Abstract
Background
Human metapneumovirus (HMPV) is an important cause of acute respiratory illnesses in children. HMPV encodes two major surface glycoproteins, fusion (F) and glycoprotein (G). The function of G has not been fully established, though it is dispensable for in vitro and in vivo replication.
Methods
We analyzed 87 full-length HMPV G sequences from isolates collected over 20 years.
Results
The G sequences fell into four subgroups with a mean 63% amino acid identity (minimum 29%). The length of G varied from 217 to 241 residues. Structural features such as proline content and N- and O-glycosylation sites were present in all strains but quite variable between subgroups. There was minimal drift within the subgroups over 20 years. The estimated time to the most recent common ancestor was 215 years.
Conclusions
HMPV G was conserved within lineages over 20 years, suggesting functional constraints on diversity. However, G was poorly conserved between subgroups, pointing to potentially distinct roles for G among different viral lineages.
Keywords: metapneumovirus, Paramyxoviridae infections/virology, respiratory tract respiratory tract, metapneumovirus/genetics
Introduction
Human metapneumovirus (HMPV) is a paramyxovirus associated with acute lower respiratory infection [1]. HMPV is a leading cause of LRI in young children worldwide and is associated with severe disease in immunocompromised hosts or persons with underlying conditions [2–15]. The virus most closely related genetically to HMPV is avian metapneumovirus type C (AMPV-C) [1]. AMPV is an emerging pathogen of poultry first identified in 1978 [16]. Subtypes AMPV-A and AMPV-B circulate in Europe and Africa, while AMPV-C was discovered in Minnesota and has been detected in the US and Korea [17, 18]. Productive experimental infection of poultry with HMPV has not been successful, and serological studies have failed to detect evidence of human infection by AMPV [1]. Thus, HMPV infection of humans does not appear to be a recent zoonosis, but likely diverged from AMPV-C [19, 20]. An understanding of the molecular origin and evolution of HMPV sequences has implications for other emerging zoonotic paramyxoviruses such as Hendra and Nipah viruses.
HMPV encodes two major surface glycoproteins, the fusion (F) and glycoprotein (G) proteins. HMPV F is a class I viral fusion protein that mediates entry and is similar to other paramyxovirus F proteins [21–24]. HMPV G protein is a predicted type II transmembrane protein that may contribute to attachment [25]. However, the function of HMPV G is not clear. The G protein is dispensable for in vitro replication, though viruses lacking G are attenuated in vivo [26, 27]. Some data suggest that G suppresses host immune responses; however, these studies used a single isolate of HMPV [28, 29]. We sought to identify regions of G that were highly conserved over time, as might be expected for critical functional domains.
We analyzed full-length G gene sequences from 87 isolates of HMPV collected over a 20-year period from children with respiratory disease. The evolutionary rate of HMPV was similar to that of other RNA viruses; however, HMPV G gene sequences did not display progressive drift over time, and the four lineages were preserved. The time to the most recent common ancestor suggested recent divergence from AMPV-C. There were distinct amino acid differences in G that are conserved within subgroups and preserved over time. We found substantial diversity between major groups, with few highly conserved motifs in the ectodomain.
Materials and methods
HMPV isolates
Virus sequences were derived from specimens collected over a twenty-year period from 1982 to 2001 in the Vanderbilt Vaccine Clinic, as described previously [14, 15]. Nasal wash specimens were collected from children <5 years of age with acute respiratory tract illness. Written informed consent was obtained from the parent/guardian of each child in the original study. The Vanderbilt Institutional Review Board approved the study. We extracted RNA from these samples and used quantitative real-time RT-PCR to test for HMPV by detection of nucleoprotein gene sequences [15]. Specimens that tested positive for HMPV were subjected to nested RT-PCR for the G gene as described below. Viral nomenclature used in this study uses a letter code representing the geographic site of isolation (CAN = Canada, JP = Japan, NL= Netherlands, and TN = Tennessee) followed by the year of isolation. Published HMPV and AMPV-C G sequences included in the analyses were obtained from GenBank (accession nos. AF371337.2, AJ811991, AJ811992, AJ811993, AY198394, AY297749, AY485232, AY485233, AY485243, AY485251, AY485252, AY525843, AY530089, AY530091, AY530092, AY530094, AY590691, AY590692, AY590693, EF199771.1, EF199772.1, FJ168778, FJ168779, GU126687, and NC_007652.1).
RNA extraction, RT-PCR, and sequencing of G genes
RNA was extracted from 220 µl of nasal wash sample on a QIAGEN BioRobot 9604 Workstation using a QIAamp Viral RNA Kit (QIAGEN), as described [15]. Amplification of the entire G open reading frame (ORF) was carried out by RT-PCR followed by nested PCR. Primer sequences are shown in Supplemental Table 1. A Thermoscript /Platinum Taq Polymerase Kit (Invitrogen) was used in a 50-µL RT-PCR reaction with 10 µL of diluted RNA as template. The RT-PCR was carried out at 50°C for 50 min and 95°C for 3 min, followed by 5 cycles of 94°C for 30 s, 50°C for 60 s, and 68°C for 3 min, and an additional 30 cycles of 94°C for 30 s, 55°C for 60 s, and 68°C for 3 min. For nested PCR, 2 µL of RT-PCR products was added to a 50-µL reaction using Platinum PCR Supermix (Invitrogen). The reaction was incubated at 95°C for 3 min followed by 5 cycles of 94°C for 30 s, 50°C for 30 s, and 68°C for 2 min, and an additional 30 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 2 min. For all reactions, a final extension at 68°C for 7 min was included.
Table 1.
Nucleotide and amino acid sequence identity of HMPV G genes within major and minor groups.
| Gene | A | B | ALL | ||||
|---|---|---|---|---|---|---|---|
| Group | All A | A1 (n=13) |
A2 (n=24) |
All B | B1 (n=14) |
B2 (n=36) |
n=87 |
| Minimum % nt identity | 74.5 | 91.8 | 85.2 | 76.4 | 86.7 | 84.4 | 51.9 |
| Mean % nt identity | 85.9 | 94.7 | 91.6 | 87.8 | 93.2 | 95.0 | 71.9 |
| Maximum % nt identity | 99.9 | 99.9 | 99.8 | 99.9 | 99.9 | 99.9 | 99.9 |
| Minimum % aa identity | 60.2 | 85.2 | 77.5 | 62.2 | 82.2 | 75.4 | 29.3 |
| Mean % aa identity | 80.3 | 92.6 | 88.9 | 83.6 | 90.6 | 91.4 | 63.3 |
| Maximum % aa identity | 99.6 | 99.6 | 99.5 | 99.6 | 99.6 | 99.6 | 99.6 |
Sequencing reactions were carried out using an ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems). Eight sequencing primers were used for each fragment to ensure a twofold coverage of the open reading frame. Sequencing primers are available upon request. The end products were processed by capillary electrophoresis using a 3730 DNA Analyzer (Applied Biosystems) and analyzed using DNA Sequencing Analysis (Applied Biosystems) and Sequencher (Gene Codes Corp.). Rare nucleotide ambiguities between forward and reverse sequences were resolved by sequencing additional RT-PCR clones.
Sequence alignment and phylogenetic analysis
Final sequences were edited and aligned using the ClustalW algorithm in MacVector version 11.0 (MacVector) and MEGA version 5.0 [30]. Sequences identified in this study have been submitted to GenBank under accession numbers JF929831-JF929903. Pairwise sequence alignment, multiple sequence alignment, and percent nucleotide identity calculations were performed using MacVector version 11.1 (MacVector). Inference of phylogeny and overall rates of evolutionary change (nucleotide substitutions per site per year) and the time to the most recent common ancestor (tMRCA) were estimated using the Bayesian Markov chain Monte Carlo (MCMC) approach available in the BEAST package (http://www.evolve.zoo.ox.ac.uk/Beast/) [31]. We employed the HKY model of nucleotide substitution [32] with empirical base frequencies with unlinked substitution, rate heterogeneity and base frequencies at all three codon positions, as this model yielded the best 95% highest posterior density (HPD) out of all models tested (not shown). Datasets were analyzed under demographic models of constant population size, exponential population growth, and expansion population growth. For all models, we used both strict and relaxed (uncorrelated logarithmic) molecular clocks. In each case, MCMC chains were run for 10 million steps with a burn-in rate of 10%, and five separate runs were combined using the Log Combiner program (http://www.evolve.zoo.ox.ac.uk/), with uncertainty in parameter estimates reflected in the 95% HPD. Output sets of trees were combined using Log Combiner, analyzed with the TreeAnnotator program and visualized with FigTree [31].
Results
Comparison of sequence identity between subgroups
Full-length G gene sequences were obtained for 73 Tennessee strains of HMPV and assigned to one of the four proposed lineages (A1, A2, B1, or B2) based on phylogenetic analysis, discussed further below [33]. Of the 73 TN strains sequenced, 9 (12%) were of the A1, 20 (27%) A2, 10 (14%) B1 and 34 (46%) B2 lineage. Sequences obtained in this study were compared to 14 published full-length G gene sequences. The overall mean nucleotide sequence identity between all 87 isolates was 71.9%, with a minimum identity of 51.9% (Table 1). The nucleotide sequence identity within major groups was higher, with a mean of 85.9% (minimum 74.5%) between A1 and A2, and mean 87.8% (minimum 76.4%) between B1 and B2. Most of the nucleotide changes were non-synonymous, and thus the amino acid sequence identity was more divergent than the nucleotide sequence identity between and within all groups, with overall minimum identity of 29.3% and mean identity of 63.3%. The mean amino acid sequence identity within major groups was 80.3%, and the minimum was 60.2% for A1 and A2, and the mean was 83.6% and the minimum was 62.2% for B1 and B2.
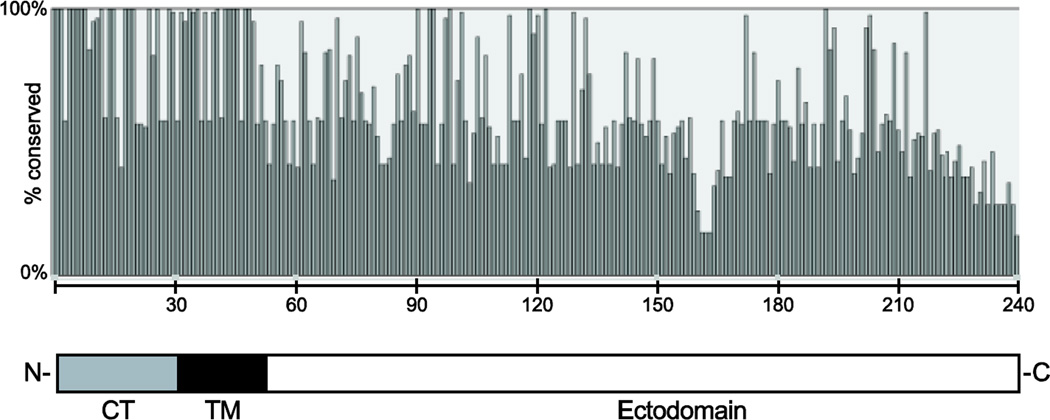
The length of the predicted protein varied considerably between groups due to truncations at the C-terminal end, though all of the A1 sequences were the same length: A1, 236 aa; A2, 217–231 aa; B1, 224–241 aa; and B2, 221–238 aa. Thus, HMPV G, at 217–241 aa, is shorter than RSV G, at 298 aa [34, 35]. Amino acid conservation was greatest in the predicted cytoplasmic tail and transmembrane domains, with progressively more variation in the ectodomain (Figure 1). The cytoplasmic tail was 89% conserved among all isolates at the amino acid level (98% within subgroups), while the transmembrane domain was 86% conserved overall (97% within subgroups).
Figure 1. Amino acid conservation in HMPV G protein.
The figure represents analysis of 87 aligned full-length G protein sequences. The y-axis indicates % identity among the strains. The x-axis indicates specific residue positions from the N- to the C-terminus. Note that the length of the G protein ranged from 217 to 241 amino acids, and thus, the percent identity of the extreme C-terminal end is affected by the length. The schematic diagram below indicates N- and C-termini with domains of G. CT, cytoplasmic tail; TM, transmembrane region.
Conserved and divergent features of the G protein
Despite the diversity overall in the ectodomain, certain motifs were variably present in most groups. One N-glycosylation site was absolutely conserved: 30–32 NAS/T (Table 2). All subgroups had four to seven additional N-glycosylation sites, though conserved N-glycosylation sites varied between subgroups. Similarly, serines and threonines, potential O-linked glycosylation acceptor sites, were present in all strains but quite variable. The number of serines ranged from 15 to 42, with group A strains containing more serine residues than B strains (Table 2). Conversely, threonine residues ranged from 35 to 54, with more threonines in group B strains. Thus, all strains had a similar number of potential O-linked acceptor sites. All strains had a high proline content, but group A viruses generally encoded more proline residues. A cysteine within the cytoplasmic tail was conserved in all strains, but only group B viruses had an additional conserved cysteine in the ectodomain. Thus, the conserved “cysteine noose” present in RSV G [34] was not present in any of the HMPV G proteins.
Table 2.
Characteristics of length and specific amino acid motifs among HMPV G proteins by subgroup.
| All | A1 | A2 | B1 | B2 | |
|---|---|---|---|---|---|
| Length (residues) | 217–241 | 236 | 217–231 | 224–241 | 219–231 |
| N-glycosylation | 30–32 NAS/T | 52–55 NYT (most) | 52–55 NYT (all) | 20–23 NRT (TN/87-27 only) | 58–60 NMT (most) |
| 58–60 NTS all | 58–60 NTS (few) | 101–103 NST (all) | 163–165 NTT (TN/91–316 only) | ||
| 226–229 NTS few | 180–182 NST (TN/92–357 only) | 169–171 NQT (all) | 166–168 NQT (few) | ||
| 233–235 NQT all | 193–196 NSS (JPS/03–178 only) | 181–183 NTT (all) | 178–180 NTT (all except TN/99–419) | ||
| 184–186 NQT (TN/982-42 only) | |||||
| 223–226 NNT (few) | 188–190 NAS (all except TN/982-42) | 181–183 NQT (all) | |||
| 199–201 NAT (few) | |||||
| 190–192 NET (TN/982-42 only) | 206–208 NQT (TN/89-356 only) | ||||
|
Mean no. (range) |
Mean no. (range) | Mean no. (range) | Mean no. (range) | Mean no. (range) | |
| Serine | 26 (15–42) | 39 (36–42) | 34 (32–37) | 17 (15–18) | 20 (17–22) |
| Threonine | 46 (35–54) | 41 (39–43) | 38 (35–43) | 49 (48–52) | 51 (47–54) |
| O-glycosylation | 73 (64–84) | 80 (77–84) | 72 (67–78) | 66 (65–69) | 72 (64–76) |
| Proline | 15 (10–23) | 20 (19–23) | 17 (15–21) | 14 (12–17) | 12 (10–14) |
Alternate start codons that produce secreted forms of RSV G exist [36]. The HMPV A1 subgroup had two AUG codons in the putative ectodomain (residues 55 and 71), the B group three (residues 59, 68, and 79), but the A2 subgroup lacked alternate start codons that would be expected to produce a secreted form. Further, none of these putative alternate start codons was preceded by an apparent signal sequence. All of the B1 G sequences had an apparent insertion sequence K/EKGKE at residues 162–166, similar to a predicted heparin-binding site [25], but this motif was absent in other subgroups. No HMPV G sequences had large duplications such as described in some RSV strains [37].
Phylogenetic diversity and evolution over time
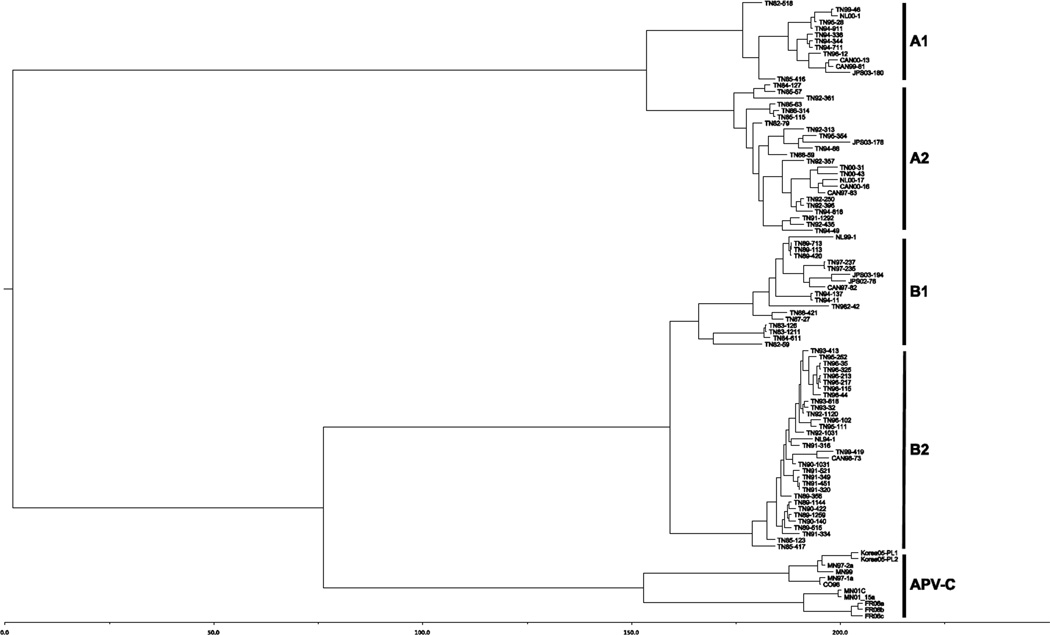
We performed phylogenetic and evolutionary analysis of the aligned full-length G sequences with six different models using the BEAST program suite [31]. Comparison of the output of each model showed that the relaxed clock, exponential population growth model had the highest posterior density (HPD) (not shown). The phylogenetic tree representing the sequence relationships by nucleotide substitutions suggested four genetic subgroups (Figure 2), consistent with previous analyses [19, 20, 33]. The four distinct subgroups remained stable over time, and viruses within these lineages were closely related genetically despite being isolated over a twenty-year period. Thus, the clustering based on sequence did not correlate with the chronological origin of the sequences. For example, one clade in the A2 lineage contained closely related sequences from Tennessee in 1991, 1992, 1994 and 2000, as well as the Netherlands in 2000 and Canada in 1997 and 2000 (Figure 2). Similar clustering of chronologically and geographically disparate sequences was present within each subgroup. In the B1 subgroup, Tennessee sequences from 1989, 1994, and 1997 were closely related to a Canadian sequence from 1997 and Japanese sequences from 2002 and 2003. To examine further the evolution of HMPV G gene sequences over time, we aligned sequences within each subgroup in chronological order (Supplemental Figures 1–4). There were a few amino acid changes that persisted in chronologically later viruses; however, the majority of the changes from year to year were not preserved and reverted in subsequent isolates, showing a lack of progressive evolution over time. For example, a putative N-glycosylation site at residues 101- 103 of the A2 subgroup (Supplemental Figure 2) was variably present in isolates from alternating years. Conversely, a putative N-glycosylation site at residues 166–168 of the B2 subgroup (Supplemental Figure 4) was only present in isolates from 1983–1984. Analysis of multiple sequences collected over time allowed a molecular clock calculation of viral nucleotide changes. The evolutionary rate of HMPV G was 3.8 × 10−3 substitutions/site/year (95% HPD 2.8 × 10−3, 4.9 × 10−3). The estimated time to the most recent common ancestor (tMRCA) of HMPV and AMPV-C was 215 years (95% HPD 95–367) (Figure 2).
Figure 2. Phylogenetic tree of HMPV and AMPV-C G nucleotide sequences by tMRCA.
Phylogenetic analysis of 87 full-length HMPV G nucleotide sequences from Canada (CAN), Japan (JPS or JPY), Tennessee (TN) and the Netherlands (NL). The first two digits of the sequence names indicate the year of the isolate. The names of the AMPV sequences indicate geographic origin (CO, Colorado; FR, France; MN, Minnesota) and year. The x-axis scale is in years. The tree was constructed as described in Materials and methods.
Discussion
We analyzed 87 full-length HMPV G gene sequences obtained over a twenty-year period and confirm the four distinct genetic lineages of HMPV: A1, A2, B1 and B2 [33]. The G sequences fell into the same four clades with concordant F sequences [20]; other groups have shown that HMPV genes form a clade together [19]. These data suggest that recombination does not occur among HMPV strains, and analysis of the sequences using the Recombination Detection Program (RDP) did not identify evidence of recombination (not shown) [38].
Nucleotide sequence identity was variable among subgroups, and a high rate of non-synonymous changes led to greater amino acid sequence diversity, with a mean of only 63% amino acid sequence identity between major groups and ~80% between minor subgroups. However, the biological reason for this diversity is unclear. Two groups have suggested selective immune pressure on HMPV G based on computer analysis of partial G sequences [39, 40]. However, several lines of evidence suggest that while G induces humoral antibodies in animals and humans [41–44], G-specific antibodies do not exhibit in vitro neutralizing activity and are not protective in vivo. HMPV G expressed in a reverse-engineered PIV1 backbone, HMPV G encoded by an alphavirus replicon vector, and recombinant HMPV G protein all induced antibodies that were non-neutralizing, and all failed to induce protective immunity in different rodent models [41–43]. One putative HLA-A2-restricted epitope has been described in the highly conserved transmembrane domain of G [45, 46]; however, extensive epitope mapping in different MHC class I-restricted mouse strains in our laboratory has identified numerous CTL epitopes in several HMPV proteins, but none in G [47]. Thus, there is little published evidence for immune pressure on G.
One suggestion for selective pressure has been the preservation of potential O-linked glycosylation sites [19, 39, 40]. Our analysis shows that over 20 years and between all four lineages, the general content of putative O-linkage acceptor serines and threonines was preserved. However, the location of specific serine and threonine residues was conserved within but not between subgroups (Table 2 and Supplemental Figures 1–4). Similarly, the relative proline content, but not specific residues, was conserved within but not between lineages. HMPV G, like RSV G and other mucin-like viral proteins, is indeed both N- and O-glycosylated [24, 26, 42, 48] and binds to cell-surface glycosaminoglycans, likely as the first step in viral attachment [25]. Notably, a 159QRRGKGKE166 motif was identified as critical for binding to heparin in that study; the only sequence that nearly matched this in any of the G sequences we determined was the insertion unique to the B1 subgroup: 159QKRK/EKGKE166. Thus, the broad relevance of this motif is not clear. Recent analysis of the genomes of several RNA viruses that encode mucin-like proteins showed that selective pressure was relaxed over the mucin-like regions, suggesting that the mucin-like character was more conserved than the specific amino acid sequences [49]. Our data support this as a possible explanation for the diversity of HMPV G, since the mucin-like character of G is preserved.
Nonetheless, if conservation of the general mucin-like nature of G is all that is required for viral fitness, why do the four distinct lineages of HMPV G persist over time? Two studies have suggested a role for G in suppressing innate immunity by interacting with RIG-I and TLR4 [28, 29]. While the cytoplasmic tail and transmembrane domain of G are highly conserved, the ectodomain has few conserved stretches of consecutive residues, especially between A and B subgroups (Figure 1); thus, a putative conserved motif that interacts with host immune molecules is not apparent. The identification of specific regions of G that may interact with host proteins or other HMPV proteins would facilitate an understanding of whether these findings are applicable to all HMPV strains or whether different viruses exhibit important differences based on G. Most published epidemiologic data show no significant clinical differences between illness caused by viruses of the A or B lineages [50].
Although the four lineages were maintained over twenty years, we did not find evidence of rapid progressive evolution over time, but rather gradual changes, as seen in other paramyxoviruses [51] and in contrast to the rapid antigenic drift exhibited by human influenza viruses [52]. This is despite the high mutation rate of G at 3.8 × 10−3 substitutions/site/year, remarkably similar to the rate calculated by de Graaf et al [19]. Further, the estimated tMRCA between HMPV and AMPV-C was 215 years, very similar to that predicted based on F sequences [20, 53]. It should be noted that viruses with rapid mutation rates may confound molecular clock calculations, leading to substantial errors in estimations of virus and host co-evolution [54]. Nonetheless, zoonotic transmission of HMPV may have occurred fairly recently in human history. Productive infection of chickens and turkeys with HMPV was unsuccessful [1], although viral RNA and antigen could be detected in turkey poults inoculated with a high titer of HMPV [55]. HMPV and AMPV contain analogous open reading frames in the same order that are distinct from those of members of the genus Pneumovirus, and both metapneumoviruses lack the NS1 and NS2 genes of pneumoviruses [53]. This finding suggests that AMPV and HMPV diverged from a common progenitor, likely a pneumovirus. This suggests that other emerging zoonotic paramyxoviruses such as Hendra and Nipah viruses could become established in the human population.
Supplementary Material
Acknowledgments
Financial support:
Supported by NIH AI-82417 and AI-85062 to JVW. The Vanderbilt Vaccine Clinic was supported in part by NIH Respiratory Pathogens Research Unit N01-AI-65298 and by NIH GCRC center award RR 00095.
Footnotes
Competing interest information:
Chiaoyin K. Wang, Chin-Fen Yang, Linda D. Lintao, Alexis Liem, and Marla Chu were employees of MedImmune at the time of this study. John Williams serves on the Scientific Advisory Board of Quidel.
References
- 1.van den Hoogen BG, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7(6):719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boivin G, et al. An outbreak of severe respiratory tract infection due to human metapneumovirus in a long-term care facility. Clin Infect Dis. 2007;44(9):1152–1158. doi: 10.1086/513204. [DOI] [PubMed] [Google Scholar]
- 3.Dollner H, et al. Outbreak of human metapneumovirus infection in norwegian children. Pediatr Infect Dis J. 2004;23(5):436–440. doi: 10.1097/01.inf.0000126401.21779.74. [DOI] [PubMed] [Google Scholar]
- 4.Ebihara T, et al. Human metapneumovirus infection in Japanese children. J Clin Microbiol. 2004;42(1):126–132. doi: 10.1128/JCM.42.1.126-132.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esper F, et al. A 1-year experience with human metapneumovirus in children aged <5 years. J Infect Dis. 2004;189(8):1388–1396. doi: 10.1086/382482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gray GC, et al. Multi-year study of human metapneumovirus infection at a large US Midwestern Medical Referral Center. J Clin Virol. 2006;37(4):269–276. doi: 10.1016/j.jcv.2006.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamelin ME, et al. Human metapneumovirus infection in adults with communityacquired pneumonia and exacerbation of chronic obstructive pulmonary disease. Clin Infect Dis. 2005;41(4):498–502. doi: 10.1086/431981. [DOI] [PubMed] [Google Scholar]
- 8.Kahn JS. Epidemiology of human metapneumovirus. Clin Microbiol Rev. 2006;19(3):546–557. doi: 10.1128/CMR.00014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madhi SA, et al. Seasonality, incidence, and repeat human metapneumovirus lower respiratory tract infections in an area with a high prevalence of human immunodeficiency virus type-1 infection. Pediatr Infect Dis J. 2007;26(8):693–699. doi: 10.1097/INF.0b013e3180621192. [DOI] [PubMed] [Google Scholar]
- 10.Maggi F, et al. Human metapneumovirus associated with respiratory tract infections in a 3-year study of nasal swabs from infants in Italy. J Clin Microbiol. 2003;41(7):2987–2991. doi: 10.1128/JCM.41.7.2987-2991.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinello RA, et al. Human metapneumovirus and exacerbations of chronic obstructive pulmonary disease. J Infect. 2006;53(4):248–254. doi: 10.1016/j.jinf.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Hoogen BG, Osterhaus DM, Fouchier RA. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr Infect Dis J. 2004;23(1 Suppl):S25–S32. doi: 10.1097/01.inf.0000108190.09824.e8. [DOI] [PubMed] [Google Scholar]
- 13.Williams JV, et al. Population-based incidence of human metapneumovirus infection among hospitalized children. J Infect Dis. 2010;201(12):1890–1898. doi: 10.1086/652782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams JV, et al. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004;350(5):443–450. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams JV, et al. The role of human metapneumovirus in upper respiratory tract infections in children: a 20-year experience. J Infect Dis. 2006;193(3):387–395. doi: 10.1086/499274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buys SB, du Preez JH, Els HJ. The isolation and attenuation of a virus causing rhinotracheitis in turkeys in South Africa. The Onderstepoort journal of veterinary research. 1989;56(2):87–98. [PubMed] [Google Scholar]
- 17.Njenga MK, Lwamba HM, Seal BS. Metapneumoviruses in birds and humans. Virus Res. 2003;91(2):163–169. doi: 10.1016/s0168-1702(02)00256-3. [DOI] [PubMed] [Google Scholar]
- 18.Toquin D, et al. Subgroup C avian metapneumovirus (MPV) and the recently isolated human MPV exhibit a common organization but have extensive sequence divergence in their putative SH and G genes. J Gen Virol. 2003;84(Pt 8):2169–2178. doi: 10.1099/vir.0.19043-0. [DOI] [PubMed] [Google Scholar]
- 19.de Graaf M, et al. Evolutionary dynamics of human and avian metapneumoviruses. J Gen Virol. 2008;89(Pt 12):2933–2942. doi: 10.1099/vir.0.2008/006957-0. [DOI] [PubMed] [Google Scholar]
- 20.Yang CF, et al. Genetic diversity and evolution of human metapneumovirus fusion protein over twenty years. Virology journal. 2009;6:138. doi: 10.1186/1743-422X-6-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cseke G, et al. Integrin alphavbeta1 promotes infection by human metapneumovirus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(5):1566–1571. doi: 10.1073/pnas.0801433106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Graaf M, et al. Fusion protein is the main determinant of metapneumovirus host tropism. J Gen Virol. 2009;90(Pt 6):1408–1416. doi: 10.1099/vir.0.009688-0. [DOI] [PubMed] [Google Scholar]
- 23.Miller SA, et al. Examination of a fusogenic hexameric core from human metapneumovirus and identification of a potent synthetic peptide inhibitor from the heptad repeat 1 region. Journal of virology. 2007;81(1):141–149. doi: 10.1128/JVI.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schowalter RM, Smith SE, Dutch RE. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. Journal of virology. 2006;80(22):10931–10941. doi: 10.1128/JVI.01287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thammawat S, et al. Role of cellular glycosaminoglycans and charged regions of viral G protein in human metapneumovirus infection. Journal of virology. 2008;82(23):11767–11774. doi: 10.1128/JVI.01208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biacchesi S, et al. Recombinant human Metapneumovirus lacking the small hydrophobic SH and/or attachment G glycoprotein: deletion of G yields a promising vaccine candidate. Journal of virology. 2004;78(23):12877–12887. doi: 10.1128/JVI.78.23.12877-12887.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biacchesi S, et al. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. Journal of virology. 2005;79(19):12608–12613. doi: 10.1128/JVI.79.19.12608-12613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bao X, et al. Human metapneumovirus glycoprotein G inhibits innate immune responses. PLoS pathogens. 2008;4(5):e1000077. doi: 10.1371/journal.ppat.1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolli D, et al. Human metapneumovirus glycoprotein G inhibits TLR4-dependent signaling in monocyte-derived dendritic cells. Journal of immunology. 2011;187(1):47–54. doi: 10.4049/jimmunol.1002589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 31.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22(2):160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 33.van den Hoogen BG, et al. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis. 2004;10(4):658–666. doi: 10.3201/eid1004.030393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson PR, et al. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(16):5625–5629. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wertz GW, et al. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(12):4075–4079. doi: 10.1073/pnas.82.12.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberts SR, et al. The membrane-associated and secreted forms of the respiratory syncytial virus attachment glycoprotein G are synthesized from alternative initiation codons. J Virol. 1994;68(7):4538–4546. doi: 10.1128/jvi.68.7.4538-4546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trento A, et al. Natural history of human respiratory syncytial virus inferred from phylogenetic analysis of the attachment (G) glycoprotein with a 60-nucleotide duplication. Journal of virology. 2006;80(2):975–984. doi: 10.1128/JVI.80.2.975-984.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin DP, et al. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26(19):2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaunt ER, et al. Molecular epidemiology and evolution of human respiratory syncytial virus and human metapneumovirus. PloS one. 2011;6(3):e17427. doi: 10.1371/journal.pone.0017427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padhi A, Verghese B. Positive natural selection in the evolution of human metapneumovirus attachment glycoprotein. Virus research. 2008;131(2):121–131. doi: 10.1016/j.virusres.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skiadopoulos MH, et al. Individual contributions of the human metapneumovirus F, G, and SH surface glycoproteins to the induction of neutralizing antibodies and protective immunity. Virology. 2006;345(2):492–501. doi: 10.1016/j.virol.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 42.Ryder AB, et al. Soluble recombinant human metapneumovirus G protein is immunogenic but not protective. Vaccine. 2010;28(25):4145–4152. doi: 10.1016/j.vaccine.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mok H, et al. An alphavirus replicon-based human metapneumovirus vaccine is immunogenic and protective in mice and cotton rats. Journal of virology. 2008;82(22):11410–11418. doi: 10.1128/JVI.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Endo R, et al. Detection of four genetic subgroup-specific antibodies to human metapneumovirus attachment (G) protein in human serum. The Journal of general virology. 2008;89(Pt 8):1970–1977. doi: 10.1099/vir.0.83679-0. [DOI] [PubMed] [Google Scholar]
- 45.Herd KA, et al. Cytotoxic T-lymphocyte epitope vaccination protects against human metapneumovirus infection and disease in mice. Journal of virology. 2006;80(4):2034–2044. doi: 10.1128/JVI.80.4.2034-2044.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herd KA, et al. Major histocompatibility complex class I cytotoxic T lymphocyte immunity to human metapneumovirus (hMPV) in individuals with previous hMPV infection and respiratory disease. The Journal of infectious diseases. 2008;197(4):584–592. doi: 10.1086/526536. [DOI] [PubMed] [Google Scholar]
- 47.Erickson JJ, et al. Viral acute lower respiratory infections impair CD8+ T cells through PD-1. The Journal of clinical investigation. 2012;122(8):2967–2982. doi: 10.1172/JCI62860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu L, Bastien N, Li Y. Intracellular processing, glycosylation, and cell surface expression of human metapneumovirus attachment glycoprotein. Journal of virology. 2007;81(24):13435–13443. doi: 10.1128/JVI.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wertheim JO, Worobey M. Relaxed selection and the evolution of RNA virus mucin-like pathogenicity factors. Journal of virology. 2009;83(9):4690–4694. doi: 10.1128/JVI.02358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schildgen V, et al. Human Metapneumovirus: lessons learned over the first decade. Clinical microbiology reviews. 2011;24(4):734–754. doi: 10.1128/CMR.00015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCarthy AJ, Goodman SJ. Reassessing conflicting evolutionary histories of the Paramyxoviridae and the origins of respiroviruses with Bayesian multigene phylogenies. Infection, genetics and evolution : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2010;10(1):97–107. doi: 10.1016/j.meegid.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 52.Rambaut A, et al. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453(7195):615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van den Hoogen BG, et al. Analysis of the genomic sequence of a human metapneumovirus. Virology. 2002;295(1):119–132. doi: 10.1006/viro.2001.1355. [DOI] [PubMed] [Google Scholar]
- 54.Sharp PM, Simmonds P. Evaluating the evidence for virus/host co-evolution. Current opinion in virology. 2011;1(5):436–441. doi: 10.1016/j.coviro.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 55.Velayudhan BT, et al. Human metapneumovirus in turkey poults. Emerg Infect Dis. 2006;12(12):1853–1859. doi: 10.3201/eid1212.060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.