL, Fortier I, et al. Size matters: just how big is BIG?: Quantifying realistic sample size requirements for human genome epidemiology. Int J Epidemiol. 2009;38:263–73. doi: 10.1093/ije/dyn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voight BF, Kang HM, Ding J, et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet. 2012;8:e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 22.Executive Summary of the Third Report of the National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III) JAMA. 2001;285:2486–97. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 23.Genuth S, Alberti KG, Bennett P, et al. Follow-up report on the diagnosis of diabetes mellitus. Diabetes Care. 2003;26:3160–7. doi: 10.2337/diacare.26.11.3160. [DOI] [PubMed] [Google Scholar]
- 24.Rotger M, Gsponer T, Martinez R, et al. Impact of single nucleotide polymorphisms and of clinical risk factors on new-onset diabetes mellitus in HIV-infected individuals. Clin Infect Dis. 2010;51:1090–8. doi: 10.1086/656630. [DOI] [PubMed] [Google Scholar]
- 25.Friis-Moller N, Thiebaut R, Reiss P, et al. Predicting the risk of cardiovascular disease in HIV-infected patients: the data collection on adverse effects of anti-HIV drugs study. Eur J Cardiovasc Prev Rehabil. 2010;17:491–501. doi: 10.1097/HJR.0b013e328336a150. [DOI] [PubMed] [Google Scholar]
- 26.White IR, Royston P, Wood AM. Multiple imputation using chained equations: Issues and guidance for practice. Stat Med. 2011;30:377–99. doi: 10.1002/sim.4067. [DOI] [PubMed] [Google Scholar]
- 27.Clayton D, Hills M. Statistical models in epidemiology. New York, NY: Oxford University Press; 1993. [Google Scholar]
- 28.Ortiz M, Poloni ES, Furrer H, et al. No longitudinal mitochondrial DNA sequence changes in HIV-infected individuals with and without lipoatrophy. J Infect Dis. 2011;203:620–4. doi: 10.1093/infdis/jiq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berg AO, Baird MA, Botkin JR, et al. National Institutes of Health State-of-the-Science Conference Statement: Family History and Improving Health. Ann Intern Med. 2009;151:872–7. doi: 10.7326/0003-4819-151-12-200912150-00165. [DOI] [PubMed] [Google Scholar]
- 30.Chow CK, Islam S, Bautista L, et al. Parental history and myocardial infarction risk across the world: the INTERHEART Study. J Am Coll Cardiol. 2011;57:619–27. doi: 10.1016/j.jacc.2010.07.054. [DOI] [PubMed] [Google Scholar]
- 31.Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Triant VA. HIV infection and coronary heart disease: an intersection of epidemics. J Infect Dis. 2012;205(suppl 3):S355–61. doi: 10.1093/infdis/jis195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Periard D, Telenti A, Sudre P, et al. Atherogenic dyslipidemia in HIV-infected individuals treated with protease inhibitors. The Swiss HIV Cohort Study. Circulation. 1999;100:700–5. doi: 10.1161/01.cir.100.7.700. [DOI] [PubMed] [Google Scholar]
- 34.Sabin C, Worm SW, Law M, et al. on behalf of the D:A:D study group. Association between markers of immunosuppression and the risk of cardiovascular disease (CVD): the D:A:D study. 2012 19th Conference on Retroviruses and Opportunistic Infections, Seattle, WA, Poster 822. [Google Scholar]
- 35.Klein DL, Leyden WA, Xu L, et al. Contribution of immunodeficiency to CHD: cohort study of HIV+ and HIV– Kaiser Permanente members. 2011 18th Conference on Retroviruses and Opportunistic Infections, Boston, MA Poster 810. [Google Scholar]
- 36.Lang S, Mary-Krause M, Simon A, et al. HIV replication and immune status are independent predictors of the risk of myocardial infarction in HIV-infected individuals. Clin Infect Dis. 2012;55:600–7. doi: 10.1093/cid/cis489. [DOI] [PubMed] [Google Scholar]
- 37.Hasse B, Ledergerber B, Furrer H, et al. Morbidity and aging in HIV-infected persons: the Swiss HIV cohort study. Clin Infect Dis. 2011;53:1130–9. doi: 10.1093/cid/cir626. [DOI] [PubMed] [Google Scholar]
- 38.Thanassoulis G, Vasan RS. Genetic cardiovascular risk prediction: will we get there? Circulation. 2010;122:2323–34. doi: 10.1161/CIRCULATIONAHA.109.909309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duprez DA, Neuhaus J, Kuller LH, et al. Inflammation, coagulation and cardiovascular disease in HIV-infected individuals. PLoS One. 2012;7:e44454. doi: 10.1371/journal.pone.0044454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.