Abstract
Objective:
To determine the possible interaction of curcumin with P-glycoprotein (P-gp) expression and function by in vitro and in silico studies.
Materials and Methods:
In this study, curcumin was compared for its potential to modulate the expression and function of P-gp in Y79 RB cells by western blot, RT-PCR (reverse transcription polymerase chain reaction) and functional assay. Further, in silico molecular modeling and docking simulations were performed to deduce the inhibitory binding mode of curcumin.
Results:
Western blot and RT-PCR analysis decreased the expression of P-gp in a dose-dependent manner. The effect of curcumin on P-gp function was demonstrated by Rhodamine 123 (Rh123) accumulation and efflux study. Curcumin increased the accumulation of Rh123 and decreased its efflux in retinoblastoma (RB) cells. In addition, curcumin inhibited verapamil stimulated ATPase activity and photoaffinity labeling study showed no effect on the binding of 8-azido-ATP-biotin, indicating its interaction at the substrate binding site. Moreover, molecular docking studies concurrently infer the binding of curcumin into the substrate binding site of P-gp with a binding energy of -7.66 kcal/mol.
Conclusion:
These findings indicate that curcumin suppresses the MDR1 expression and function, and therefore may be useful as modulators of multidrug resistance in RB tumor.
Keywords: Multidrug resistance, p-glycoprotein, retinoblastoma, Y79
INTRODUCTION
Retinoblastoma (RB) is the most common primary intraocular malignancy in children. Chemotherapy plays an important role in the treatment of these RB tumors.[1] Most RB shows an initial response to chemotherapy, but later develops resistance to anticancer drugs due to the expression of multidrug resistance (MDR) protein. Over-expression of P-glycoprotein (P-gp), multidrug resistance-associated protein (MRP1) and lung resistance protein (LRP) is associated with development of drug resistance in most of the cancer cells.[2]
MDR1/P-gp, a 170 kDa plasma membrane protein was the first human ATP-binding cassette transporter to be identified and has the ability to mediate drug resistance in cancer chemotherapy. It transports variety of anti-cancer drugs out of cells and found to be over-expressed in drug resistant cell lines.[3] Chan et al. have reported the increased expression of P-gp in RB cell lines and tumors, while drug-sensitive cell lines were negative for P-gp.[4] Similarly, in our earlier study we showed that, prior to chemotherapy, RB tumor expresses P-gp (38%) and the vault LRP (58%) protein.[1]
For the past few decades, extensive study has been done to solve the problem of MDR. Various chemo-agents have been identified to reverse MDR and sensitize tumor cells to chemotherapeutic agents. These include Elacridar (GF120918), Tariquidar (XR9576) and Valspodar (PSC833) which belong to the second and third-generation modulators.[5] However, MDR in vivo also results due to various other mechanisms which are not caused by P-gp alone. It has been reported that these chemo-agents also interact with the P-gp of healthy organs, resulting in more toxic effects of given anti-cancer drug.[6] Many groups have studied that naturally occurring polyphenols can interact with membrane transport proteins thereby reversing MDR phenotype even at low concentrations. Curcumin is a natural phenolic coloring compound that is found in the rhizomes of Curcuma longa, commonly called as turmeric. Many studies have shown that curcumin exerts various pharmacological properties like anti-inflammatory, anti-oxidant, anti-viral and anti-cancer activities.[7] Thus, in the present study, we attempt to determine the possible interaction of curcumin with P-gp expression and function by in vitro and in silico studies.
MATERIALS AND METHODS
Reagents
Curcumin (Sigma) in pure form was dissolved in dimethyl sulphoxide (DMSO) and stored at -20°C. Cell culture materials were purchased from Invitrogen (Carlsbad, CA, USA). All other chemicals and reagents were of the highest grade commercially available.
Cell lines and culture conditions
The Y79 cell line was obtained from the American Type Culture Collection (Manassas, VA, USA) and maintained in a RPMI-1640 medium supplemented with 20% fetal bovine serum with 50 ng/ml of streptomycin and 1.25 ng/ml of Amphotericin B at 37°C in a humidified atmosphere with 95% O2 and 5% CO2.
Transient transfection with pMDR1-EGFP in RB cell line
Transient transfection were performed by using Lipofectamine 2000 and OPTI-MEM I reduced serum medium (Invitrogen, Carlsbad, CA), following the manufacturer's protocol. Y79 RB cells were plated in 12-well plates at 2 × 105 cells/well and incubated for 24 hours and then transfected with pMDR1-EGFP for 48 hours. After transfection, the efficiency of over-expression of P-gp was measured by western blot.
Effects of curcumin on P-gp expression in RB cell line
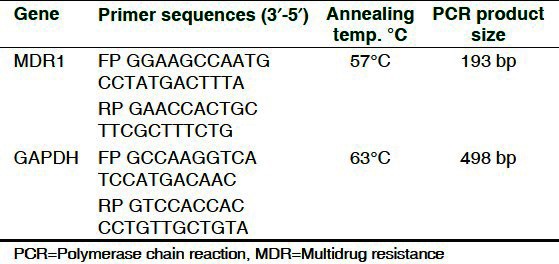
RT-PCR studies for P-gp on curcumin-treated (2-10 μM) and -untreated transfected Y79 RB cells were performed with 2 μg of total RNA from each sample. The cDNA products were amplified for 35 cycles using MDR1- and GAPDH-specific primers with the sense and antisense primer sequence in an Eppendorf PCR system using following cycles: MDR1: 95°C - 45 sec, 57°C - 50 sec, 72°C - 50 sec and final elongation with 72°C - 1 min and GAPDH: 94°C - 5 min, 94°C - 45 sec, 63°C - 45 sec, 72°C - 45 sec and final elongation 72°C - 2 min. PCR product of 193 bp encoding MDR1 and 498 bp encoding GAPDH was fractionated by electrophoresis using 2% agarose gel containing 0.5% ethidium bromide [Table 1]. Total 100 base pair DNA ladder was used to confirm the size of the resultant product (Genei, India). Further, for protein expression studies, the curcumin-treated and -untreated cell lysates (50 μg) were loaded onto 8% polyacrylamide gels, and the western blot for P-gp (1:4000) and β-actin (1:4000) was performed. Protein bands were visualized using a chemiluminescence kit. Molecular weight markers ranging from 220 to 14.3 kDa were used to identify the corresponding bands of P-gp (Genei, India). The relative amount of P-gp mRNA and protein expression was determined by densitometer.
Table 1.
The PCR primers used in the gene expression studies
Accumulation and efflux of rhodamine 123 by flow cytometry
Y79-MDR1 cells (3 × 105/well) were incubated with 1 μg/ml of Rh123 in the presence of curcumin in the dark at 37°C in 5% CO2 for 3 hours. After incubation cells were washed twice with ice-cold HBSS with 10% FBS, and were analyzed using FACScan flow cytometer (Becton-Dickinson) equipped with 488 nm argon laser. The green fluorescence of Rh123 was measured at 530 nm. A minimum of 10,000 events were collected for each sample. For determination of Rh123 efflux, cells were treated with Rh123 for 60 min in the absence of curcumin, and then the medium was replaced with a drug-free medium containing curcumin. After the incubation, the cells were washed twice with ice-cold HBSS and analyzed under flow cytometry.
ATPase activity of P-gp
Y79-MDR1 cells expressing P-gp were incubated at 37°C for 30 min with varying concentrations of curcumin in the presence and absence of sodium orthovanadate (Vi, 0.3 mM) in ATPase assay buffer to measure the amount of inorganic phosphate that is being released. Membrane extracts were incubated with increasing concentration of curcumin in the presence and absence of verapamil. The reaction was initiated by the addition of 5 mM ATP and incubated for 20 min at 37°C. SDS (0.1 ml of 5% SDS) solution was added to terminate the reaction, and the amount of inorganic phosphate released was quantified with a colorimetric reaction.
Photo cross-linking of P-gp with 8-azido-ATP-biotin
Crude membrane of P-gp expressing in transfected Y79-MDR1 cells (100 μg protein) were incubated with 5 mM MgCl2 and 100 μM 8-azido-ATP-biotin in the dark on ice for 5 min in the presence and absence of 10 μM curcumin. The mixture was photolinked using UV light for 10 min. Blotting was performed on the photolinked sample using P-gp monoclonal antibody. The protein band was visualized with streptavidin horseradish peroxidase.
Statistical analysis
All data were expressed as the mean ± standard deviation (SD) from triplicate samples of 3 independent experiments. Comparison of the mean values was performed using the student's unpaired t-test. The results were considered to be statistically significant when P < 0.05.
In silico analysis
Three-dimensional structure of P-gp is yet to be elucidated. Hence, homology search was performed for P-gp (Accession ID: P08183) using BLASTp against PDB with optimal parameters to identify suitable templates for homology modeling. Further, homology-based structure prediction was attempted to determine the plausible 3D structure of P-gp using MODELLER 9V7 software.[8] Among the 100 models generated for P-gp, the one with significant Probability Density Function (PDF) and Discrete Optimized Potential Energy (DOPE) as calculated by the MODELLER 9V7 was chosen as the best model and was subjected to further refinement and analysis.
Further, the refined structure was geometry optimized using GROMACS 4.3.1, a molecular dynamics package.[9,10] The stereo chemical aspects of the refined model was inspected by checking the Phi-Psi angles through visualizing Ramachandran plot generated by Procheck.[11] The structure was also validated using ProSA II server.[12] The structural quality of the protein was further assessed by comparing the topology of the protein through the more sensitive TM-score calculation implemented through TM align.[13] The possible ligand binding cavities within the generated model was predicted using DEPTH.[14] The structural coordinates of curcumin (ACD0022) was retrieved from Indian Plant Anticancer Compound Database[15] and was geometry optimized using PRODRG2 sever[16] with the full charges and optimal chirality.
Molecular docking study was carried out using Auto Dock 4.2. Initially, the energy-optimized macromolecules and the ligands were prepared by adding polar hydrogen. Further, the receptor and ligand were optimized by adding Kollman united atom charges and Gasteiger charges, respectively. Flexibility of the ligand was assigned based on its torsional degrees of freedom through Autotors, with the protein fixed to rigid throughout the process of docking simulation.[17,18]
Grid box covering the entire surface of P-gp (126 × 126 × 126; 0.825 Ǻ) was constructed and used for all docking processes. Further, Grid maps were generated for each atom within the ligand of P-gp using Auto grid. Docking calculation was performed using Lamarckian genetic algorithm (LGA) with default parameters, except for the number of GA runs which was set to 100. Further, cluster analysis was performed to find the optimal binding orientation of the ligand.
Potential binding pose of curcumin with P-gp was predicted based on the binding energy and inhibitory constant. Moreover, polar and non polar interactions were also analyzed and visualized using PyMOL (www.pymol.org) and Ligplot.[19] All the molecular modeling and simulation studies were carried out on Open Discovery Linux platform.[20]
RESULTS
Expression of P-gp in transfected RB cells
The expression of P-gp protein was observed in RB cells transfected with pMDR1-EGFP and no bands were observed in untransfected RB cells by western blot [Figure 1a].
Figure 1.
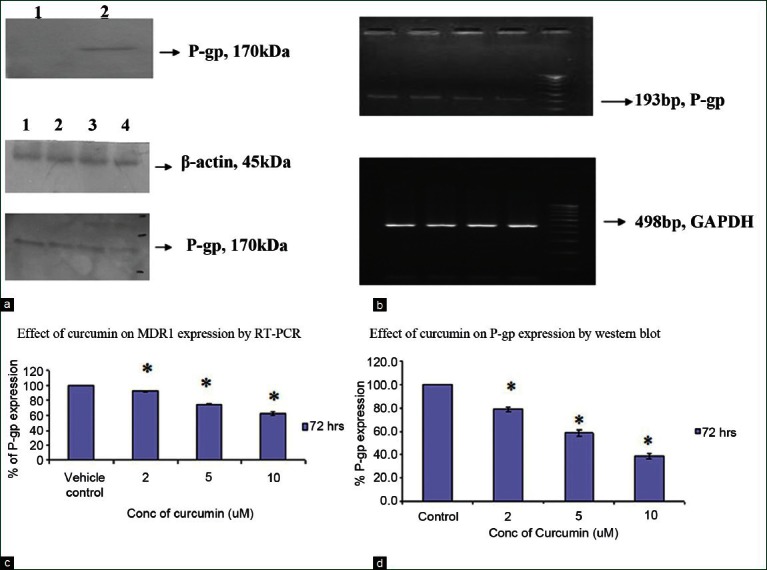
Transient transfection of pMDR1-EGFP in RB cell line: (a) RB cells transfected with MDR1-EGFP in RB cell line showed the expression of P-gp by western blot in RB cell line and the untransfected cells showed no expression of P-gp in RB cell line. (b and c) Effect of curcumin on MDR1 mRNA and protein expression in RB cell line: Dose-dependent effect of curcumin (2, 5, 10 μM) after 72 hours on the MDR1 mRNA and protein expression in Y79-MDR1 cells. Lane 1: Control cells, Lane 2: 2 μM curcumin, Lane 3: 5 μM curcumin, Lane 4: 10 μM curcumin and Lane 5: Molecular weight marker. (d) Densitometric analysis for the MDR1/P-gp expression. For MDR1 mRNA studies, data are expressed as the mean ratio of MDR/GAPDH mRNA band density and P-gp/β-actin band intensity
Effect of curcumin treatment on MDR1 expression by RT-PCR and western blot analysis in RB cells
To verify if curcumin could modulate the P-gp expression, 2-10 μM curcumin was added to the Y79-MDR1 transfected cells and examined after 72 hours. It was found that curcumin-treated cells showed decrease in the expression of MDR1 mRNA and protein expression with increase in concentration of curcumin. The P-gp at 170 kDa and β-actin (internal control) at 45 kDa was observed by western blot analysis [Figure 1b]. The MDR1 and GAPDH (housekeeping gene) mRNA expression in the Y79-MDR1 cells indicated bands at 193 bp and 498 bp, respectively [Figure 1c]. Values for the expression of MDR1 mRNA by Y79 cells after treatment with 2, 5 and 10 μM were decreased by 92%, 77% and 61%, respectively, in 3 independent experiments. Similarly the P-gp expression in Y79-MDR1 cells was decreased by 79%, 59% and 38% in curcumin-treated cells when compared with the control cells [Figure 1d].
Effect of curcumin on MDR1 function by flow cytometry
To examine the function of P-gp on curcumin-treated Y79-MDR1 cells, Rh123 accumulation and efflux were studied using flow cytometry. It was observed that curcumin-treated RB cells showed an increase in the accumulation of Rh123 in a concentration-dependent manner and also showed a significant decrease in the efflux of Rh123. The results were statistically significant when compared with the control values P < 0.05 [Figure 2].
Figure 2.
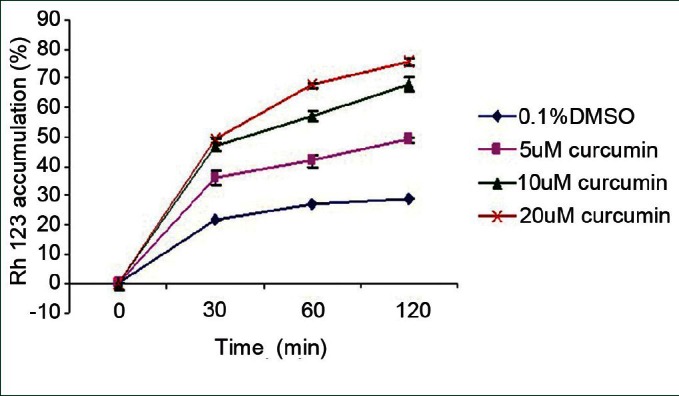
MDR1 functional study in the Y79 RB cells: Cells were treated with different concentration of curcumin (5, 10 and 20 μM) and vehicle control (0.1% DMSO). Rh123 was added and the cells were incubated for different time intervals at 37°C in the dark. Cells were then harvested and used immediately to measure Rh123 fluorescence using flow cytometry. Data obtained are the mean ± S.D from 3 independent experiments
Effect of curcumin on ATPase activity of P-gp
ATPase activity of P-gp was studied to assess the effect of curcumin on this transporter. Our results show that curcumin was able to stimulate the basal ATPase activity of P-gp at very a low concentration, but inhibited the activity at a higher concentration. This is indicative of curcumin's direct interaction with the P-gp. Moreover, we also observed the inhibition of verapamil-stimulated ATP hydrolysis by P-gp, mediated by curcumin in Y79 RB cell lines [Figure 3a].
Figure 3.
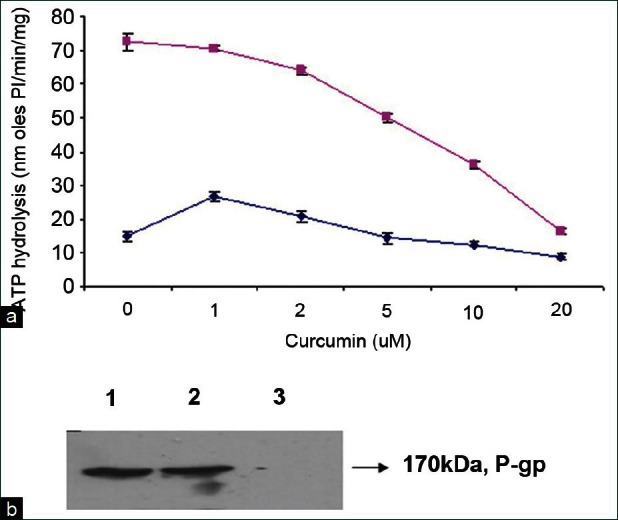
Effect of curcumin on basal and verapamil-stimulated ATPase activity and photoaffinity labeling of MDR1 with 8-azido-ATP-biotin: (a) Y79-MDR1 cells expressing P-gp (100 μg of protein/ml) were incubated with increasing concentration of curcumin in the presence of verapamil (■) or an equivalent volume of DMSO (♦) in the ATPase assay buffer. Values are mean ± SE from at least 3 independent experiments. (b) Crude membranes (100 μg protein) were incubated with 100 μM 8-azido-ATP-biotin in the dark on ice for 5 min in the presence and absence of 10 μM curcumin. The protein was electrophoresed, blotted and visualized with streptavidin horseradish peroxidase. The addition of 10 mM ATP prevented photo-labeling. Lane 1: 8-azido-ATP-biotin, Lane 2: 10 μM curcumin and Lane 3: 10 mM ATP
Effect of curcumin on photoaffinity labeling of P-gp with 8-azido-ATP-biotin
To determine the interaction sites of P-gp with curcumin, photoaffinity labeling of P-gp was performed using 8-azido-ATP-biotin. Curcumin had no effect on 8-azido-ATP-biotin at 10 μM concentration in Y79 RB cell line. The cross linking of 8-azido-ATP was inhibited in the presence of 10 mM ATP. This suggests that curcumin produces their effect mostly by interacting at the substrate binding sites rather than at the nucleotide-binding sites of this protein [Figure 3b].
Molecular modeling of P-gp
Among the experimentally determined structures of ABC transporter super family, MRP 1a) of Mus musculus (PDBID: 3G5U) was found to be an appropriate template for human P-gp. Primary sequence analysis revealed that human P-gp and MRP 1a of Mus musculus share the sequence identity of 88% and similarity of 94% with an E-value of 0.0 and query coverage of 99% (34-1280) in comparison to other hits. Hence, a 3-dimensional structure of human P-gp was generated with 3G5U as a template using MODELLER 9V7. The model was further optimized through energy minimization with a potential energy of -1.6654603e + 05 kJ/mol. Like 3G5U, the optimized 3-dimensional structure of human P-gp is predominantly helical in TM domains and β-sheets in the NBDs [Figure 4].
Figure 4.
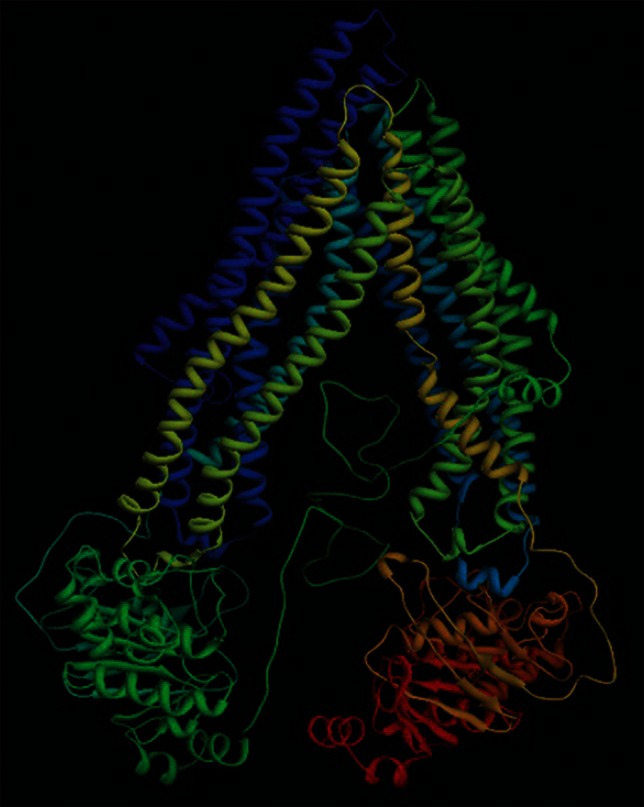
Homology-based 3-dimensional structure of P-gp
The overall stereo-chemical aspects of the model were assessed through Procheck. Psi-Phi distribution of the amino acids were inspected through Ramachandran plot, in which 88.8% of the residues in the most favored region, 9.3% in allowed region, 1.9% in additionally allowed region and no residues in disallowed region. Moreover, the overall quality of the model was evaluated by comparing the Z-score of generated model to the experimentally determined [Figure 5a]. The interaction energy of each amino acid within the generated model was validated by checking the energetic aspect of the model calculated using ProSA, by plotting the energies as the function of amino acid sequence position. The plot revealed negative energy for maximum of residues with few residues having positive energy [Figure 5b].
Figure 5.
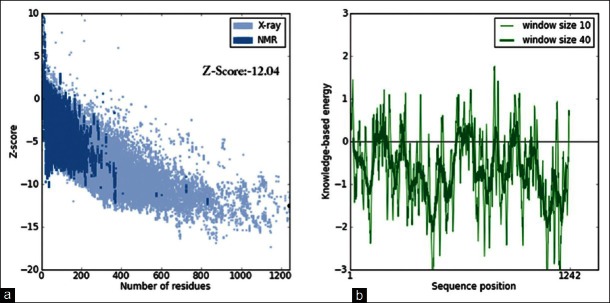
(a) Overall quality of the model assessed based on Z-score. Z-score of the generated structure is within the range observed for native set of proteins of same size. (b) Local quality of the P-gp model assessed based on the energy plot calculated using ProSA
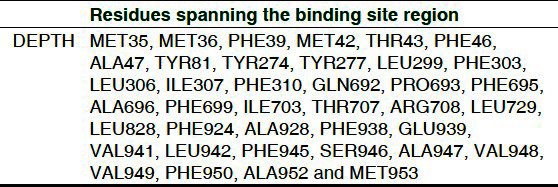
The quality of the structural fold was assessed by comparing the predicted model with the experimentally determined one through structural super positioning. RMSD of 0.47 and TM-score of 0.99830 were calculated using the more sensitive structural alignment algorithm, TM-align for the model/template. These scores were suggestive of the generated model to share the same topology with a high probability. Hence, the model validated at the geometrical and energetic aspects ensure the plausibility for future analysis. Further, binding site residues of the multi-domain protein P-gp was predicted using DEPTH with a threshold fixed to 0.50 [Table 2].
Table 2.
Binding cavity prediction using DEPTH
Molecular docking of P-gp with curcumin
Molecular docking studies were performed for the optimized 3-dimensional structures of P-gp with curcumin using LGA, to infer its inhibitory binding mode. Among the 100 conformers generated during the molecular docking analysis for P-gp, potential pose with lowest binding energy and highest binding affinity was selected for further analysis. Curcumin showed increased binding affinity towards P-gp with a binding energy of -7.66 kcal/mol and inhibitory constant of 2.42 μM by establishing a network of bonded and non-bonded interactions with Leu32, Met35, Met36, Phe39, Phe161, Leu299, Thr300, Phe303, Ile307, Ser311, Met916, and Phe945 residing in the transmembrane region and were proven to have drug interactions [Figure 6a and b].
Figure 6.
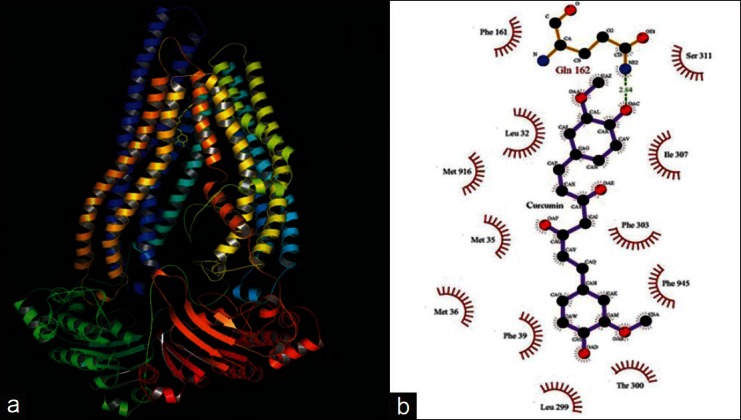
Molecular interactions observed between P-gp and curcumin in 3-dimension (a) using PyMOL and 2D (b) using LigPlot analysis
DISCUSSION
Over-expression of MDR proteins in tumors is a major obstacle for successful chemotherapy.[21] Various modulators have been identified to reverse MDR mechanisms and thus sensitize MDR cancer cells to anti-cancer agents.[22] Our recent study found that curcumin is an inhibitors of MRP1[23] and LRP,[24] which have been found to play an important role in the development of drug resistance in tumor cells. It has been reported that curcumin is able to modulate the expression and function of P-gp in human gastric carcinoma cells and primary cultures of rat hepatocytes.[25] In Y79 RB cells, P-gp is not expressed at the protein level and thus in the present study, P-gp was over-expressed by transfection in RB cells and tested for their interaction of curcumin with MDR1 protein by in vitro and in silico studies.
Concentration with minimal toxicity was chosen to study the effect of curcumin on MRP. The effect of curcumin on the MDR1 gene (P-gp) expression was determined by western blotting and RT-PCR in Y79-MDR1 RB cells. The result showed that curcumin inhibited the MDR1 gene expression at the mRNA and protein level in Y79-MDR1 cells. It has been reported earlier that the methoxy group present in curcumin might be responsible for the inhibitory effect of P-gp, thereby functioning as an MDR modulator.[26]
In the present study we also investigated the effect of curcumin on the P-gp function in Y79-MDR1 cells. Rh123 was used for the assay, which is known to be a good substrate for P-gp.[27] Curcumin caused an increase in the accumulation of Rh123, and inhibited its efflux in a concentration dependent manner, but had no effect on the untransfected RB cells. These results are quite similar to the work done by Tang et al., where they have shown modulation of P-gp expression and function by curcumin in MDR human gastric carcinoma cell line.[28] An ATPase assay was performed, which clearly demonstrated that curcumin inhibited verapamil-stimulated ATPase activity of P-gp at higher concentration. These findings were further supported by the effect of curcumin on the photoaffinity labeling study, which showed that curcumin had no effect on the binding of 8-azido-ATP-biotin. These results clearly demonstrate that curcumin interact at the substrate binding site of P-gp and not on the nucleotide binding region. Similar results were observed by Chearwae et al. where stimulation of ATPase activity of verapamil was inhibited by curcumin.[7] Further, docking studies inferred bonded and non-bonded interaction of curcumin with the TMDs of P-gp.
CONCLUSION
The present study suggested that curcumin modulated the expression and function of the MDR1 in the Y79 RB cells, which is similar to our earlier study on MRP1.[23] However, extensive studies on the bioavailability of curcumin have to be done to know its distribution in tissues after administration at pharmacological dose. Thus, curcumin may be used as a chemosensitizer in cancer therapy to reverse the MDR in cancer cells.
ACKNOWLEDGMENT
Grant from the Indian Council of Medical Research (5/13/25/04/NCD-III). The pMDR1-EGFP were a generous gift from Dr. S. Simon, The Rockefeller University, New York.
Footnotes
Source of Support: Grant from the Indian Council Medical Research (5/13/25/04/NCD-III)
Conflict of Interest: None declared.
REFERENCES
- 1.Krishnakumar S, Mallikarjuna K, Desai N, Muthialu A, Venkatesan N, Sundaram A, et al. Multidrug resistant proteins: P-glycoprotein and lung resistance protein expression in retinoblastoma. Br J Ophthalmol. 2004;88:1521–6. doi: 10.1136/bjo.2004.047928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Van Brussel JP, Van Steenbrugge GJ, Romijn JC, Schroder FH, Mickisch GH. Chemosensitivity of prostate cancer cell lines and expression of multidrug resistance-related proteins. Eur J Cancer. 1999;35:664–71. doi: 10.1016/s0959-8049(98)00435-3. [DOI] [PubMed] [Google Scholar]
- 3.Choi CH. ABC transporters as multidrug resistance mechanisms and the development of chemosensitizers for their reversal. Cancer Cell Int. 2005;5:30. doi: 10.1186/1475-2867-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan HS, Thorner PS, Haddad G, Gallie B. Multidrug-resistant phenotype in retinoblastoma correlates with P-glycoprotein expression. Ophthalmology. 1991;98:1425–31. doi: 10.1016/s0161-6420(91)32134-1. [DOI] [PubMed] [Google Scholar]
- 5.Tan B, Piwnica-Worms D, Rater L. Multidrug resistance transporters and modulation. Curr Opin Oncol. 2000;12:450–8. doi: 10.1097/00001622-200009000-00011. [DOI] [PubMed] [Google Scholar]
- 6.Oh KT, Baik HJ, Lee AH, Oh YT, Youn YS, Lee ES. The reversal of drug-resistance in tumors using a drug-carrying nanoparticular system. Int J Mol Sci. 2009;10:3776–92. doi: 10.3390/ijms10093776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chearwae W, Anuchapreeda S, Nandigama K, Ambudkar SV, Limtrakul P. Biochemical mechanism of modulation of human P-glycoprotein (ABCB1) by curcumin I, II, and III purified from Turmeric powder. Biochem Pharmacol. 2004;68:2043–52. doi: 10.1016/j.bcp.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Dawson RJ, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–5. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- 9.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–59. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 10.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 11.Summa CM, Levitt M. Near-native structure refinement using in vacuo energy minimization. Proc Natl Acad Sci USA. 2007;104:3177–82. doi: 10.1073/pnas.0611593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–91. [Google Scholar]
- 13.Wiederstein M, Sippl MJ. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–10. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu J, Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics. 2010;26:889–95. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan KP, Varadarajan R, Madhusudhan MS. DEPTH: A web server to compute depth and predict small-molecule binding cavities in proteins. Nucleic Acids Res. 2011;39:W242–8. doi: 10.1093/nar/gkr356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vetrivel U, Subramanian N, Pilla K. InPACdb: Indian plant anticancer compounds database. Bioinformation. 2009;5:71–4. doi: 10.6026/97320630004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schüttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–63. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 18.Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: Applications of AutoDock. J Mol Recognit. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Corbeil CR, Englebienne P, Moitessier N. Docking ligands into flexible and solvated macromolecules. 1. Development and validation of FITTED 1.0. J Chem Inf Model. 2007;47:435–49. doi: 10.1021/ci6002637. [DOI] [PubMed] [Google Scholar]
- 20.Vetrivel U, Pilla K. Open discovery: An integrated live Linux platform of Bioinformatics tools. Bioinformation. 2008;3:144–6. doi: 10.6026/97320630003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arceci RJ. Tumor cell survival and resistance to therapy. Curr Opin Hematol. 1996;3:279–87. doi: 10.1097/00062752-199603040-00006. [DOI] [PubMed] [Google Scholar]
- 22.Anuchapreeda S, Leechanachai P, Smith MM, Ambudkar SV, Limtrakul PN. Modulation of P-glycoprotein expression and function by curcumin in multidrug-resistant human KB cells. Biochem Pharmacol. 2002;64:573–82. doi: 10.1016/s0006-2952(02)01224-8. [DOI] [PubMed] [Google Scholar]
- 23.Sreenivasan S, Ravichandran S, Vetrivel U, Krishnakumar S. In vitro and In silico studies on inhibitory effects of curcumin on multi-drug resistance associated protein (MRP1) in retinoblastoma cells. Bioinformation. 2012;8:13–9. doi: 10.6026/97320630008013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thiyagarajan S, Thirumalai V, Nirmala S, Biswas J, Krishnakumar S. Effect of curcumin on lung resistance-related protein (LRP) in retinoblastoma cells. Curr Eye Res. 2009;34:845–51. doi: 10.3109/02713680903125329. [DOI] [PubMed] [Google Scholar]
- 25.Romiti N, Tongiani R, Cervelli F, Chieli E. Effects of curcumin on P-glycoprotein in primary cultures of rat hepatocytes. Life Sci. 1998;62:2349–58. doi: 10.1016/s0024-3205(98)00216-1. [DOI] [PubMed] [Google Scholar]
- 26.Ammon HP, Wahl MA. Pharmacology of Curcuma longa. Planta Med. 1991;57:1–7. doi: 10.1055/s-2006-960004. [DOI] [PubMed] [Google Scholar]
- 27.Chen LM, Liang YJ, Ruan JW, Ding Y, Wang XW, Shi Z, et al. Reversal of P-gp mediated multidrug resistance in vitro and in vivo by FG020318. J Pharm Pharmacol. 2004;56:1061–6. doi: 10.1211/0022357043879. [DOI] [PubMed] [Google Scholar]
- 28.Tang XQ, Bi H, Feng JQ, Cao JG. Effect of curcumin on multidrug resistance in resistant human gastric carcinoma cell line SGC7901/VCR. Acta Pharmacol Sin. 2005;26:1009–16. doi: 10.1111/j.1745-7254.2005.00149.x. [DOI] [PubMed] [Google Scholar]