Abstract
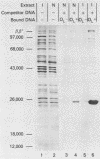
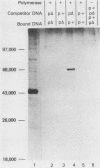
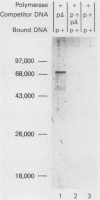
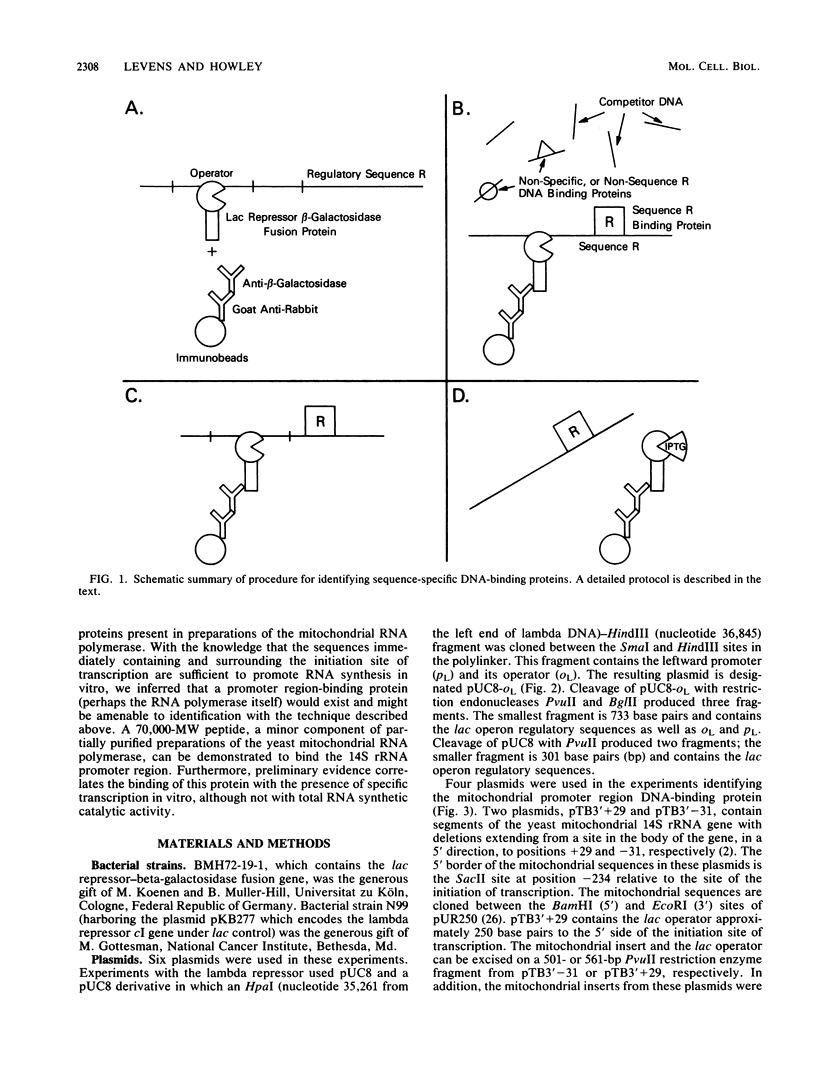
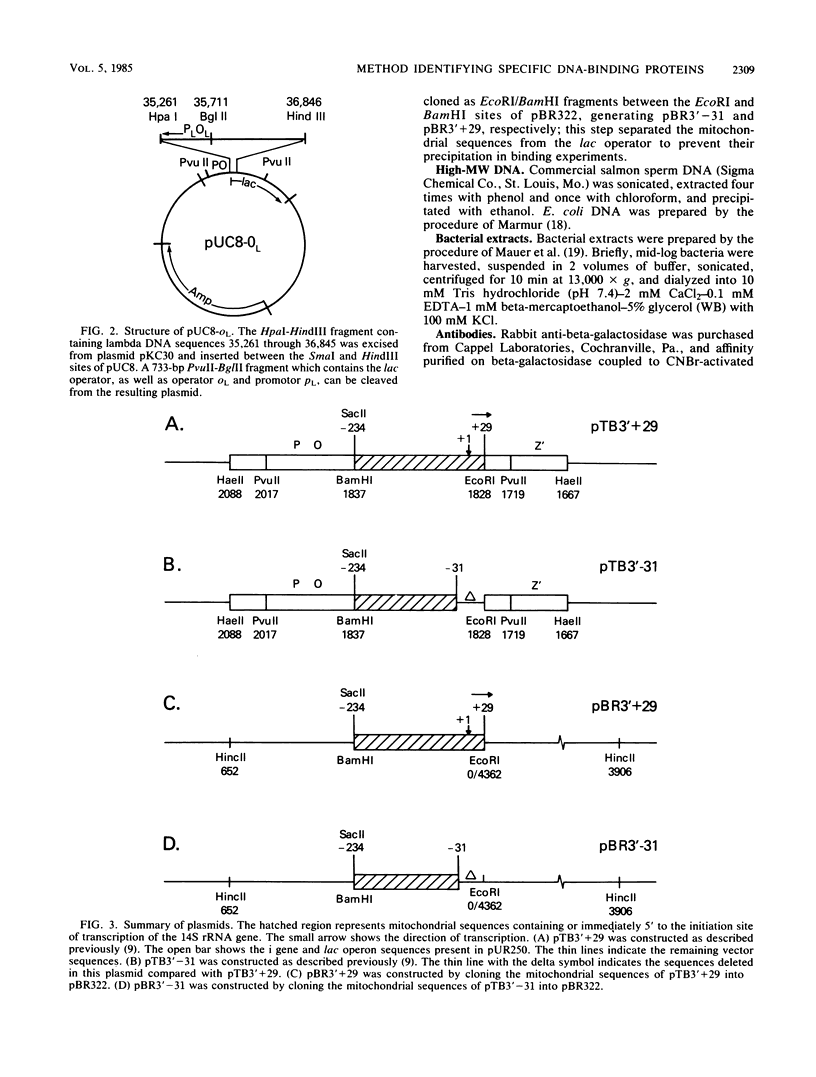
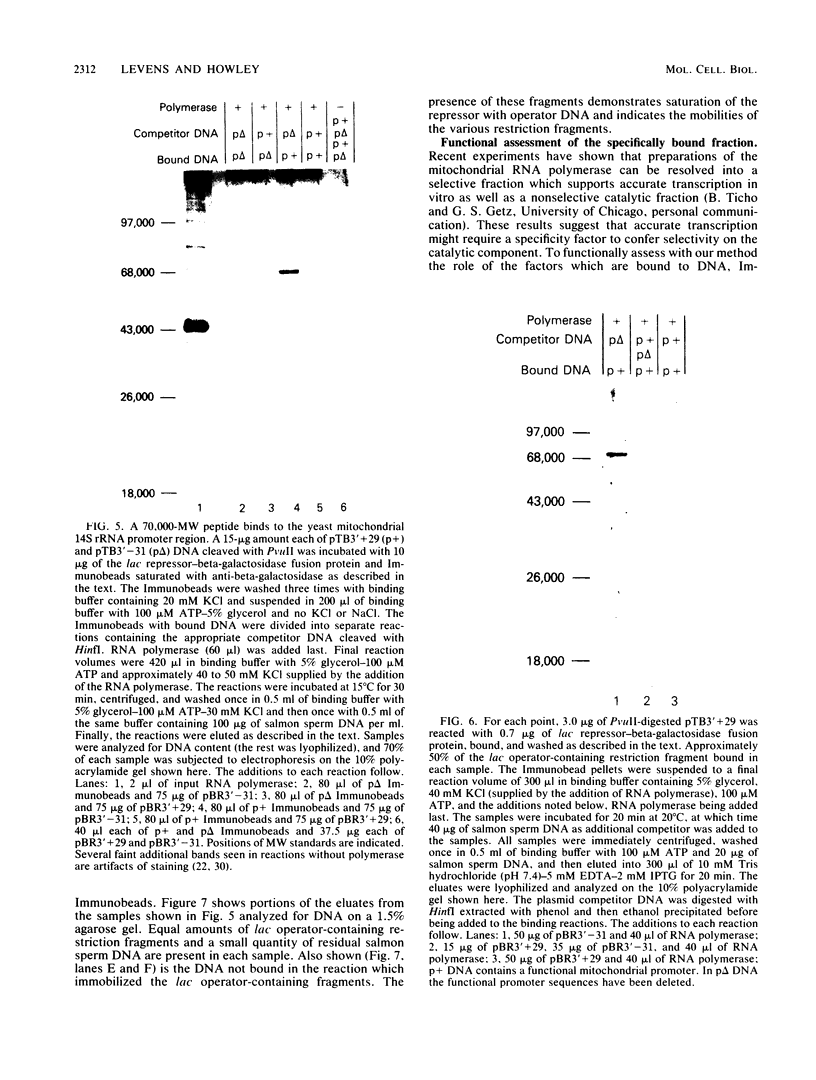
We developed a general method for the enrichment and identification of sequence-specific DNA-binding proteins. A well-characterized protein-DNA interaction is used to isolate from crude cellular extracts or fractions thereof proteins which bind to specific DNA sequences; the method is based solely on this binding property of the proteins. The DNA sequence of interest, cloned adjacent to the lac operator DNA segment is incubated with a lac repressor-beta-galactosidase fusion protein which retains full operator and inducer binding properties. The DNA fragment bound to the lac repressor-beta-galactosidase fusion protein is precipitated by the addition of affinity-purified anti-beta-galactosidase immobilized on beads. This forms an affinity matrix for any proteins which might interact specifically with the DNA sequence cloned adjacent to the lac operator. When incubated with cellular extracts in the presence of excess competitor DNA, any protein(s) which specifically binds to the cloned DNA sequence of interest can be cleanly precipitated. When isopropyl-beta-D-thiogalactopyranoside is added, the lac repressor releases the bound DNA, and thus the protein-DNA complex consisting of the specific restriction fragment and any specific binding protein(s) is released, permitting the identification of the protein by standard biochemical techniques. We demonstrate the utility of this method with the lambda repressor, another well-characterized DNA-binding protein, as a model. In addition, with crude preparations of the yeast mitochondrial RNA polymerase, we identified a 70,000-molecular-weight peptide which binds specifically to the promoter region of the yeast mitochondrial 14S rRNA gene.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backman K., Ptashne M. Maximizing gene expression on a plasmid using recombination in vitro. Cell. 1978 Jan;13(1):65–71. doi: 10.1016/0092-8674(78)90138-1. [DOI] [PubMed] [Google Scholar]
- Biswas T. K., Edwards J. C., Rabinowitz M., Getz G. S. Characterization of a yeast mitochondrial promoter by deletion mutagenesis. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1954–1958. doi: 10.1073/pnas.82.7.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brake A. J., Fowler A. V., Zabin I., Kania J., Müller-Hill B. beta-Galactosidase chimeras: primary structure of a lac repressor-beta-galactosidase protein. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4824–4827. doi: 10.1073/pnas.75.10.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T., Rabinowitz M. Identification of multiple transcriptional initiation sites on the yeast mitochondrial genome by in vitro capping with guanylyltransferase. J Biol Chem. 1983 Nov 25;258(22):14025–14033. [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Edwards J. C., Levens D., Rabinowitz M. Analysis of transcriptional initiation of yeast mitochondrial DNA in a homologous in vitro transcription system. Cell. 1982 Dec;31(2 Pt 1):337–346. doi: 10.1016/0092-8674(82)90127-1. [DOI] [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fowler A. V., Zabin I. Purification, structure, and properties of hybrid beta-galactosidase proteins. J Biol Chem. 1983 Dec 10;258(23):14354–14358. [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Meyer B. J., Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D., Pabo C. O., Sauer R. T. Bacteriophage lambda repressor and cro protein: interactions with operator DNA. Methods Enzymol. 1980;65(1):839–856. doi: 10.1016/s0076-6879(80)65078-2. [DOI] [PubMed] [Google Scholar]
- Levens D., Lustig A., Rabinowitz M. Purification of mitochondrial RNA polymerase from Saccharomyces cerevisiae. J Biol Chem. 1981 Feb 10;256(3):1474–1481. [PubMed] [Google Scholar]
- Lin S. Y., Riggs A. D. Lac repressor binding to non-operator DNA: detailed studies and a comparison of eequilibrium and rate competition methods. J Mol Biol. 1972 Dec 30;72(3):671–690. doi: 10.1016/0022-2836(72)90184-2. [DOI] [PubMed] [Google Scholar]
- Manley J. L., Fire A., Cano A., Sharp P. A., Gefter M. L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer R., Meyer B., Ptashne M. Gene regulation at the right operator (OR) bacteriophage lambda. I. OR3 and autogenous negative control by repressor. J Mol Biol. 1980 May 15;139(2):147–161. doi: 10.1016/0022-2836(80)90302-2. [DOI] [PubMed] [Google Scholar]
- McKay R. D. Binding of a simian virus 40 T antigen-related protein to DNA. J Mol Biol. 1981 Jan 25;145(3):471–488. doi: 10.1016/0022-2836(81)90540-4. [DOI] [PubMed] [Google Scholar]
- Müller-Hill B., Kania J. Lac repressor can be fused to beta-galactosidase. Nature. 1974 Jun 7;249(457):561–563. doi: 10.1038/249561a0. [DOI] [PubMed] [Google Scholar]
- Ochs D. Protein contaminants of sodium dodecyl sulfate-polyacrylamide gels. Anal Biochem. 1983 Dec;135(2):470–474. doi: 10.1016/0003-2697(83)90714-5. [DOI] [PubMed] [Google Scholar]
- Penefsky H. S. Reversible binding of Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1977 May 10;252(9):2891–2899. [PubMed] [Google Scholar]
- Ptashne M., Backman K., Humayun M. Z., Jeffrey A., Maurer R., Meyer B., Sauer R. T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976 Oct 8;194(4261):156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- Riggs A. D., Newby R. F., Bourgeois S. lac repressor--operator interaction. II. Effect of galactosides and other ligands. J Mol Biol. 1970 Jul 28;51(2):303–314. doi: 10.1016/0022-2836(70)90144-0. [DOI] [PubMed] [Google Scholar]
- Rüther U. pUR 250 allows rapid chemical sequencing of both DNA strands of its inserts. Nucleic Acids Res. 1982 Oct 11;10(19):5765–5772. doi: 10.1093/nar/10.19.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver N., Gruss P., Law M. F., Khoury G., Howley P. M. Bovine papilloma virus deoxyribonucleic acid: a novel eucaryotic cloning vector. Mol Cell Biol. 1981 Jun;1(6):486–496. doi: 10.1128/mcb.1.6.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Folkmanis A., Echols H. Cro regulatory protein specified by bacteriophage lambda. Structure, DNA-binding, and repression of RNA synthesis. J Biol Chem. 1977 Sep 10;252(17):6177–6183. [PubMed] [Google Scholar]
- Tasheva B., Dessev G. Artifacts in sodium dodecyl sulfate-polyacrylamide gel electrophoresis due to 2-mercaptoethanol. Anal Biochem. 1983 Feb 15;129(1):98–102. doi: 10.1016/0003-2697(83)90057-x. [DOI] [PubMed] [Google Scholar]