Abstract
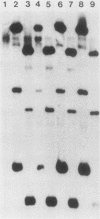
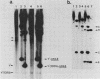
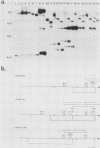
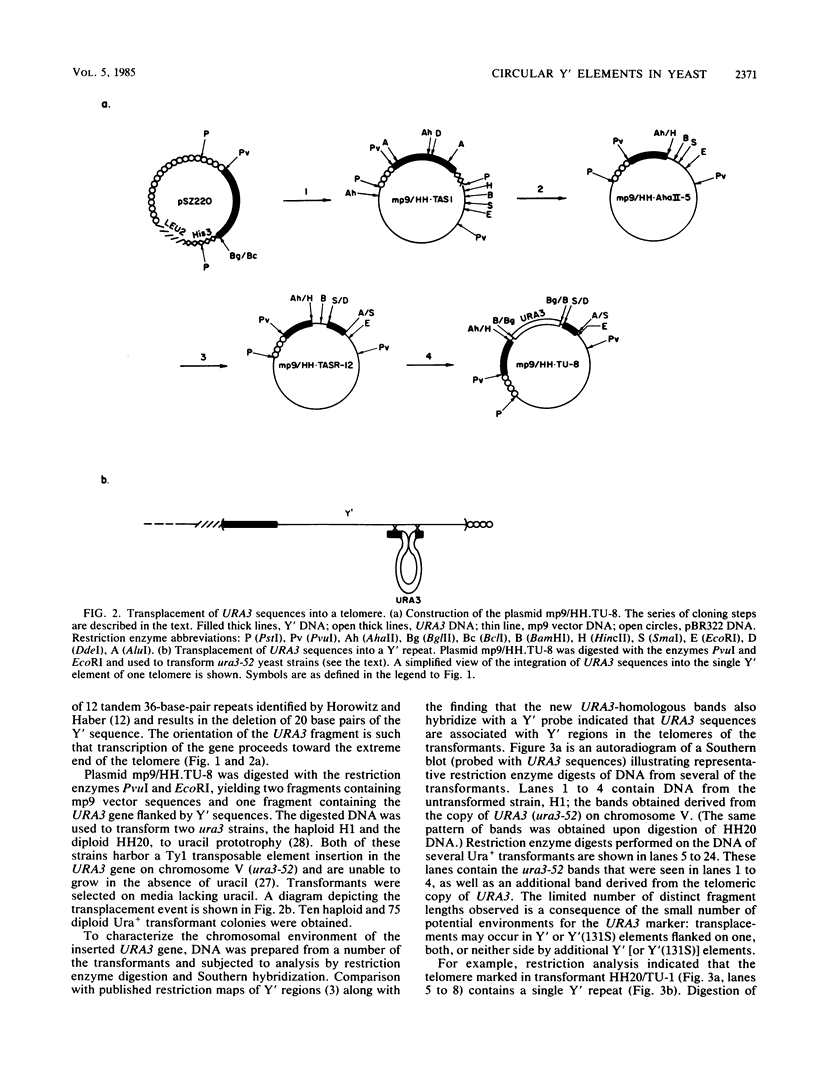
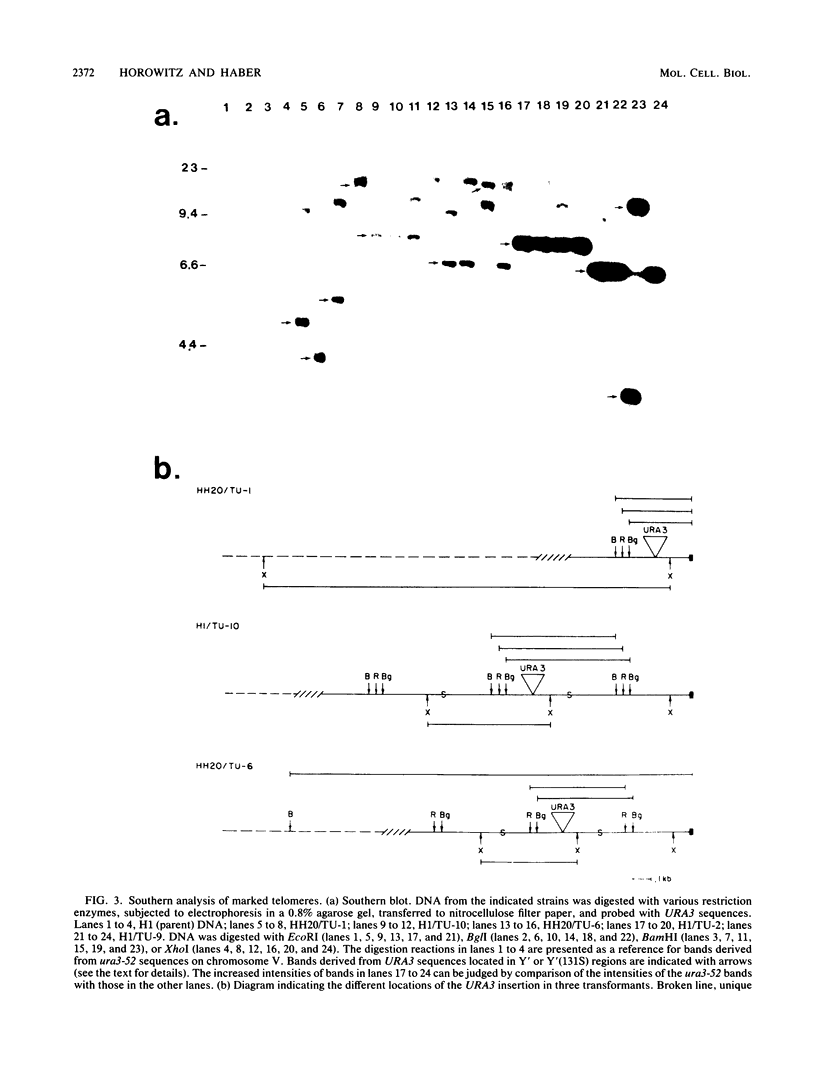
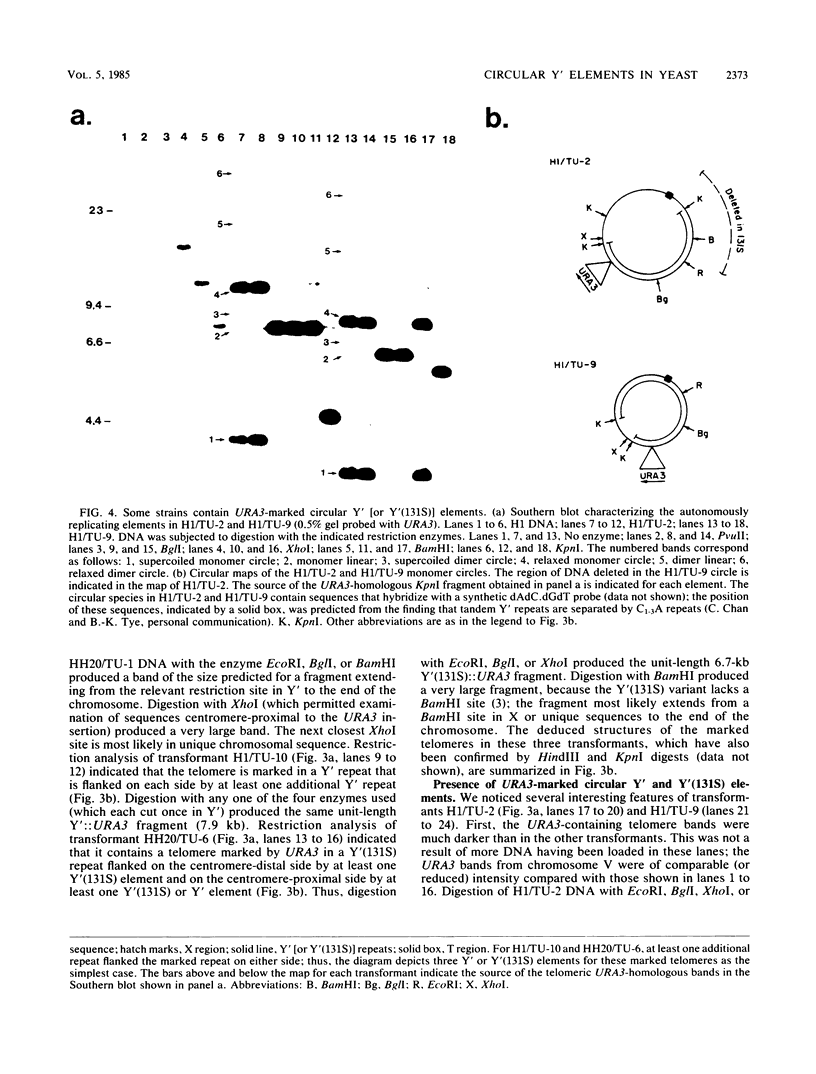
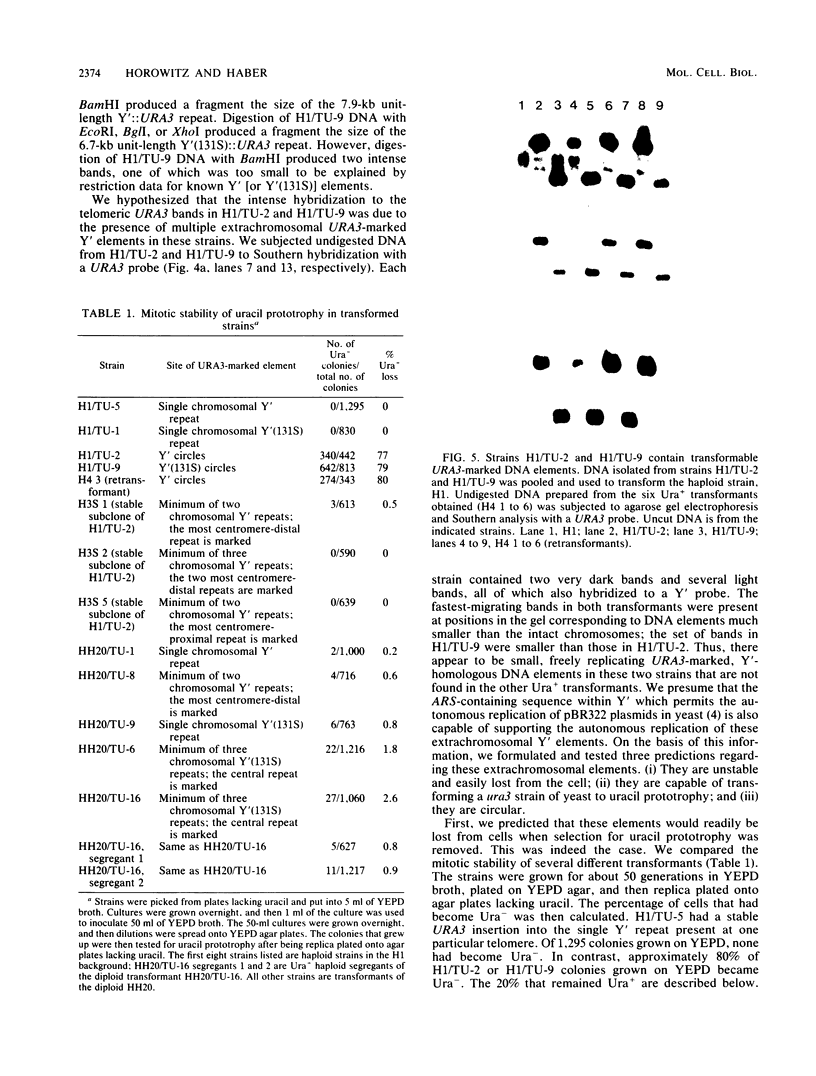
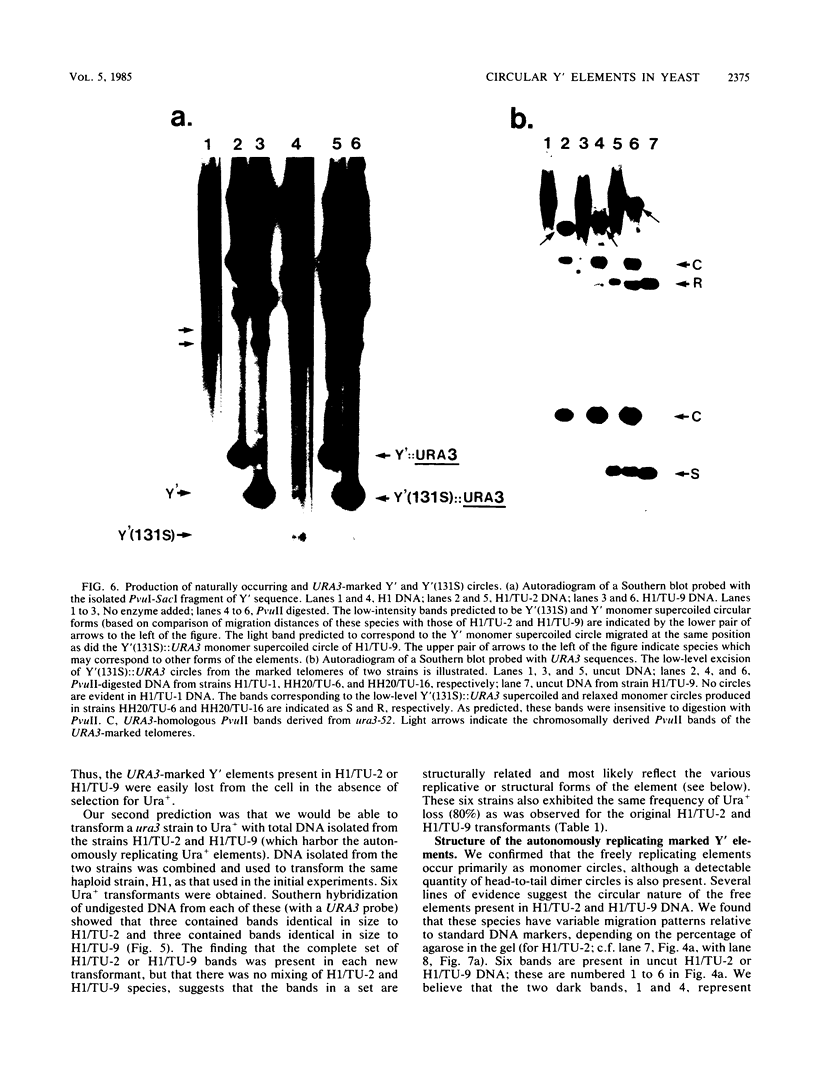
We marked a large number of yeast telomeres within their Y' regions by transforming strains with a fragment of Y' DNA into which the URA3 gene had been inserted. A few of the Ura+ transformants obtained were very unstable and were found to contain autonomously replicating URA3-marked circular Y' elements in high copy number. These marked extrachromosomal circles were capable of reintegrating into the chromosome at other telomeric locations. In contrast, most of the Ura+ transformants obtained were quite stable mitotically and were marked at bona fide chromosomal ends. These stable transformants gave rise to mitotically unstable URA3-marked circular Y' elements at a low frequency (up to 2.5%). The likelihood that such excisions and integrations represent a natural process in Saccharomyces cerevisiae is supported by our identification of putative Y' circles in untransformed strains. The transfer of Y' information among telomeres via a circular intermediate may be important for homogenizing the sequences at the ends of yeast chromosomes and for generating the frequent telomeric rearrangements that have been observed in S. cerevisiae.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ballario P., Filetici P., Junakovic N., Pedone F. Ty1 extrachromosomal circular copies in Saccharomyces cerevisiae. FEBS Lett. 1983 May 8;155(2):225–229. doi: 10.1016/0014-5793(82)80608-x. [DOI] [PubMed] [Google Scholar]
- Blackburn E. H. Telomeres: do the ends justify the means? Cell. 1984 May;37(1):7–8. doi: 10.1016/0092-8674(84)90295-2. [DOI] [PubMed] [Google Scholar]
- Chan C. S., Tye B. K. A family of Saccharomyces cerevisiae repetitive autonomously replicating sequences that have very similar genomic environments. J Mol Biol. 1983 Aug 15;168(3):505–523. doi: 10.1016/s0022-2836(83)80299-x. [DOI] [PubMed] [Google Scholar]
- Chan C. S., Tye B. K. Organization of DNA sequences and replication origins at yeast telomeres. Cell. 1983 Jun;33(2):563–573. doi: 10.1016/0092-8674(83)90437-3. [DOI] [PubMed] [Google Scholar]
- Clark-Walker G. D., Azad A. A. Hybridizable sequences between cytoplasmic ribosomal RNAs and 3 micron circular DNAs of Saccharomyces cerevisiae and Torulopsis glabrata. Nucleic Acids Res. 1980 Mar 11;8(5):1009–1022. doi: 10.1093/nar/8.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Walker G. D., Miklos G. L. Localization and quantification of circular DNA in yeast. Eur J Biochem. 1974 Jan 16;41(2):359–365. doi: 10.1111/j.1432-1033.1974.tb03278.x. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Devenish R. J., Newlon C. S. Isolation and characterization of yeast ring chromosome III by a method applicable to other circular DNAs. Gene. 1982 Jun;18(3):277–288. doi: 10.1016/0378-1119(82)90166-4. [DOI] [PubMed] [Google Scholar]
- Dunn B., Szauter P., Pardue M. L., Szostak J. W. Transfer of yeast telomeres to linear plasmids by recombination. Cell. 1984 Nov;39(1):191–201. doi: 10.1016/0092-8674(84)90205-8. [DOI] [PubMed] [Google Scholar]
- Filetici P., Junakovic N., Ballario P. Rapid alkaline preparation for yeast circular covalently closed DNA molecules. Curr Genet. 1985;9(2):123–126. doi: 10.1007/BF00436959. [DOI] [PubMed] [Google Scholar]
- Haber J. E., Thorburn P. C. Healing of broken linear dicentric chromosomes in yeast. Genetics. 1984 Feb;106(2):207–226. doi: 10.1093/genetics/106.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz H., Haber J. E. Subtelomeric regions of yeast chromosomes contain a 36 base-pair tandemly repeated sequence. Nucleic Acids Res. 1984 Sep 25;12(18):7105–7121. doi: 10.1093/nar/12.18.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz H., Thorburn P., Haber J. E. Rearrangements of highly polymorphic regions near telomeres of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2509–2517. doi: 10.1128/mcb.4.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov V. L., Grishin A. V., Smirnov M. N. 3 micron DNA - an extrachromosomal ribosomal DNA in the yeast Saccharomyces cerevisiae. Gene. 1980 Dec;12(1-2):41–49. doi: 10.1016/0378-1119(80)90014-1. [DOI] [PubMed] [Google Scholar]
- McClintock B. The Fusion of Broken Ends of Chromosomes Following Nuclear Fusion. Proc Natl Acad Sci U S A. 1942 Nov;28(11):458–463. doi: 10.1073/pnas.28.11.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics. 1941 Mar;26(2):234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker J. H., Haber J. E. Evidence of Chromosomal Breaks near the Mating-Type Locus of SACCHAROMYCES CEREVISIAE That Accompany MATalpha xMATalpha Matings. Genetics. 1981 Nov;99(3-4):383–403. doi: 10.1093/genetics/99.3-4.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Meyerink J. H., Klootwijk J., Planta R. J., van der Ende A., van Bruggen E. F. Extrachromosomal circular ribosomal DNA in the yeast Saccharomyces carlsbergensis. Nucleic Acids Res. 1979 Sep 11;7(1):69–76. doi: 10.1093/nar/7.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A., Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976 Jan 16;143(2):119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- Rose M., Winston F. Identification of a Ty insertion within the coding sequence of the S. cerevisiae URA3 gene. Mol Gen Genet. 1984;193(3):557–560. doi: 10.1007/BF00382100. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Shampay J., Szostak J. W., Blackburn E. H. DNA sequences of telomeres maintained in yeast. Nature. 1984 Jul 12;310(5973):154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Blackburn E. H. Cloning yeast telomeres on linear plasmid vectors. Cell. 1982 May;29(1):245–255. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- Walmsley R. M., Szostak J. W., Petes T. D. Is there left-handed DNA at the ends of yeast chromosomes? Nature. 1983 Mar 3;302(5903):84–86. doi: 10.1038/302084a0. [DOI] [PubMed] [Google Scholar]
- Walmsley R. W., Chan C. S., Tye B. K., Petes T. D. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984 Jul 12;310(5973):157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]