Abstract
A single somatic FOXL2 mutation (FOXL2C134W) was identified in almost all granulosa cell tumor (GCT) patients. In the pituitary, FOXL2 and Smad3 coordinately regulate activin stimulation of follistatin transcription. We explored whether a similar regulation occurs in the ovary, and whether FOXL2C134W has altered activity. We show that in primary granulosa cells, GDF-9 and activin increase Smad3-mediated follistatin transcription. In contrast to findings in the pituitary, FOXL2 negatively regulates GDF-9 and activin-stimulated follistatin transcription in the ovary. Knockdown of endogenous FOXL2 confirmed this inhibitory role. FOXL2C134W displayed enhanced inhibitory activity, completely ablating GDF-9 and activin-induced follistatin transcription. GDF-9 and activin activity was lost when either the smad binding element or the forkhead binding element were mutated, indicating that both sites are required for Smad3 actions. This study highlights that FOXL2 negatively regulates follistatin expression within the ovary, and that the pathogenesis of FOXL2C134W may involve an altered interaction with Smad3.
Keywords: FOXL2, Follistatin, Granulosa cell tumor, Ovary, GDF-9, Activin
1. Introduction
FOXL2 is a member of the forkhead transcription factor family required for normal ovary and eyelid development [1]. Like other members of the forkhead family, it is comprised of the characteristic 100 amino acid DNA-binding forkhead domain in addition to a polyalanine tract at the C-terminus [1]. Histological studies in humans, mice and goats have detected FOXL2 mRNA and protein in the mesenchyme of developing eyelids, in fetal and adult granulosa cells (GCs) of the ovary, and in embryonic as well as adult gonadotrope and thyrotrope cells of the pituitary [1–7]. Within the ovary, FOXL2 has been shown to be essential for ovary development and function [8]. Germline FOXL2 mutations are present in blepharophimosis-ptosis-epicanthus inversus syndrome (BPES) patients, an autosomal dominant condition characterized by defects in eyelid development with (type I) or without (type II) premature ovarian failure [1,9–16]. BPES FOXL2 mutations can induce cytoplasmic mislocalization as well as cytoplasmic or nuclear aggregation [15,17], which most likely contributes to altered function in some patients.
In addition to germline FOXL2 mutations associated with BPES, recent studies have shown that a single somatic FOXL2 mutation (FOXL2C134W) is present in almost all adult GC tumors (GCTs) [18–20]. GCTs account for around 5% of ovarian cancers, with an overall frequency of 1 per 100,000 women [21]. The majority of GCTs are in adults, with only around 5% presenting in pre- or peri-pubertal patients [22]. FOXL2 mutations have not been detected in juvenile GCT patients; however, FOXL2 expression is markedly reduced in more than half of juvenile GCTs [23]. These findings provide further evidence that FOXL2 plays a key role in regulating normal GC growth and function. Interestingly, although germline FOXL2 mutations are present in BPES patients, GCTs do not develop in these patients. Therefore, in type I patients the predicted loss-of-function mutations that are present during ovary development may prevent aberrant GC proliferation and tumor development [23]. In type II BPES patients, as FOXL2 mutations do not appear to disrupt ovarian function they likely exhibit normal regulation of ovarian gene targets. FOXL2 is implicated in a number of cellular processes, including cell cycle arrest, apoptosis, cell adhesion, genome stability and reactive oxygen species detoxification, which overall appear to unite in tumor suppressor activity [24]. However, at present of the few known ovarian FOXL2 targets evaluated, any differences between FOXL2wt and FOXL2C134W activity have been modest at best and seem insufficient to explain GCT formation. Further, there appears to be disparities depending on the cell line utilized, which may relate to the species the cells/cell line is derived from, or inherent differences of primary GCs and tumor cells.
FOXL2 and Smad2/3 cooperatively regulate activin stimulation of follistatin [25], the β-subunit of follicle-stimulating hormone (FSH) [26–28] and gonadotropin releasing hormone receptor (GnRHR) [29] transcription in mouse pituitary cells. Potential cooperation between Smad2/3 and FOXL2 in GCs is feasible as they are co-expressed in GCs of ovarian follicles [30] [6,7] in which growth and differentiation factor-9 (GDF-9) and activin, the two principal ovarian Smad2/3 signaling TGF-β superfamily members, are known to play essential roles [31–33]. The 134Cys residue that is mutated in GCTs is located within the forkhead domain that is required for DNA binding as well as binding to other cofactors; modeling suggests that the mutation does not disrupt folding in this domain, interaction of FOXL2 with DNA or cellular distribution [18]. Smad3 is known to require an intact forkhead domain in FOXL2 for binding [25]. Therefore, if FOXL2 is involved in Smad2/3 signaling in the ovary, a loss of FOXL2 interaction with Smad2/3 as a result of FOXL2 mutations has the potential to significantly disrupt ovarian function. There is some evidence to suggest that the mutation changes FOXL2 interaction with Smad3, with altered activity of FOXL2C134W shown on the artificial GRAS luciferase reporter consisting of AP-1, FOXL2 and Smad binding sites [29,34]. Thus, altered interaction of FOXL2C134W with Smad3 may contribute to GCT formation.
In addition to actions in the pituitary, follistatin also plays an important role in the ovary. In mammalian ovaries, follistatin is highly expressed in GCs of developing follicles [35–38] and has been shown to bind activin A, AB and B with high affinities [39–42], and upon binding effectively block activin activity [43–48]. Follistatin is also able to bind some BMPs, including BMP-2, -4, -7 and BMP-15, and inhibit the biological activities of these proteins [49–51]. As GDF-9, like the BMPs and activin, is a member of the TGF-β superfamily, it seems likely follistatin similarly binds and negates GDF-9 actions, though to date this has not been established. Therefore, we hypothesized that if activin and GDF-9 stimulate GC follistatin gene expression, this may constitute a negative feedback loop with follistatin down-regulating their bioavailability. Due to the importance of these factors in the local regulation of follicular recruitment and development, this feedback system if present is most likely crucial in the balance needed for controlled folliculogenesis. As follistatin null mice die within hours of birth due to widespread defects [52] it is impractical to directly study in vivo the role of follistatin in folliculogenesis. However, as follistatin has been identified as one of the few known FOXL2 targets in pituitary cells, it is reasonable to hypothesize that FOXL2 also regulates ovarian follistatin expression.
The goal of this work was to confirm whether GDF-9 and activin regulate follistatin expression in GCs, and secondly determine whether FOXL2 cooperates with Smad3 to mediate GDF-9 and activin actions in GCs as seen in the pituitary. As an altered interaction with co-regulatory factors, such as Smad3, is considered a potential mechanism for the pathogenicity of the FOXL2C134W mutation linked to GCTs, we also evaluated whether FOXL2C134W showed altered activity compared with FOXL2wt, to determine whether dysregulation of the anti-proliferative follistatin may be involved in GC tumor formation. In this study we focused on activin and GDF-9 signals as they utilize the Smad3 signaling pathway and play essential roles in regulating GC function.
2. Materials and Methods
2.1. Plasmids and Reagents
Activin A [53] and recombinant human follistatin 288 [54] were generated by our laboratory as previously described, rmGDF-9 was purchased from R & D Systems (Minneapolis, MN). Phospho- Smad2 (#3101), phospho–Smad3 (#9520), total Smad2/3 (#3102), and β-actin (#4967) antibodies used for western blots were purchased from Cell Signaling (Danvers, MA). The FOXL2 antibody (IMG-3228) used for western blots was purchased from Imgenex (San Diego, CA). Diethylstilbestrol (DES) and the Flag antibody (F1804) used for western blots were purchased from Sigma-Aldrich Co. (St. Louis, MO). Female Sprague Dawley rats were obtained from Charles River Laboratories (Wilmington, MA).
The activin-responsive p3TP-lux reporter plasmid [55], which contains three copies of a 12-Otetraphorbol acetate-responsive element and the promoter of the human plasminogen activator inhibitor-1-linked luciferase reporter gene, was a gift from Dr. Joan Massague. Rat follistatin-luciferase (rFS-luc) reporter plasmids, N-terminally Flag-tagged hFOXL2wt and N-terminally Myc–tagged hSmad2, -3 and -4 expression plasmids were kindly provided by Dr. Louise Bilezikjian of the Salk Institute [25]. The rFS(2.9i)-luc and rFS(2.9)-luc plasmids incorporate the −2864/+136 fragment of rat follistatin with or without the entire first intron (+227/+2097) ligated to the 3’ end respectively. The rFS(0.3Ex45)-luc plasmid contains the +1784/+1912 section of intron 1 downstream of a −312/+136 minimal promoter. The +1784/+1912 section of intron 1 contains a forkhead-binding element (FBE) just downstream of Smadbinding element (SBE). From the hFOXL2wt expression plasmid, we generated the GCT mutant FOXL2C134W expression plasmid by site directed mutagenesis using two sets of primers in a two-step PCR. Briefly, primers A (5’-TACGTGGCGCTCATCGCCATGGCGATC-3’) and D (5’-AGCGCCATGCTCTGCACGCGTGTGTAC-3’) contained Sty I and Mlu I restriction sites respectively, while primers B (5’-CTCGAACATGTCTTCCCAGGCCGGGTCCAG-3’) and C (5’-CTGGACCCGGCCTGGGAAGACATGTTCGAG-3’) included the 402 C to G (or C402G) mutation that corresponds to the C134W amino acid change. The resulting mutant FOXL2 was ligated to the pFlaghFoxl2/pCS2+ construct using standard procedures. The SBE and FBE mutant forms of the rFS(0.3Ex45)-luc reporter were generated by PCR using primers to introduce the substitutions into the rFS(0.3Ex45)-luc reporter. The SBE mutation was the same as used by Blount et. al. [25]. The FBE mutation was a composite of smaller mutations shown to prevent FOXL2 action on this reporter in pituitary cells [25]. A β-galactosidase reporter plasmid driven by the Herpes virus thymidine kinase promoter was used as an internal control for transfection efficiency.
2.2. Harvest and culture of primary rat GCs
For harvest of primary rat GCs, female Sprague Dawley rats (24 days old) were implanted with silastic implants (Dow Corning, Corp., Midland, MI) containing 10 mg DES to stimulate GC proliferation. Four days after DES implantation GCs were harvested from the ovaries as previously described [53]. Briefly, ovarian follicles were punctured with needles to release GCs and oocytes, oocytes were subsequently removed using a 40 µm cell strainer . GCs were cultured in serum-free McCoys 5A culture media containing antibiotics. The Institutional Animal Care and Use Committee at the University of California, San Diego, approved all animal protocols.
2.3. RNA extractions and quantitative real-time PCR
GCs were plated in 12-well tissue culture plates at a density of 1 × 106 cells/well. Cells were treated with indicated doses of activin A or GDF-9 for 15 hr before media was removed and total RNA collected and extracted using RNeasy mini kit (Qiagen, Valencia, CA) as per manufactures instructions. RNA (1 µg) was reverse transcribed using the iScript cNDA synthesis kit (Biorad, Hercules, CA) before quantitation of mRNA expression by quantitative real-time PCR on a Biorad MyIQ iCycler. Primer sequences used were (Forward, 5’-TGCTTAGGCTACAGAAGAGG-3’) and (Reverse, 5’-CAGCTTCCTGATCTGTTGAC-3’) for L19, (Forward, 5’-ATGTAAAGAGCAGCCGGAAC-3’) and (Reverse, 5’-ATTGGTCTGATCCACCACAC-3’) for follistatin. Expression levels of the target gene were normalized to L19 expression.
2.4. Transfections and Luciferase Reporter Assays
For follistatin luciferase reporter assays, GCs were plated into serum-coated 12-well tissue culture plates at a density of 5 × 105 cells/well. The next day media were replaced with fresh media containing 10% FBS and transfected with indicated plasmids for 4 hr using Lipofectamine Ltx transfection reagent (Invitrogen, Carlsbad, CA) as per manufactures instructions. At the end of the transfection, the media containing transfection reagents was removed and replaced with fresh serum-free culture media and transfected cells cultured for a further 24 hr with vehicle, activin A, or GDF-9 at described doses. Cells were lysed and luciferase reporter activity measured using a dual-light reporter assay (Applied Biosystems, Bedford, MA) on a Veritas Microplate luminometer. Luciferase activity was normalized to the β-gal internal control to correct for transfection efficiency.
2.5. Western Blots
For evaluation of protein expression, GCs were plated into serum-coated 12-well tissue culture plates at a density of 1 × 106 cells/well in 1 ml serum-free culture medium. For measurement of activin and GDF-9 induction of phospho-Smad2 and phospho-Smad3, cells were left to attach before treatment for 15, 60 or 120 minutes. For evaluation of FOXL2 protein levels, cells were transfected and cultured as described in 2.4. Total protein was collected by lysis of cells in RIPA buffer containing a protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Cell lysates were added to 4X SDS sample buffer and heated to 95°C for 5 min to denature proteins. Cell lysates were separated on 10% SDS-PAGE gels before transfer to nitrocellulose membranes. Membranes were incubated with primary antibodies overnight at 4°C, washed then incubated with horseradish peroxidase-conjugated secondary antibodies. Chemiluminescence detection was performed using SuperSignal West Pico and Femto reagents (Thermo, Rockford IL).
2.6. FOXL2 siRNA
For siRNA experiments, GCs were plated into serum-coated 12-well tissue culture plates at a density of 1 × 106 cells/well. The same day cells were transfected with 30 nM of scramble (All Stars Negative Control siRNA, QIAGEN, Cambridge, MA) or FOXL2 (Custom siRNA, sense strand sequence with a TT overhang 5’-GGCAUCUACCAGUACAUCATT-3’, QIAGEN, Cambridge, MA) siRNA using Invitrogen RNAiMax as per manufactures instructions. Cells were cultured overnight, then after a media change transfected with luciferase reporters as in 2.4. At the end of the transfection, media containing transfection reagents was removed and replaced with fresh serum-free culture media containing vehicle, activin A or GDF-9 and cells cultured for a further 48 hr before determining luciferase reporter activity as in 2.4.
2.7. Statistic analysis
Data analysis was performed using StatPlus (Chicago, IL) software using one-way ANOVA or two-way ANOVA (as required), supplemented with Tukey-Kramer post hoc tests. Significance was set at P < 0.05. Values or figures are all presented as the mean ± SEM.
3. Results
3.1. GDF-9 and Activin A regulation of follistatin mRNA expression
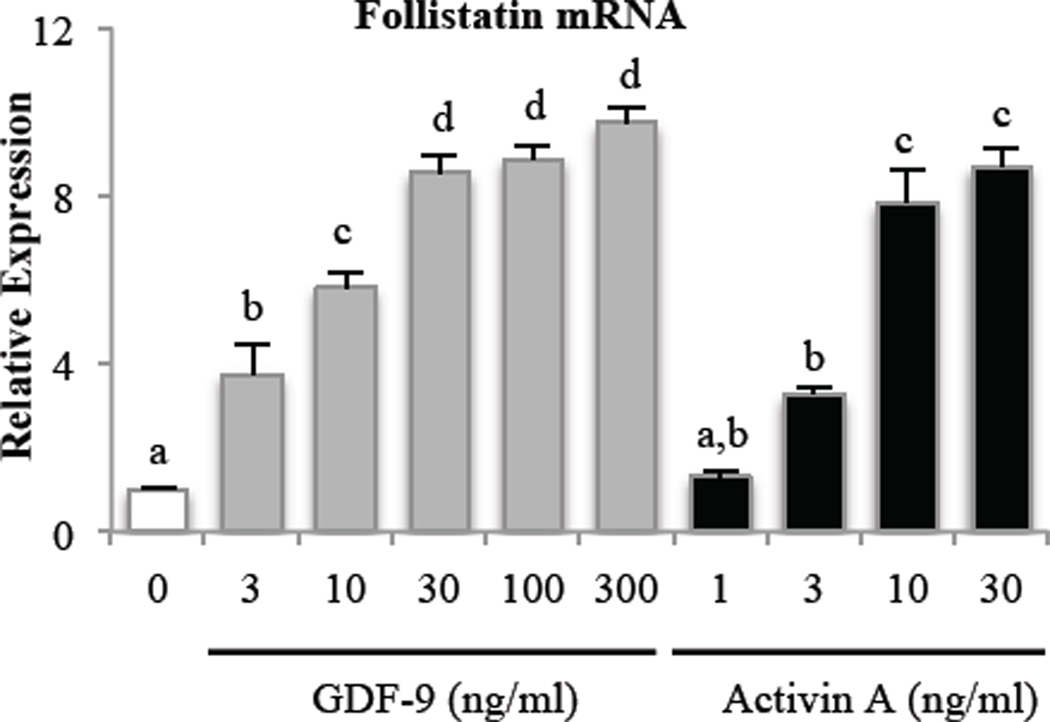
As activin has been shown to stimulate follistatin expression in the pituitary [56], and GCs of the ovary express follistatin [36] and activin [57], initial experiments evaluated whether a similar regulation occurs in the ovary. In addition to testing activin, we also evaluated oocyte-derived GDF-9, as it is another predominant ovarian Smad2/3 signaling factor. To evaluate whether activin and GDF-9 regulate follistatin in primary GCs, we treated primary rat GCs with increasing doses of activin A and GDF-9. As shown in Fig. 1, both GDF-9 and activin A dose-dependently induced follistatin mRNA expression in GCs within 15 hr of treatment.
Figure 1. GDF-9 and activin dose-dependently increase follistatin mRNA expression in primary rat GCs.
GCs were treated with increasing doses of GDF-9 or activin A for 15 hr before evaluation of follistatin mRNA expression. Follistatin mRNA levels were measured by quantitative real-time PCR and normalized to L19 mRNA levels. Results were evaluated by a one-way ANOVA followed by Tukey’s posthoc analysis. Different letters indicate a significant difference (p < 0.05) between groups within each treatment. Values shown are mean ± SEM. N = 3.
3.2. Ability of follistatin to antagonize GDF-9 and Activin A
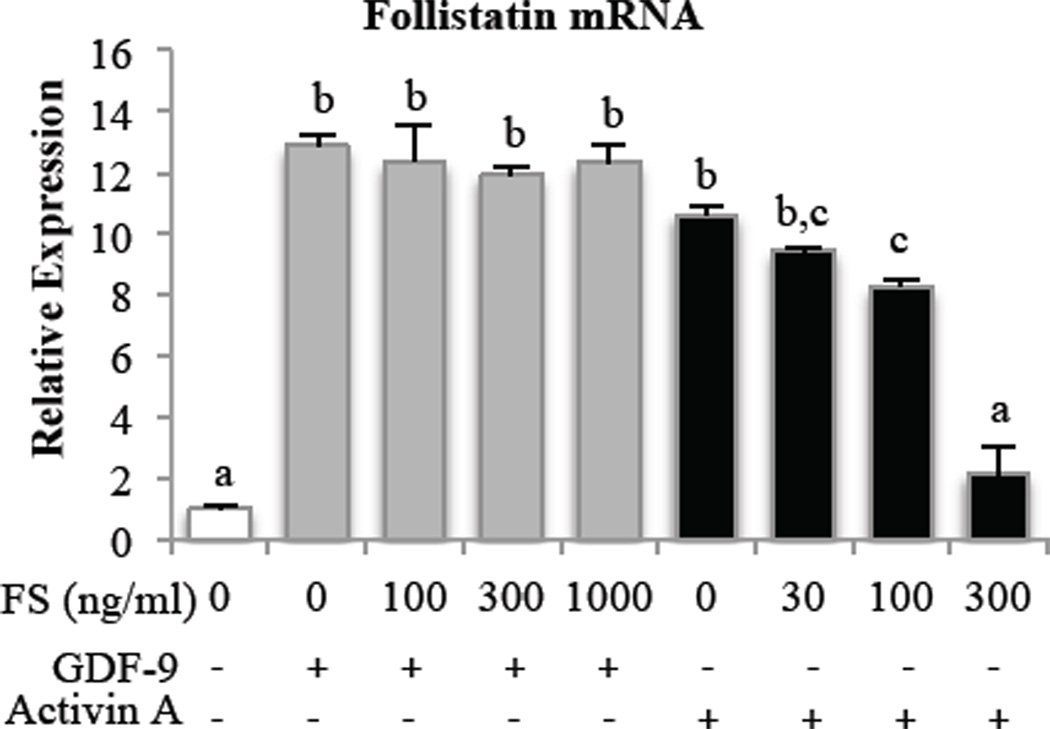
The ability of follistatin to bind to and inhibit activin actions is well known. However, the capacity for follistatin to similarly prevent GDF-9 actions has not been described. We pre-incubated GCs with follistatin at increasing doses before treatment with a fixed dose of either activin A or GDF-9 and evaluated the effects on follistatin mRNA expression. While follistatin inhibited activin A induction of follistatin mRNA, follistatin had no effect on the ability of GDF-9 to increase follistatin mRNA (Fig. 2). We achieved the same result (data not shown) when GDF-9 was pre-incubated with follistatin for 30 minutes before adding to cells.
Figure 2. Pre-incubation with follistatin negates activin- but not GDF-9-induced follistatin mRNA expression in primary rat GCs.
GCs were pretreated with increasing doses of follistatin (FS) for 1 hr before treatment with 30 ng/ml GDF-9 or 10 ng/ml activin A for 15 hr followed by measurement of follistatin mRNA expression. Follistatin mRNA levels were measured by quantitative real-time PCR and normalized to L19 mRNA levels. Results were evaluated by a one-way ANOVA followed by Tukey’s posthoc analysis. Different letters indicate a significant difference (p < 0.05) between groups within each treatment. Values shown are mean ± SEM. N = 3.
3.3. GDF-9 and Activin A regulation of follistatin transcription
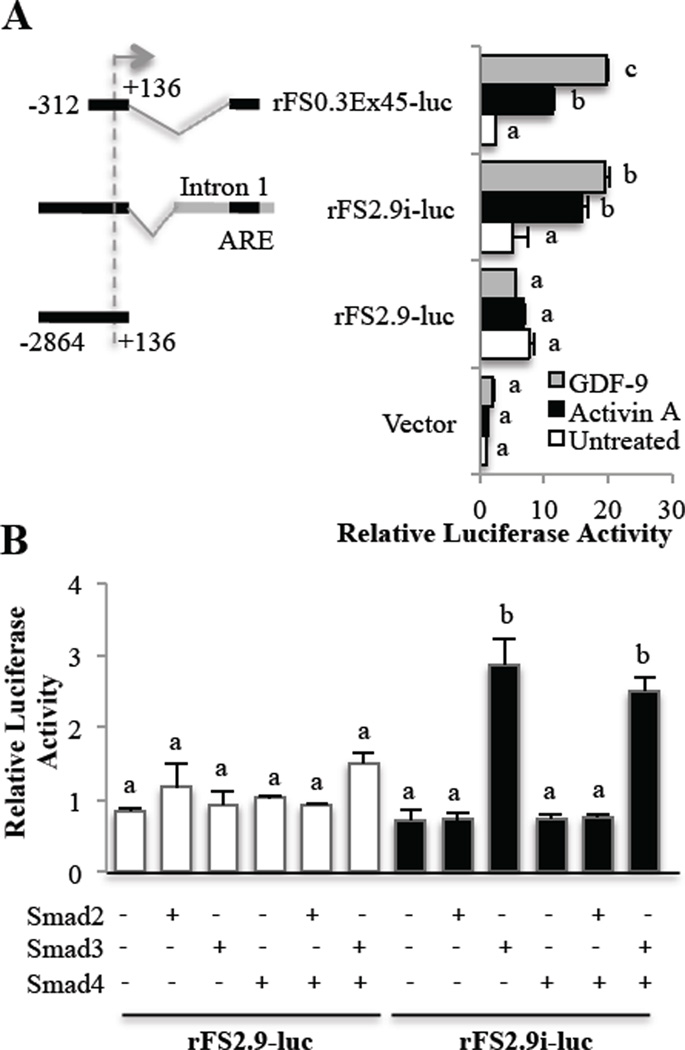
To confirm GDF-9 and activin regulation of follistatin was at the level of transcription, GCs were transfected with rat follistatin luciferase reporter constructs then treated with activin A or GDF-9. We compared three previously described [25] rat follistatin luciferase constructs. In pituitary cells, it was shown that activin responsiveness required the presence of small section of the first intron, named the activin responsive element (ARE) that includes a region for Smad binding just upstream of a region for forkhead binding. GDF-9- and activin A-induced activity was dependent on the presence of the ARE of intron 1 that contains the FBE and SBE (Fig. 3A). Activin A and GDF-9 exerted similar effects in the presence of the ARE of intron 1, regardless of whether a 0.3 or 2.9 kb promoter sequence was used, indicating that the ARE rather than the promoter is required for activin A and GDF-9 transcriptional activation of follistatin.
Figure 3. GDF-9- and activin-driven Smad3 regulation of follistatin transcription in primary rat GCs.
Numbers shown on the schematics of the depicted reporter plasmids indicate the nucleotide positions relative to the transcription start site (indicated by an arrow) of the rat Fst gene. AGCs transfected with either empty vector (vector), a rat follistatin-luciferase reporter with a minimal promoter and the activin-responsive element (ARE) of Intron 1 which contains both a Smad and a FOXL2 binding site (rFS0.3Ex45), or a follistatin-luciferase reporter with a 2.9 kb promoter with (rFS2.9i-luc) or without (rFS2.9-luc) a complete Intron 1. Following transfection, cells were treated with vehicle (untreated), 100 ng/ml activin A or 300 ng/ml GDF-9 for 24 hr before evaluation of luciferase activity. GDF-9 and activin A stimulate follistatin luciferase activity only in the presence of the ARE of intron 1. BPrimary rat GCs were transfected with Smad2, 3 and/or 4 and either the rat follistatin reporter with (rFS2.9i-luc) or without (rFS2.9-luc) Intron 1 containing the Smad binding element within the ARE. In untreated GCs, overexpression of Smad3, but not Smad2, stimulates follistatin luciferase activity in the presence of the ARE of intron 1. Data are represented as the ratio of the normalized luciferase activity to the untreated empty vector (in A) or untreated rFS2.9-luc (in B). Differences in activity were evaluated by a one-way ANOVA followed by Tukey’s posthoc analysis. Different letters indicate a significant difference (p < 0.05) between groups within each luciferase reporter. Values shown are mean ± SEM. N = 3.
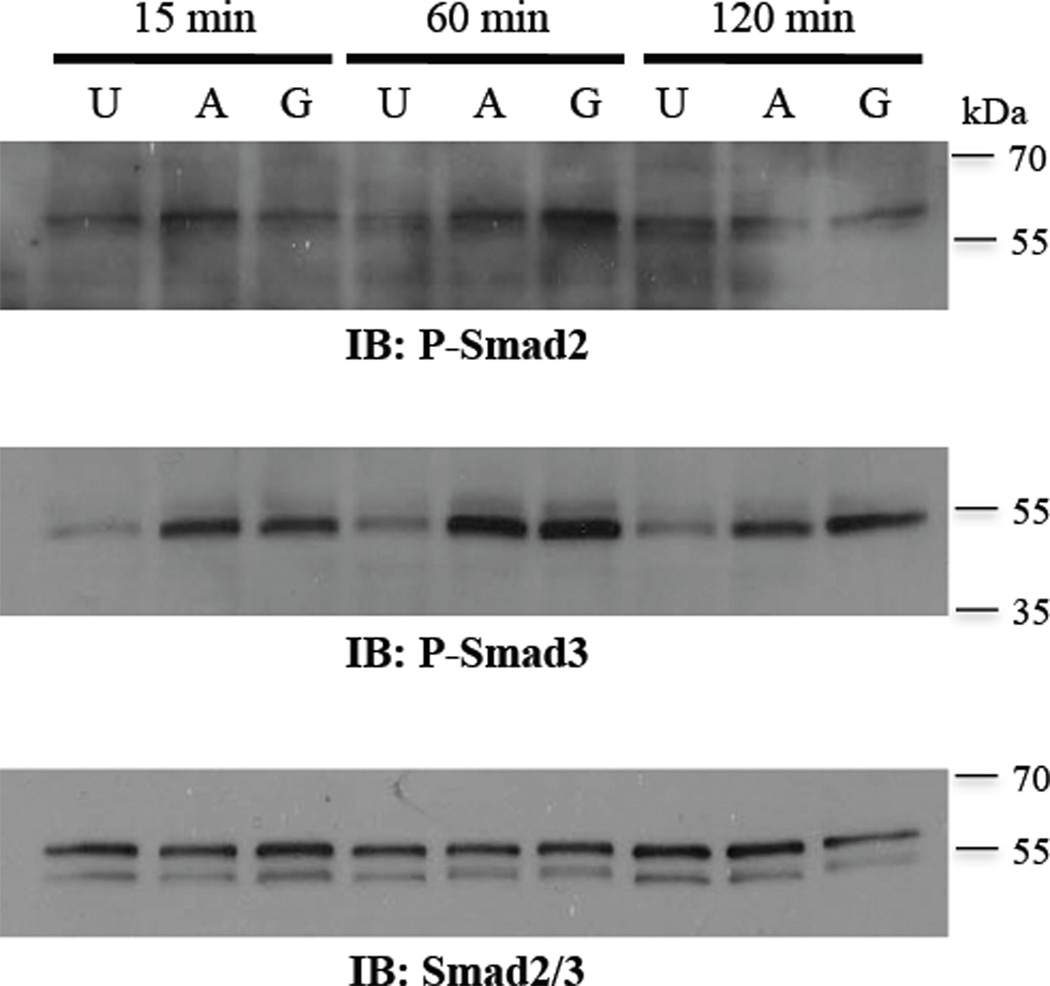
In pituitary cells, Smad3 but not Smad2 induces follistatin luciferase activity [25]. To determine whether a similar regulation of follistatin exclusively by Smad3 exists in GCs, we over-expressed Smads and evaluated follistatin luciferase activity. Consistent with findings in pituitary cells, in the absence of ligand treatments, over-expression of Smad3, but not Smad2, in GCs induced follistatin reporter luciferase activity in the presence of the first intron containing the ARE (Fig. 3B). Activity of the Smad2 expression vector was confirmed using the activin-responsive 3TP-lux reporter (Suppl. Fig. 1). Also, the addition of Smad4 did not appreciably alter the activity of Smad3. Our findings that exogenous Smad2 alone, in the absence of ligand, did not stimulate either reporter may indicate that Smad3 plays more of a role in GCs than Smad2. Further, we evaluated endogenous Smad2 and 3 in response to activin and GDF-9 in GCs, as activin and GDF-9 are known to stimulate the Smad2/3 pathway [32]. Fig. 4 confirmed that both activin A and GDF-9 induce phosphorylation of Smad2 and 3; however, the activation of Smad3 was considerably more marked. Based on previous reports that Smad3, not Smad2, interacts with FOXL2 [25], combined with our findings in GCs of greater phosphorylation of Smad3 than Smad2 by activin and GDF-9, and an effect of exogenous on the follistatin reporter by Smad3, but not Smad2, we used exogenous Smad3 in subsequent experiments.
Figure 4. Smad phosphorylation by activin and GDF-9 in primary rat GCs.
Treatment of GCs with 100 ng/ml activin A (A) or 300 ng/ml GDF-9 (G) increases phosphorylated Smad2 (P-Smad2) and Smad3 (P-Smad3) within 15 min with maximal activation at 60 min, compared with untreated (U) cells. For total Smad2/3, the antibody detected both total Smad2 (upper band, 60 kDa) and Smad3 (lower band, 52 kDa).
3.4. Effect of FOXL2 siRNA knockdown on follistatin mRNA and GDF-9 and activin A induction of follistatin transcription
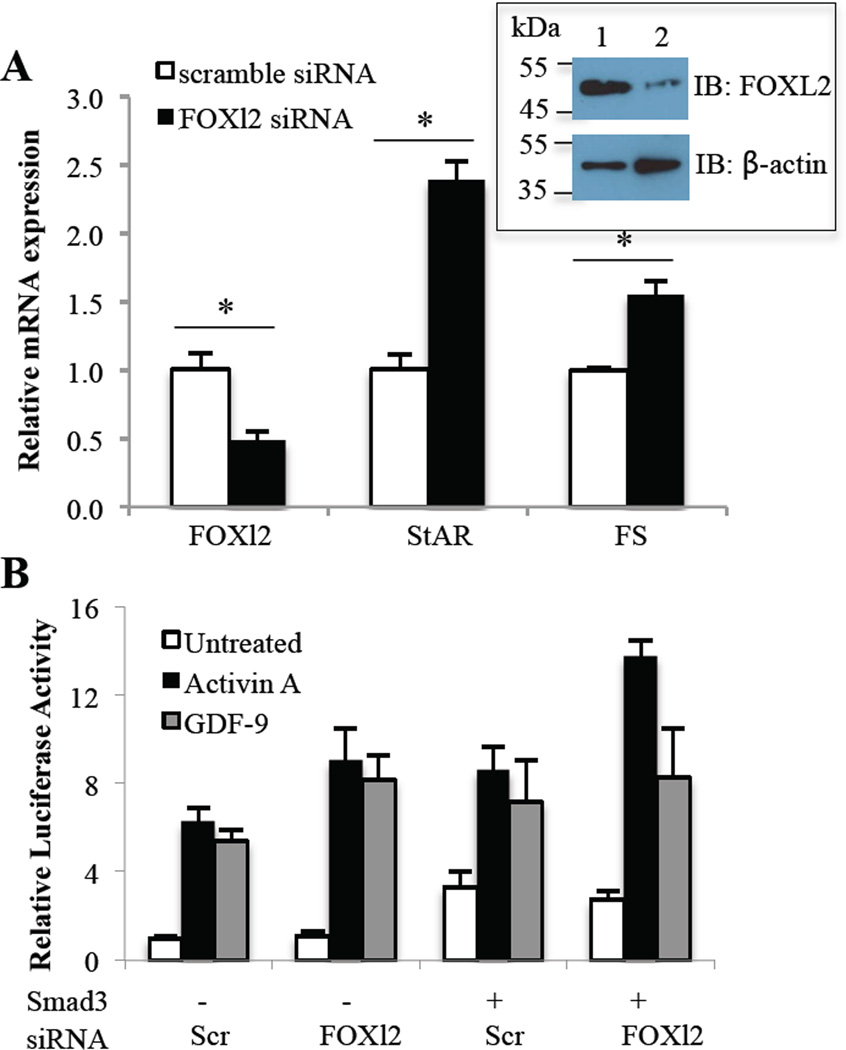
Having demonstrated that GDF-9 and activin A stimulate follistatin mRNA expression and transcription in GCs, we next sought to determine whether FOXL2 was required in this process. We utilized a FOXL2 siRNA to reduce endogenous FOXL2 levels in GCs. After testing a number of siRNA, we selected the most efficacious siRNA that achieved a 52% reduction (p<0.05) in FOXL2 mRNA levels in primary GCs along with a marked reduction in FOXL2 protein levels (Fig. 5A and inset). As StAR is an established target of FOXL2, with FOXL2 shown to inhibit StAR transcription and gene expression in other cell types [6,58], we measured StAR mRNA expression and confirmed that the reduction in FOXL2 lead to increased StAR expression in GCs (Fig. 5A). Interestingly, when we measured follistatin mRNA, we found that FOXL2 reduction by siRNA increased follistatin mRNA expression in GCs (Fig. 5A). This finding was surprising to us based on the pituitary data that showed FOXL2 supported activin-driven Smad3 stimulation of follistatin [25]. To confirm this at a transcriptional level, we transfected GCs with scramble or FOXL2 siRNA 24 hr prior to transfection with the luciferase reporter. Analysis by two-way anova indicated that FOXL2 siRNA significantly increased (P<0.05) follistatin reporter activity across all treatments compared with scramble siRNA (Fig. 5B). Post-hoc tests failed to show significance between groups, thus there was an overall small enhancement of follistatin reporter activity in the presence of FOXL2 suppression. Based on the increase in follistatin mRNA levels by FOXL2 knockdown, we expected to see a similar rise in the basal activity of the follistatin promoter in the presence of FOXL2 siRNA. This disparity most likely reflects different experimental methodologies in the two studies. There was also a significant effect of treatment (P<0.001) on follistatin reporter activity, with activin A and GDF-9 increasing follistatin reporter activity as described in 3.3. Thus, these results in GCs contrast earlier findings in the pituitary, with FOXL2 inhibiting follistatin transcription and expression in the ovary.
Figure 5. Effect of FOXL2 siRNA on follistatin mRNA expression and transcription in primary rat GCs.
A, GCs were transfected with 30 nM negative control scramble siRNA or FOXL2 siRNA and cultured for a further 63 hr prior to evaluation of FOXl2, StAR and Follistatin (FS) mRNA expression. FOXl2, StAR and FS mRNA levels were measured by quantitative real-time PCR and normalized to L19 mRNA levels. Differences in activity were evaluated by a one-way ANOVA. An asterisk indicates a significant difference (p < 0.05) between the indicated scramble siRNA and FOXL2 siRNA groups. Inset shows FOXL2 and control β-actin protein expression in primary rat GCs transfected with scramble siRNA (1) or FOXL2 siRNA (2). B, GCs were transfected with 30 nM control scramble (Scr) or FOXL2 siRNA for 24 hr before transfection with a rat follistatin-luciferase reporter (rFS2.9i-luc) and a Smad3 expression plasmid where indicated, then treated with vehicle (untreated), 100 ng/ml activin A or 300 ng/ml GDF-9 for 48 hr before evaluating luciferase activity. Data are represented as the ratio of the normalized luciferase activity to the untreated no-expression plasmid group. Differences in activity were evaluated by a two-way ANOVA followed by Tukey’s posthoc analysis.
3.5. Comparison of FOXL2C134W and FOXL2wt transcriptional regulation of follistatin
Smad3 is one of the few identified co-regulatory factors known to bind FOXL2 through the forkhead domain, where the C134W mutation is located, and the FOXL2C134W mutant has altered activity on a FOXL2-Smad3 dependent artificial promoter compared with FOXL2wt [29,34]. Thus, we wanted to determine whether this mutation altered GDF-9 and activin A regulation of follistatin in GCs. To achieve this, we over-expressed FOXL2wt or FOXL2C134W. While over-expression is not an ideal system, we feel, in light of findings by other groups that showed disparate regulation of other target genes by FOXL2 when different cell lines were compared, that it is most appropriate to determine FOXL2 function in primary GCs that would contain all the potential co-regulatory factors that may contribute to FOXL2 activity.
We determined by quantitative real-time PCR that FOXL2 mRNA levels were increased around 7 fold in FOXL2 transfected cells, with equivalent levels of FOXL2wt and FOXL2C134W overexpression (Suppl. Fig. 2A). However, when we evaluated FOXL2 protein levels in GCs by immunoblotting with a FOXL2 antibody, we did not detect a dramatic change in total FOXL2 protein levels above the high background of endogenous FOXL2 (Suppl. Fig. 2B). Nonetheless, we confirmed equivalent exogenous FOXL2wt and FOXL2C134W protein expression by immunoblotting against the Flag tag that is present on both the FOXL2wt and FOXL2C134W expression plasmids (Suppl. Fig. 2B). We feel that the vector-derived FOXL2 is translated very poorly and so even though the mRNA levels increased 7 fold, FOXL2 protein levels were only slightly increased, and thus not easily detected by western blotting. We attribute this low level expression of exogenous protein to the fact that they are primary cells (poor transfection efficiency) and cultured in serum free conditions. Therefore, it is possible that poorly transfected GCs masked the expression of exogenous FOXL2.
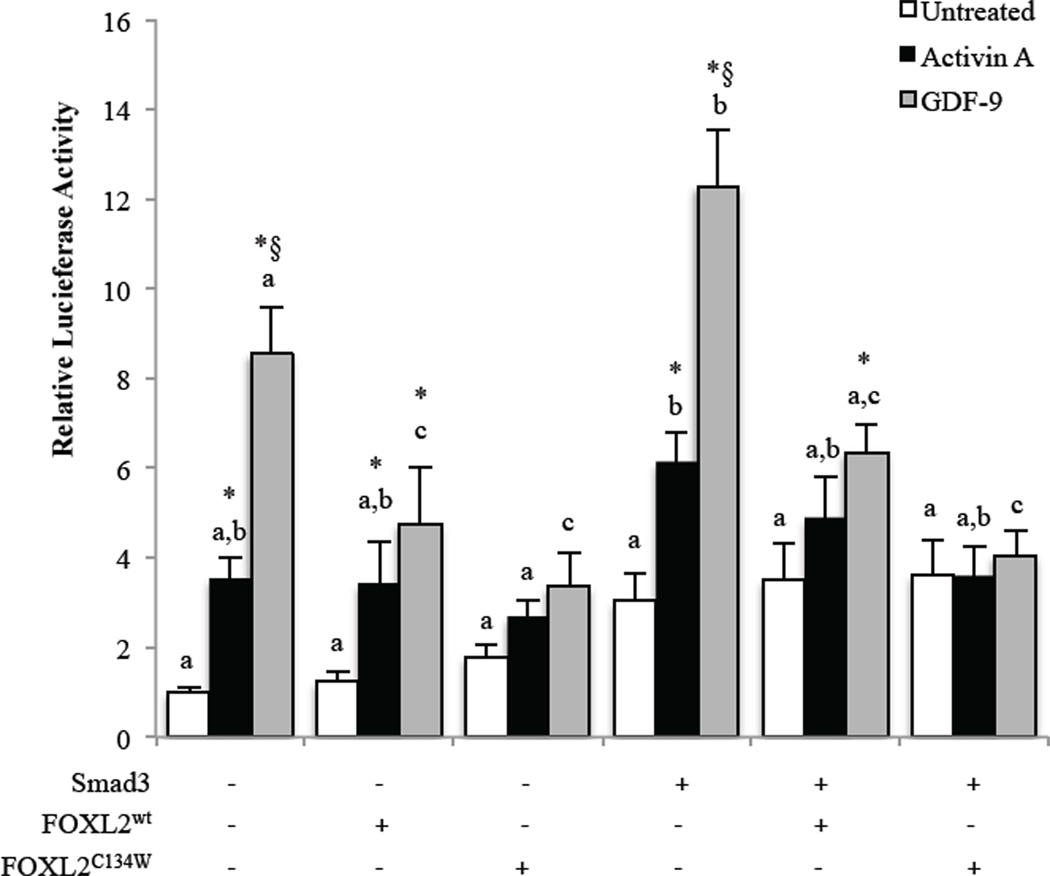
Nevertheless, we have obtained interesting effects of FOXL2 expression in primary rat GCs (Fig. 6), suggesting that the degree of over-expression was small but sufficient to have activity on the luciferase reporter system utilized. As described in 3.3, activin A and GDF-9 stimulate follistatin transcription in GCs (Fig. 6). In the presence of FOXL2wt overexpression, GDF-9 activity was reduced compared with the absence of exogenous FOXL2 expression, but GDF-9 retained stimulatory activity as did activin A compared with untreated cells with FOXL2wt overexpression. However, when FOXL2C134W was expressed the stimulatory activity of both activin A and GDF-9 was lost compared with the matched untreated cells, indicating that the mutant exhibits enhanced suppressive activity compared with FOXL2wt. Overexpression of Smad3 alone, compared with the control, increased follistatin luciferase activity overall (Fig. 6). Under these conditions, FOXL2wt significantly reduced GDF-9-induced follistatin luciferase activity, but the effect of GDF-9 treatment on the activity remained, whereas activin A activity was lost. Also, in the presence of Smad3 overexpression, FOXL2C134W completely ablated the effects of both activin A and GDF-9, indicating again that the mutant has enhanced inhibitory activity when compared with FOXL2wt.
Figure 6. Effect of FOXL2wt and FOXL2C134W on Smad3 and activin or GDF-9-induced follistatin transcription in primary rat GCs.
GCs were transfected with a rat follistatin-luciferase reporter (rFS2.9i-luc) and the indicated FOXL2, FOXL2C134W mutant or Smad3 expression plasmids, then cultured overnight with fresh media before treatment the next day with vehicle (untreated), 100 ng/ml activin A or 300 ng/ml GDF-9 for 8 hr. Data are represented as the ratio of the normalized luciferase activity to the untreated no-expression plasmid group. Differences in activity were evaluated by a two-way ANOVA followed by Tukey’s posthoc analysis. * indicates a significant (p < 0.05) effect of treatment compared with untreated within each transfection condition. § indicates a significant (p < 0.05) difference between activin A and GDF-9 treatment within each transfection condition. Different letters indicate a significant difference (p < 0.05) between transfection conditions within each treatment. Each experiment contained triplicate wells and values shown are mean ± SEM of 5 replicate experiments.
4. Discussion
Follistatin is highly expressed in GCs of developing follicles [35–38] and binds activin [39,43–48] and some BMPs including BMP-7, and BMP-15 [49–51]. Thus, in so doing follistatin inhibits the actions of these GC mitogens [59–61] making follistatin an anti-proliferative factor. Recent findings that FOXL2 and Smad3 coordinately regulate activin induction of follistatin expression in the pituitary drove us to question whether the same regulation occurs in GCs of the ovary; as GCs express follistatin, Smad3 and FOXL2 and are known to respond to several Smad3 signaling factors including activin and GDF-9. An inverted FOXL2-binding site (anti-sense strand, 5′-ATCAATGT-3′), implicated in Smad3-FOXL2 co-regulation of activin-induced follistatin transcription, was identified just downstream of a SBE, within the intronic enhancer of the rat follistatin gene [25]. We analyzed the human follistatin gene and can confirm that this sequence is completely conserved in humans and similarly located within first intron. If the FOXL2C134W mutant exhibits altered interaction with Smad3, as has been suggested [29,34], this could have an effect on GC follistatin expression and ultimately, via altered growth factor availability, on TGF-β superfamily member signaling in the ovary. Thus, dysregulation of follistatin by the GCT FOXL2C134W mutation could result in increased GC proliferation.
Our analysis confirmed that both GDF-9 and activin A similarly induce follistatin mRNA expression. Follistatin acts to negate activin actions [43–48], completing a negative feedback loop between activin and follistatin. In contrast, we found that GDF-9 induction of follistatin was not affected by co-incubation with follistatin indicating that it does not act to regulate GDF-9 actions. We confirmed that GDF-9 and activin A regulation of follistatin was at the level of transcription, and that this is mediated via activation of Smad3. GDF-9 appeared to be slightly more potent than activin A in inducing follistatin reporter activity, which may reflect inherent differences in type I or type II receptor levels for the respective ligands or differences in post-receptor signaling.
To our surprise, in contrast to the supporting role FOXL2 plays in the pituitary, we found that in GCs FOXL2 negatively regulates GDF-9- and activin A-induced follistatin activity. This is a significant and novel finding, suggesting that FOXL2 actions are cell type specific which may reflect the absence/presence of un-identified co-regulatory factors between these two cell types. These results conflict with those described in the mouse GC tumor cell line, KK1 [62], which may reflect species-specific differences or indicate that the KK1 cell line has inherent differences to primary GCs. Cell line-specific FOXL2 activity has been demonstrated for other target genes, with FOXL2 negatively-regulating STAR in two non-GCT cell lines, CHO [6] and COS7 [63], but not in the GCT cell line, COV434 [63]. Similarly, FOXL2 suppressed CYP19A1 promoter activity in CHO cells [64], but enhanced PKA/PKC-stimulated CYP19A1 activity in COS7 and COV434 cells [63]. Cell type differences in follistatin regulation have also been described previously [25]. HepG2 and HEK293 cells that do not express FOXL2 exhibit activin responsiveness in the absence of the intronic enhancer containing the SBE and FBE. Further, in HepG2 and HEK293 cell lines activin responsiveness is linked to both Smad2 and Smad3. Thus, the nature of FOXL2 involvement and the requirement for the intronic SBE and FBE enhancer for activin responsiveness is dependent on the tissue being studied. We propose that cell-specific expression of co-regulatory proteins is a mechanism for controlling sensitivity to stimulators of follistatin production such as activin. Thus, to gain an understanding of FOXL2 actions in the ovary and determine how the FOXL2C134W mutant alters GC function to drive tumor formation, we feel that where possible it is important to evaluate FOXL2 actions in primary GCs to provide the most biologically relevant results, as findings in ovarian and non-ovarian tumor cell lines may be distorted by altered expression of co-regulatory factors.
As a logical next step to determine whether these FOXL2 inhibitory effects required the FBE, we generated follistatin reporters with mutations in the SBE or FBE. Unfortunately, activin A and GDF-9 activity were lost regardless of whether the SBE, FBE or both sites were mutated (data not shown), indicating that both sites are important for activin A- and GDF-9-driven Smad3 signaling. Presumably as a result of this, neither FOXL2wt nor FOXL2C134W exerted any activity when either the SBE or the FBE was mutated. This suggested that although exogenous FOXL2 exerted an inhibitory effect on follistatin transcription, binding of FOXL2 or another unknown forkhead factor to the FBE is required for Smad activity in GCs. However, there could be other explanations, such as unbound FOXL2 sequestering Smad3. Identification of potential co-regulatory factors associated with the Smad3-FOXL2 complex in pituitary versus granulosa cells may shed some light on how FOXL2 is stimulatory in the pituitary and inhibitory in GCs. The exact nature of this relationship and the identity of any other involved GC-specific co-factors remain questions for future studies.
We have shown that FOXL2C134W completely inhibits GDF-9 and activin A transcriptional activation of follistatin in primary rat GCs. Thus, FOXL2C134W exhibits enhanced activity compared with FOXL2wt with respect to follistatin transcription in GCs. Our finding of altered activity of FOXL2C134W on the Smad3 responsive follistatin promoter is consistent with previous findings of altered activity on the artificial GRAS luciferase reporter, which contains AP-1, FOXL2 and Smad binding sites [29,34]. The C134W mutation is located within the DNA-binding forkhead domain. Modeling suggests that the mutation does not disrupt the interaction of FOXL2 with DNA [18]. The enhanced activity of FOXL2C134W may instead reflect an altered interaction with Smad3, which prevents the transcriptional activity of Smad3, or as yet unknown other co-regulators. Exploration of the transcriptional activity of FOXL2C134W on known ovarian FOXL2 targets have shown that other than enhancing PKA/PKC-driven up-regulation of CYP19A1 [63], FOXL2C134W typically exhibits similar transcriptional activity to FOXL2wt [34]. Reduced follistatin in GCs expressing the somatic FOXL2C134W mutation could contribute to tumor formation as a direct result of the loss of activin antagonism. However, as the change in the activity of the FOXL2C134W mutant, with respect to FOXL2wt, on activin A and GDF-9 induction of follistatin transcriptional activity in GCs was relatively small, it seems unlikely that this is a significant mechanism contributing to GCT formation in women.
5. Conclusions
In summary, in this study we have shown that FOXL2 inhibits activin A and GDF-9 induction of follistatin, which is in contrast to previous reports in pituitary cells. Therefore, we propose that FOXL2 transcriptional activity is cell-type dependent, which may reflect a different sub-population of transcriptional co-factors. These novel findings reinforce the importance of studying the role of signaling factors in the cell of interest, preferably in primary cells that should most closely reflect what happens in vivo. Previous studies identifying that a somatic FOXL2 mutation is associated with adult GCTs, supports the view that FOXL2 is involved in the regulation of GC proliferation. In this study, we show that the FOXL2C134W has enhanced activity compared with FOXL2wt, resulting in a more complete inhibition of activin A and GDF-9 induction of anti-proliferative follistatin. This supports a role for FOXL2C134W in promoting GC proliferation, although the small dysregulation of follistatin transcription observed here is unlikely to be a main driving factor in GCT development. Achieving a greater understanding of how FOXL2 acts in the ovary and determining the molecular mechanisms that underlie the pathogenesis of the FOXL2C134W mutation in GCTs should facilitate the development of novel therapeutic and diagnostic options for GCT patients.
Supplementary Material
Highlights.
GDF-9 and activin A stimulate follistatin transcription in primary granulosa cells
Follistatin does not antagonize GDF-9 activity
FoxL2 suppresses GDF-9 and activin A-induced follistatin transcription in the ovary
FoxL2C134W exerts enhanced suppressive activity compared with FoxL2wt
GDF-9 and activin A activity requires intact smad and forkhead binding cis-elements
Acknowledgements
We thank Dr. Louise Bilezikjian for providing us with the follistatin luciferase reporters and Flag-tagged hFOXL2 and Myc-tagged hSmad2, 3 and 4 expression plasmids. We also thank Dr. Rik Derynck for the original hSmad2 and 3 and Dr. Scott Kern for the original hSmad4 cDNA clones. This work was supported in part by the National Institutes of Health (NIH) Grants R01 HD41494 and R21 HD060032, and by the National Institute of Child Health and Human Development/NIH through a cooperative agreement (U54 HD012303) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure summary: The authors have nothing to declare.
References
- 1.Crisponi L, Deiana M, Loi A, Chiappe F, Uda M, Amati P, Bisceglia L, Zelante L, Nagaraja R, Porcu S, Ristaldi MS, Marzella R, Rocchi M, Nicolino M, Lienhardt-Roussie A, Nivelon A, Verloes A, Schlessinger D, Gasparini P, Bonneau D, Cao A, Pilia G. The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat Genet. 2001;27:159–166. doi: 10.1038/84781. [DOI] [PubMed] [Google Scholar]
- 2.Cocquet J, De Baere E, Gareil M, Pannetier M, Xia X, Fellous M, Veitia RA. Structure, evolution and expression of the FOXL2 transcription unit. Cytogenet Genome Res. 2003;101:206–211. doi: 10.1159/000074338. [DOI] [PubMed] [Google Scholar]
- 3.Cocquet J, Pailhoux E, Jaubert F, Servel N, Xia X, Pannetier M, De Baere E, Messiaen L, Cotinot C, Fellous M, Veitia RA. Evolution and expression of FOXL2. J Med Genet. 2002;39:916–921. doi: 10.1136/jmg.39.12.916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellsworth BS, Egashira N, Haller JL, Butts DL, Cocquet J, Clay CM, Osamura RY, Camper SA. FOXL2 in the pituitary: molecular, genetic, and developmental analysis. Mol Endocrinol. 2006;20:2796–2805. doi: 10.1210/me.2005-0303. [DOI] [PubMed] [Google Scholar]
- 5.Loffler KA, Zarkower D, Koopman P. Etiology of ovarian failure in blepharophimosis ptosis epicanthus inversus syndrome: FOXL2 is a conserved, early-acting gene in vertebrate ovarian development. Endocrinology. 2003;144:3237–3243. doi: 10.1210/en.2002-0095. [DOI] [PubMed] [Google Scholar]
- 6.Pisarska MD, Bae J, Klein C, Hsueh AJ. Forkhead l2 is expressed in the ovary and represses the promoter activity of the steroidogenic acute regulatory gene. Endocrinology. 2004;145:3424–3433. doi: 10.1210/en.2003-1141. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt D, Ovitt CE, Anlag K, Fehsenfeld S, Gredsted L, Treier AC, Treier M. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development. 2004;131:933–942. doi: 10.1242/dev.00969. [DOI] [PubMed] [Google Scholar]
- 8.Pisarska MD, Barlow G, Kuo FT. Minireview: Roles of the Forkhead Transcription Factor FOXL2 in Granulosa Cell Biology and Pathology. Endocrinology. 2011 doi: 10.1210/en.2010-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser IS, Shearman RP, Smith A, Russell P. An association among blepharophimosis, resistant ovary syndrome, and true premature menopause. Fertil Steril. 1988;50:747–751. doi: 10.1016/s0015-0282(16)60309-6. [DOI] [PubMed] [Google Scholar]
- 10.Raile K, Stobbe H, Trobs RB, Kiess W, Pfaffle R. A new heterozygous mutation of the FOXL2 gene is associated with a large ovarian cyst and ovarian dysfunction in an adolescent girl with blepharophimosis/ptosis/epicanthus inversus syndrome. Eur J Endocrinol. 2005;153:353–358. doi: 10.1530/eje.1.01974. [DOI] [PubMed] [Google Scholar]
- 11.De Baere E, Copelli S, Caburet S, Laissue P, Beysen D, Christin-Maitre S, Bouchard P, Veitia R, Fellous M. Premature ovarian failure and forkhead transcription factor FOXL2: blepharophimosis-ptosis-epicanthus inversus syndrome and ovarian dysfunction. Pediatr Endocrinol Rev. 2005;2:653–660. [PubMed] [Google Scholar]
- 12.Beysen D, De Paepe A, De Baere E. FOXL2 mutations and genomic rearrangements in BPES. Hum Mutat. 2009;30:158–169. doi: 10.1002/humu.20807. [DOI] [PubMed] [Google Scholar]
- 13.Meduri G, Bachelot A, Duflos C, Bstandig B, Poirot C, Genestie C, Veitia R, De Baere E, Touraine P. FOXL2 mutations lead to different ovarian phenotypes in BPES patients: Case Report. Human Reproduction. 2010;25:235–243. doi: 10.1093/humrep/dep355. [DOI] [PubMed] [Google Scholar]
- 14.Nallathambi J, Moumne L, De Baere E, Beysen D, Usha K, Sundaresan P, Veitia RA. A novel polyalanine expansion in FOXL2: the first evidence for a recessive form of the blepharophimosis syndrome (BPES) associated with ovarian dysfunction. Hum Genet. 2007;121:107–112. doi: 10.1007/s00439-006-0276-0. [DOI] [PubMed] [Google Scholar]
- 15.Dipietromaria A, Benayoun BA, Todeschini AL, Rivals I, Bazin C, Veitia RA. Towards a functional classification of pathogenic FOXL2 mutations using transactivation reporter systems. Hum Mol Genet. 2009;18:3324–3333. doi: 10.1093/hmg/ddp273. [DOI] [PubMed] [Google Scholar]
- 16.De Baere E, Fellous M, Veitia RA. The transcription factor FOXL2 in ovarian function and dysfunction. Folia Histochem Cytobiol. 2009;47:S43–S49. doi: 10.2478/v10042-009-0062-7. [DOI] [PubMed] [Google Scholar]
- 17.Beysen D, Moumne L, Veitia R, Peters H, Leroy BP, De Paepe A, De Baere E. Missense mutations in the forkhead domain of FOXL2 lead to subcellular mislocalization, protein aggregation and impaired transactivation. Hum Mol Genet. 2008;17:2030–2038. doi: 10.1093/hmg/ddn100. [DOI] [PubMed] [Google Scholar]
- 18.Shah SP, Kobel M, Senz J, Morin RD, Clarke BA, Wiegand KC, Leung G, Zayed A, Mehl E, Kalloger SE, Sun M, Giuliany R, Yorida E, Jones S, Varhol R, Swenerton KD, Miller D, Clement PB, Crane C, Madore J, Provencher D, Leung P, DeFazio A, Khattra J, Turashvili G, Zhao Y, Zeng T, Glover JN, Vanderhyden B, Zhao C, Parkinson CA, Jimenez-Linan M, Bowtell DD, Mes-Masson AM, Brenton JD, Aparicio SA, Boyd N, Hirst M, Gilks CB, Marra M, Huntsman DG. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med. 2009;360:2719–2729. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 19.Gershon R, Aviel-Ronen S, Korach J, Daniel-Carmi V, Avivi C, Bar-Ilan D, Barshack I, Meirow D, Ben-Baruch G, Cohen Y. FOXL2 C402G mutation detection using MALDI-TOF-MS in DNA extracted from Israeli granulosa cell tumors. Gynecol Oncol. 2011;122:580–584. doi: 10.1016/j.ygyno.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Jamieson S, Butzow R, Andersson N, Alexiadis M, Unkila-Kallio L, Heikinheimo M, Fuller PJ, Anttonen M. The FOXL2 C134W mutation is characteristic of adult granulosa cell tumors of the ovary. Mod Pathol. 2010;23:1477–1485. doi: 10.1038/modpathol.2010.145. [DOI] [PubMed] [Google Scholar]
- 21.Schumer ST, Cannistra SA. Granulosa cell tumor of the ovary. J Clin Oncol. 2003;21:1180–1189. doi: 10.1200/JCO.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Kalfa N, Veitia RA, Benayoun BA, Boizet-Bonhoure B, Sultan C. The new molecular biology of granulosa cell tumors of the ovary. Genome Med. 2009;1:81. doi: 10.1186/gm81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalfa N, Philibert P, Patte C, Ecochard A, Duvillard P, Baldet P, Jaubert F, Fellous M, Sultan C. Extinction of FOXL2 expression in aggressive ovarian granulosa cell tumors in children. Fertil Steril. 2007;87:896–901. doi: 10.1016/j.fertnstert.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 24.Benayoun BA, Kalfa N, Sultan C, Veitia RA. The forkhead factor FOXL2: A novel tumor suppressor? Biochim Biophys Acta. 2010;1805:1–5. doi: 10.1016/j.bbcan.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Blount AL, Schmidt K, Justice NJ, Vale WW, Fischer WH, Bilezikjian LM. FoxL2 and Smad3 coordinately regulate follistatin gene transcription. J Biol Chem. 2009;284:7631–7645. doi: 10.1074/jbc.M806676200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corpuz PS, Lindaman LL, Mellon PL, Coss D. FoxL2 Is required for activin induction of the mouse and human follicle-stimulating hormone beta-subunit genes. Molecular Endocrinology. 2010;24:1037–1051. doi: 10.1210/me.2009-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamba P, Fortin J, Tran S, Wang Y, Bernard DJ. A novel role for the forkhead transcription factor FOXL2 in activin A-regulated follicle-stimulating hormone beta subunit transcription. Mol Endocrinol. 2009;23:1001–1013. doi: 10.1210/me.2008-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamba P, Wang Y, Tran S, Ouspenskaia T, Libasci V, Hebert TE, Miller GJ, Bernard DJ. Activin A regulates porcine follicle-stimulating hormone beta-subunit transcription via cooperative actions of SMADs and FOXL2. Endocrinology. 2010;151:5456–5467. doi: 10.1210/en.2010-0605. [DOI] [PubMed] [Google Scholar]
- 29.Ellsworth BS, Burns AT, Escudero KW, Duval DL, Nelson SE, Clay CM. The gonadotropin releasing hormone (GnRH) receptor activating sequence (GRAS) is a composite regulatory element that interacts with multiple classes of transcription factors including Smads, AP-1 and a forkhead DNA binding protein. Mol Cell Endocrinol. 2003;206:93–111. doi: 10.1016/s0303-7207(03)00235-1. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, Oakley J, McGee EA. Stage-specific expression of Smad2 and Smad3 during folliculogenesis. Biology of Reproduction. 2002;66:1571–1578. doi: 10.1095/biolreprod66.6.1571. [DOI] [PubMed] [Google Scholar]
- 31.Pangas SA, Matzuk MM. Genetic models for transforming growth factor beta superfamily signaling in ovarian follicle development. Molecular and Cellular Endocrinology. 2004;225:83–91. doi: 10.1016/j.mce.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 32.Shimasaki S, Moore RK, Otsuka F, Erickson GF. The bone morphogenetic protein system in mammalian reproduction. Endocrine Reviews. 2004;25:72–101. doi: 10.1210/er.2003-0007. [DOI] [PubMed] [Google Scholar]
- 33.Knight PG, Glister C. TGF-beta superfamily members and ovarian follicle development. Reproduction. 2006;132:191–206. doi: 10.1530/rep.1.01074. [DOI] [PubMed] [Google Scholar]
- 34.Benayoun BA, Caburet S, Dipietromaria A, Georges A, D'Haene B, Pandaranayaka PJ, L'Hote D, Todeschini AL, Krishnaswamy S, Fellous M, De Baere E, Veitia RA. Functional exploration of the adult ovarian granulosa cell tumor-associated somatic FOXL2 mutation p.Cys134Trp (c.402C>G) PLoS One. 2010;5:e8789. doi: 10.1371/journal.pone.0008789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimasaki S, Koga M, Esch F, Cooksey F, Mercado M, Koba A, Ueno N, Ying S-Y, Ling N. Primary structure of the human follistatin precursor and its genomic organization. Proceedings of the National Academy of Sciences, USA. 1988;85:4218–4222. doi: 10.1073/pnas.85.12.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimasaki S, Koga M, Buscaglia ML, Simmons DM, Bicsak TA, Ling N. Follistatin gene expression in the ovary and extragonadal tissues. Molecular Endocrinology. 1989;3:651–659. doi: 10.1210/mend-3-4-651. [DOI] [PubMed] [Google Scholar]
- 37.Nakatani A, Shimasaki S, DePaolo LV, Erickson GF, Ling N. Cyclic changes in follistatin messenger ribonucleic acid and its protein in the rat ovary during the estrous cycle. Endocrinology. 1991;129:603–611. doi: 10.1210/endo-129-2-603. [DOI] [PubMed] [Google Scholar]
- 38.Roberts VJ, Barth S, El-Roeiy A, Yen SS. Expression of inhibin/activin subunits and follistatin messenger ribonucleic acids and proteins in ovarian follicles and the corpus luteum during the human menstrual cycle. Journal of Clinical Endocrinology and Metabolism. 1993;77:1402–1410. doi: 10.1210/jcem.77.5.8077341. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura T, Takio K, Eto Y, Shibai H, Titani K, Sugino H. Activin-binding protein from rat ovary is follistatin. Science. 1990;247:836–838. doi: 10.1126/science.2106159. [DOI] [PubMed] [Google Scholar]
- 40.Shimonaka M, Inouye S, Shimasaki S, Ling N. Follistatin binds to both activin and inhibin through the common β-subunit. Endocrinology. 1991;128:3313–3315. doi: 10.1210/endo-128-6-3313. [DOI] [PubMed] [Google Scholar]
- 41.Kogawa K, Nakamura T, Sugino K, Takio K, Titani K, Sugino H. Activin-binding protein is present in pituitary. Endocrinology. 1991;128:1434–1440. doi: 10.1210/endo-128-3-1434. [DOI] [PubMed] [Google Scholar]
- 42.Schneyer AL, Rzucidlo DA, Sluss PM, Crowley WF., Jr Characterization of unique binding kinetics of follistatin and activin or inhibin in serum. Endocrinology. 1994;135:667–674. doi: 10.1210/endo.135.2.8033815. [DOI] [PubMed] [Google Scholar]
- 43.DePaolo LV, Bicsak TA, Erickson GF, Shimasaki S, Ling N. Follistatin and activin: A potential intrinsic regulatory system within diverse tissues. Proceedings of the Society for Experimental Biology and Medicine. 1991;198:500–512. doi: 10.3181/00379727-198-43286a. [DOI] [PubMed] [Google Scholar]
- 44.Xiao S, Findlay JK. Interaction between activin and follicle-stimulating hormone-suppressing protein and their mechanisms of action on cultured rat granulosa cells. Molecular and Cellular Endocrinology. 1991;79:99–107. doi: 10.1016/0303-7207(91)90100-7. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura T, Hasegawa Y, Sugino K, Kogawa K, Titani K, Sugino H. Follistatin inhibits activin-induced differentiation of rat follicular granulosa cells in vitro. Biochim Biophys Acta. 1992;1135:103–109. doi: 10.1016/0167-4889(92)90173-9. [DOI] [PubMed] [Google Scholar]
- 46.Xiao S, Robertson DM, Findlay JK. Effects of activin and follicle-stimulating hormone (FSH)-suppressing protein/follistatin on FSH receptors and differentiation of cultured rat granulosa cells. Endocrinology. 1992;131:1009–1016. doi: 10.1210/endo.131.3.1505447. [DOI] [PubMed] [Google Scholar]
- 47.Cataldo NA, Rabinovici J, Fujimoto VY, Jaffe RB. Follistatin antagonizes the effects of activin-A on steroidogenesis in human luteinizing granulosa cells. Journal of Clinical Endocrinology and Metabolism. 1994;79:272–277. doi: 10.1210/jcem.79.1.7517947. [DOI] [PubMed] [Google Scholar]
- 48.Erämaa M, Hildén K, Tuuri T, Ritvos O. Regulation of inhibin/activin subunit messenger ribonucleic acids (mRNAs) by activin A and expression of activin receptor mRNAs in cultured human granulosa-luteal cells. Endocrinology. 1995;136:4382–4389. doi: 10.1210/endo.136.10.7664658. [DOI] [PubMed] [Google Scholar]
- 49.Yamashita H, ten Dijke P, Huylebroeck D, Sampath TK, Andries M, Smith JC, Heldin C-H, Miyazono K. Osteogenic protein-1 binds to activin type II receptors and induces certain activin-like effects. Journal of Cell Biology. 1995;130:217–226. doi: 10.1083/jcb.130.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iemura S, Yamamoto TS, Takagi C, Uchiyama H, Natsume T, Shimasaki S, Sugino H, Ueno N. Direct binding of follistatin to a complex of bone morphogenetic protein and its receptor inhibits ventral and epidermal cell fates in early Xenopus embryo. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9337–9342. doi: 10.1073/pnas.95.16.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otsuka F, Moore RK, Iemura S-I, Ueno N, Shimasaki S. Follistatin inhibits the function of the oocyte-derived factor BMP-15. Biochemical and Biophysical Research Communication. 2001;289:961–966. doi: 10.1006/bbrc.2001.6103. [DOI] [PubMed] [Google Scholar]
- 52.Matzuk MM, Kumar TR, Vassalli A, Bickenbach JR, Roop DR, Jaenisch R, Bradley A. Functional analysis of activins during mammalian development. Nature. 1995;374:354–356. doi: 10.1038/374354a0. [DOI] [PubMed] [Google Scholar]
- 53.Otsuka F, Shimasaki S. A negative feedback system between oocyte bone morphogenetic protein 15 and granulosa cell kit ligand: its role in regulating granulosa cell mitosis. Proceedings of the National Academy of Sciences U S A. 2002;99:8060–8065. doi: 10.1073/pnas.122066899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inouye S, Guo Y, DePaolo L, Shimonaka M, Ling N, Shimasaki S. Recombinant expression of human follistatin with 315 and 288 amino acids: Chemical and biological comparison with native procine follistatin. Endocrinology. 1991;129:815–822. doi: 10.1210/endo-129-2-815. [DOI] [PubMed] [Google Scholar]
- 55.Hashimoto O, Kawasaki N, Tsuchida K, Shimasaki S, Hayakawa T, Sugino H. Difference between follistatin isoforms in the inhibition of activin signalling: activin neutralizing activity of follistatin isoforms is dependent on their affinity for activin. Cell Signal. 2000;12:565–571. doi: 10.1016/s0898-6568(00)00099-1. [DOI] [PubMed] [Google Scholar]
- 56.Blount AL, Vaughan JM, Vale WW, Bilezikjian LM. A Smad-binding element in intron 1 participates in activin-dependent regulation of the follistatin gene. J Biol Chem. 2008;283:7016–7026. doi: 10.1074/jbc.M709502200. [DOI] [PubMed] [Google Scholar]
- 57.Woodruff TK, D'Agostino J, Schwartz NB, Mayo KE. Dynamic changes in inhibin messenger RNAs in rat ovarian follicles during the reproductive cycle. Science. 1988;239:1296–1299. doi: 10.1126/science.3125611. [DOI] [PubMed] [Google Scholar]
- 58.Kuo FT, Fan K, Bentsi-Barnes I, Barlow GM, Pisarska MD. Mouse Forkhead L2 (FOXL2) maintains repression of FSH-dependent genes in the granulosa cell. Reproduction. 2012 doi: 10.1530/REP-11-0259. [DOI] [PubMed] [Google Scholar]
- 59.Miró F, Hillier SG. Modulation of granulosa cell deoxyribonucleic acid synthesis and differentiation by activin. Endocrinology. 1996;137:464–468. doi: 10.1210/endo.137.2.8593790. [DOI] [PubMed] [Google Scholar]
- 60.Otsuka F, Yao Z, Lee TH, Yamamoto S, Erickson GF, Shimasaki S. Bone morphogenetic protein-15: Identification of target cells and biological functions. Journal of Biological Chemistry. 2000;275:39523–39528. doi: 10.1074/jbc.M007428200. [DOI] [PubMed] [Google Scholar]
- 61.Lee M-J, Yang CW, Jin DC, Chang YS, Bang BK, Kim Y-S. Bone morphogenetic protein-7 inhibits constitutive and interleukin-1b-induced monocyte chemoattractant protein-1 expression in human mesangial cells: Role for JNK/AP-1 pathway. Journal of Immunology. 2003;170:2557–2563. doi: 10.4049/jimmunol.170.5.2557. [DOI] [PubMed] [Google Scholar]
- 62.Kashimada K, Pelosi E, Chen H, Schlessinger D, Wilhelm D, Koopman P. FOXL2 and BMP2 act cooperatively to regulate follistatin gene expression during ovarian development. Endocrinology. 2011;152:272–280. doi: 10.1210/en.2010-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fleming NI, Knower KC, Lazarus KA, Fuller PJ, Simpson ER, Clyne CD. Aromatase is a direct target of FOXL2: C134W in granulosa cell tumors via a single highly conserved binding site in the ovarian specific promoter. PLoS One. 2010;5:e14389. doi: 10.1371/journal.pone.0014389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bentsi-Barnes IK, Kuo FT, Barlow GM, Pisarska MD. Human forkhead L2 represses key genes in granulosa cell differentiation including aromatase, P450scc, and cyclin D2. Fertility and Sterility. 2010;94:353–356. doi: 10.1016/j.fertnstert.2009.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.