Abstract
Advances in molecular biology in the early 1970s have revolutionized research strategies for studying complex biological processes, which in turn created a high demand for new means to visualize these dynamic biological changes non-invasively and in real-time. In that respect, magnetic resonance imaging (MRI) technology was a perfect fit, due the versatile possibility to alter the different contrast mechanisms. Genetic manipulations are now being translated to MRI trough the development of reporters and sensors, as well as imaging transgenic and knockout mice. In the past few years, a new molecular biology toolset, namely optogenetics, has emerged, which allows for the manipulation of cellular behavior using light. This technology provides a few particularly attractive features for combination with newly developed MRI techniques for probing in vivo cellular, and in particular neural, processes – specifically the ability to control focal, genetically-defined cellular populations with high temporal resolution using equipment that is magnetically inert and does not interact with radiofrequency pulses. Recent works demonstrate that the combination of optogenetics and functional MRI (fMRI) can provide an appropriate platform to investigate in vivo, at the cellular and molecular levels, the neuronal basis of fMRI signals. In addition, this novel combination of optogenetics with fMRI has the potential to resolve pre-synaptic vs. post-synaptic changes of neuronal activity and changes in the activity of large neuronal networks in the context of plasticity associated with development, learning and pathophysiology.
Keywords: optogenetics, fMRI, cortex, animal models, somatosensory cortex, reporter genes, MRI sensors
The molecular biology revolution
The revolution of molecular biology started in the early 1970s with the development of new technologies for transferring genetic material (e.g. genes) from one organism to another (1). This turned out to be a one of the most valuable tools for studying complex biological processes. Throughout the years, many of these principles were applied for developing research tools that can be used to study a variety of genetic and epigenetic events. This created a high demand for new means to visualize these dynamic genetic changes non-invasively and in real-time. In that sense, magnetic resonance imaging (MRI) technology was a perfect fit, due the versatility in altering the different contrast mechanisms. Genetic manipulations were initially used for altering the NMR signal, using magnetic resonance spectroscopy (2–4). Later on, these manipulations were translated to MRI trough development of reporters (5–15) and sensors (16,17), as well as imaging transgenic and knock out mice (18–20). Recently, imaging of transgenic enzymes that convert substrates with artificially elongated T1 by nuclear hyperpolarization was introduced (21). More about reporter genes for MR can be found on this issue (ref Neeman in this issue).
MRI contrast mechanisms
Keeping in mind that MRI has been widely used to characterize genetic changes, (for example to visualize differences in phenotype in transgenic mice), we will focus here only on genetic changes that were made to influence the MRI contrast, and primarily the proton contrast. In principles, three basic contrast mechanisms can be enhanced using genetic tools: (i) spin lactic, longitudinal (T1) relaxation time, (ii) the transverse (T2/T2*) relaxation time and (iii) magnetization transfer / chemical exchange saturation transfer (CEST).
The longitudinal (T1) relaxation of the water protons varies between tissues and can be modified (usually shortened) in the presence of a given “agent” (22,23). This will lead to signal enhancement or brightening of the MR image. In general, genetic alternations that affect T1 contrast are associated with binding of paramagnetic ions or complexes, or changes in the water content of the tissue.
On the other hand, changes in the transverse relaxation (T2) time can either enhance the contrast or reduce it. Form the genetic point of view, T2 relaxation is mostly manipulated by the accumulation of iron (Fe), or changes in the iron oxidation states from diamagnetic to paramagnetic. This was used for certain reporter genes such as ferritin (7–9,19,24,25), the Transferrin receptor (26), melanin (27) and MagA (28).
Genetic manipulations can also be used to indirectly affect blood oxygenation level dependent (BOLD) contrast. In addition to changes in blood volume and blood flow associated with neuronal function, the BOLD contrast is sensitive to changes in the ratio between the oxygenated hemoglobin (which is diamagnetic) and deoxygenated hemoglobin (which is paramagnetic). Therefore, hemoglobin reduction will accelerate MR transverse relaxation (T2 and T2*) due to susceptibility effect. T2 and T2* shortening would reduce the MR signal intensity and consequently enhance the MR contrast (29). BOLD MRI, also known as functional MRI (fMRI), is primarily used for measuring brain activity in human and in small animal models (30). It has been also applied for measuring tumor oxygenation (31) and formation of reactive oxygen species in cancer treatment (32).
CEST contrast, which relies on the proton exchange of the probe or protein with the protons of the surrounding water, reduces the signal intensity and has been used to detect genetically encoded artificial proteins (10) as well as enzymes (15).
Optogenetics for MR physicists: how does it work?
Optogenetics is a new toolset in the molecular biology toolbox that allows for manipulation of cellular behavior using light. As we describe below, this field provides a few particularly attractive features for combination with newly developed MRI techniques for probing in vivo cellular, and in particular neural, processes – namely the ability to control focal, genetically-defined cellular populations with high temporal resolution using equipment that is magnetically inert and does not interact with radiofrequency pulses.
The first major optogenetic protein used to study cellular physiology, Channelrhodopsin-2 (ChR2), takes principally blue light and transduces that light energy into opening its cation channel, thereby inducing cellular depolarization. This protein can be used to make neurons generate an action potential in response to a pulse of blue light, allowing direct control of neuronal action on the millisecond timescale most relevant for neural computation (33). ChR2 forms the prototype of optogenetic molecular devices as it:
-
-
Transduces applied light energy to allow control of a specific cellular action;
-
-
Is expressed as a single genetic element, allowing selection of genetically-specified cellular populations for optical manipulation through the use of molecular genetic tools; and
-
-
Does not require the application of exogenous chemical cofactors or reagents, other than the one-time introduction of the gene encoding the optogenetic device to the cell of interest.
Since the development and publication of ChR2, numerous variant optogenetic devices have been developed, with in-depth reviews of their individual characteristics available (34). Here, we focus on the key features of optogenetic devices and their applications that are useful in combination with MRI experiments (35). In general, the experimental design strategy under consideration will need to take into account the body/brain region and cell-type of interest, as well as, of course, the scientific question under consideration. It is important to keep in mind that the use of optogenetic devices requires the delivery of light to the tissue in question, which generally requires the implantation of optical hardware directed towards the region of interest.
The first choice a researcher interested in using an optogenetics approach for their experiment must make is: which cellular action do I want to control? The principal options currently available are membrane depolarization, e.g. to activate neurons, the exemplar of which is ChR2; membrane hyperpolarization, e.g. to silence neurons, the exemplar of which is Halorhodopsin (NpHR) (36); and modulation of intracellular biochemical signaling pathways, e.g. to mimic the action of hormones or neuromodulators on cells, the exemplars of which are the optoXRs (37). Related questions are: what wavelength of light do I want to use? And what kinetics of activation do I desire? Optogenetic molecular devices generally utilize endogenous retinoids to sense light and have peaks in their input action spectrum principally in the visible light wavelengths, ranging from far red to the far blue (34). Similarly, optogenetic devices have been engineered with a range of kinetics of on and off activation, with the generally more sensitive opsins requiring longer kinetic timescales (38). For most MRI applications, the most well characterized opsin in each family (e.g. ChR2 for neuronal activation) is generally sufficient.
The second major question to be answered is: which cell type do I want to control? The genetics of optogenetics refers to the ability to utilize these molecular devices to isolate genetically specified cellular populations for targeted control. This is usually achieved through placing the gene encoding the opsin under the control of a genetic promoter specific to the cell-type of interest; or, through cell-type specific tropism of the gene delivery method (39). As neural tissue is histologically heterogenous, being able to isolate, for instance, only the excitatory neurons of the cortex by using the CaMKIIα promoter (36), provides a distinct advantage for optogenetic stimulation in comparison to electrical stimulation, which would generally recruit excitatory as well as inhibitory cell types and surrounding glia. Similarly, optogenetic strategies have utilized similar methods to isolate control over hypocretinergic cells (40), dopaminergic cells (41), and parvalbumin-positive interneurons (42). The generation of viral vectors encoding optogenetic devices under the control of a recombinase dependent expression cassette allows for the utilization of Cre-driver mouse lines available for targeting myriad cell types (34). An additional strategy for isolating specific cell-types and inputs is to utilize the ability of neurons to transport these membrane proteins down their far projecting axons. Then, after delivery of the gene to the neuronal cell bodies in one brain region, by delivering light to a downstream brain region, only the synaptic inputs from the initial brain region to the downstream region will be recruited. Further details of targeting strategies are discussed in available technical reviews (34).
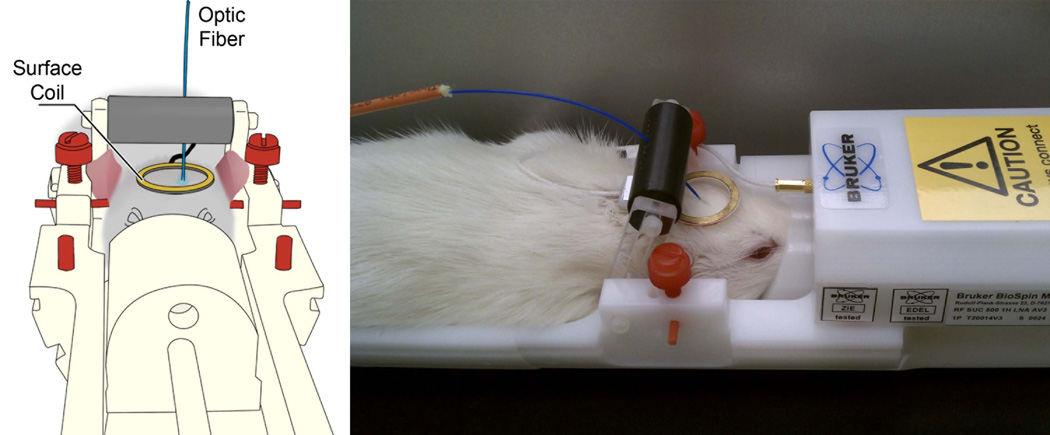
The third question to answer in designing an optogenetic experiment is: where is my region of interest and how should I deliver light there? The ability to focally control cellular populations of interest with only magnetically inert materials that do not interact significantly with radiofrequency pulses makes optogenetics incredibly attractive for combination with MRI experiments. However, visible light has relatively poor penetration through tissue. Accordingly, to generate the 1–10 mW/mm2 of light power flux necessary to activate optogenetic molecular devices, light needs to be locally delivered to the desired area. Most optogenetics experiments solve this problem by implanting a guide cannula targeted to the region of interest such that a fiber optic cable connected to a light source, such as a laser or high power LED, may be transmitted through the channel to an appropriate depth reproducibly. Considerations in this regard are whether the fiber optic system of choice has a numerical aperture and optical diameter appropriate for the geometry of the tissue region of interest. Detailed protocols for such surgery and optical stimulation are available (43). Combining optogenetics manipulations simultaneously with fMRI, to investigate the function and connectivity of brain circuits, has been performed in rats that have a specific population of (excitatory) neurons which were engineered via virus transfection to express specific opsins (ChR2 and eNpHR3), and in transgenic mice expressing ChR2. Figure 1 shows a schematic illustration of the experimental setup that we have built for the animal dedicated MRI system that includes a dedicated holder for the optic fiber and the surface coil. In order to illuminate the primary somatosensory cortex (S1), we have thinned the skull above S1 and mounted a 400 micron-diameter optic fiber coupled to a laser source over the in the center of the craniotomy. The craniotomy window was then filled with 2% agarose gel in order to minimize susceptibility artifacts. For optogenetics manipulations of deep brain areas such as the thalamus, the optic fiber was inserted through an implanted guiding cannula (44).
Figure 1. A schematic illustration and photo of the experimental setup for the animal dedicated MRI system that includes a dedicated holder for the optic fiber and the surface coil.
Putting all these considerations together towards a scientific goal may be technically challenging in initial stages, to be sure. The power of controlling a variety of cellular processes in a genetically-specific cellular population with high temporal resolution, and to do so with advanced MRI techniques as a readout of the global effects of such cellular stimulation, will allow for a revolution in our understanding of the in vivo effects of these cellular processes, as we discuss further in the next section.
Optogenetics and functional MRI
The brain has the tremendous capability to adapt itself in response to internal and external events. The ability of neurons to change their internal properties, and of neuronal networks to reshape their connections, is referred to as plasticity. The outcome of these plasticity changes can affect the time it takes the brain to process a specific stimulation, and generate a suitable response. Therefore, appropriate rewiring of neuronal connections during development and in adulthood is crucial to ensure proper and adequate propagation and processing of stimuli. One of the fundamental goals in neuroscience is to determine the genetic, epigentic, cellular, systemic and environmental basis of plasticity changes. BOLD fMRI techniques enable for the detection of hemodynamic changes due to changes in neural activity throughout the brain. In that respect, human fMRI has had a major impact in cognitive neuroscience and neurosurgery planning where an emphasis is on the role of plasticity in recovery and maintenance of brain functions in a wide range of diseases. Indeed, a revolution has occurred indicating that extensive and widespread plasticity takes place in the adult brain.
The ability to manipulate normal neuronal functions in rodents using genetic, molecular biology and neurosurgical tools and to monitor the consequent changes in neuronal behavior make rodent models the primary preference when investigating the underlying mechanisms of neuronal plasticity. Over the past decade, new developments in MRI of rodents have enabled spatial resolution of approximately 100 microns and temporal resolution for functional changes on the order of 500 ms, making MRI an emerging tool for studying plasticity in animal models. Changes in the spatial localization and the magnitude of BOLD fMRI responses were observed in rodent models following lesions in the central nervous system (45–47) and injuries of the peripheral nervous system (48–51). Similar to human fMRI studies, it is apparent that reorganization of neuronal pathways following injury in the rodent brain is reflected by the fMRI responses.
Nevertheless, the BOLD fMRI signal is an indirect measurement of neuronal activity. The exact relationship between the neuronal responses and hemodynamic responses remains unclear and under debate (52–58). Indeed in recent years there have been great efforts to resolve this controversial topic. The vast majority of the microscopic research is focused on exploring the molecular factors implicated in the underlying neurovascular coupling, such as vasoactive ions, vasoactive factors related to energy metabolism, vasoactive factors/neurotransmitters released by neuronal activation, and the role astrocytes play in neurovascular coupling (59–62). The majority of the macroscopic research is focused on investigating the temporal correlation between the magnitude of the neuronal responses and the hemodynamic responses. Studies have demonstrated that the BOLD fMRI responses are tightly correlated to increases in local field potential (LFP) and spiking activity (63–67); on the other hand, dissociations between LFP and spiking activity to hemodynamic responses have been reported (68–70). Understanding this relationship is crucial to define boundaries of cortical representations of any stimulus or response. Such knowledge, for example, would be useful in determining the degree of plasticity associated with development and injury.
Currently, electrical stimulation via electrodes is used to map and modulate the stimulus response in the brain in combination with fMRI measurements in non-primates and rodents (71–73). However, stimulating electrodes to induce specific neuronal responses or measure connectivity of brain regions can confound functional mapping since the electrical activation recruits a heterogeneous population of excitatory, inhibitory and modulatory cells. In addition, using stimulation electrodes in combination of MRI often generate susceptibility artifacts in the MR image. As discussed previously, optogenetic tools now enable neuronal manipulations that are precise, reversible and cell specific. In combination with MRI, optogenetics emerges as a powerful tool to investigate detailed neuronal mechanisms associated with brain function. For example, optogenetic tools are being utilized to address questions regarding the basis of the underlying neuronal activity in BOLD fMRI signals that were challenging to measure before.
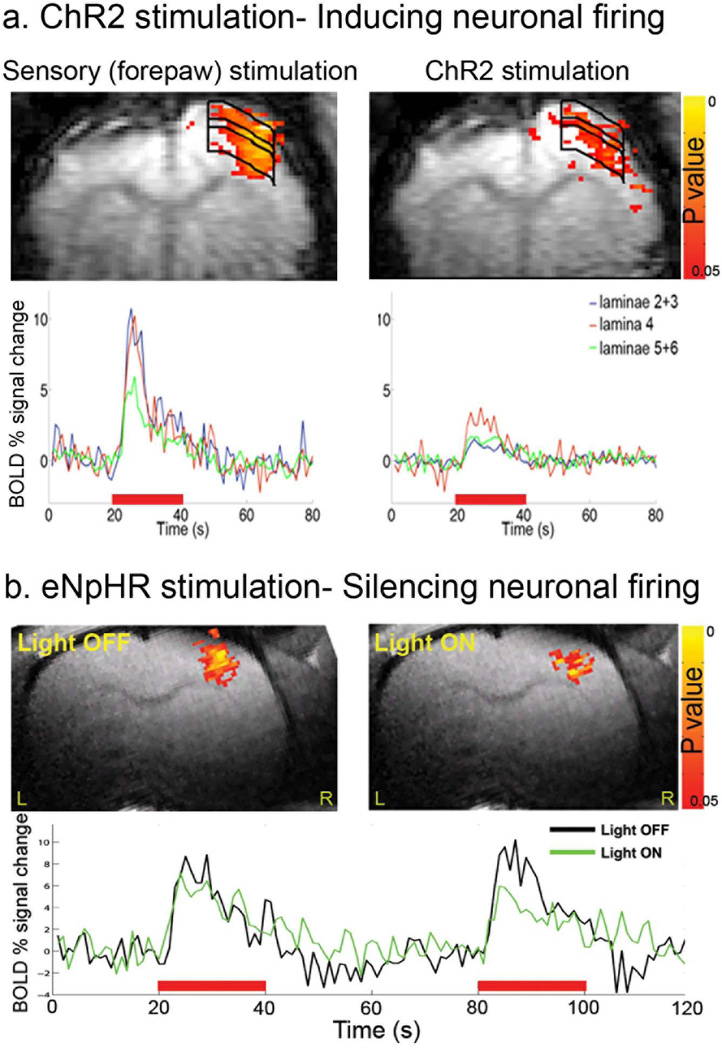
In a recent study, light stimulation of ChR2 expressing excitatory neurons in the primary motor cortex (M1) and thalamus during fMRI acquisition was performed in rats. The optogenetics stimulation have generated BOLD fMRI responses in the light stimulated areas whose time courses matched responses of conventional sensory-evoked stimulation, demonstrating that firing of excitatory neurons are involved in the neurovascular coupling driving the hemodynamic responses (44). As shown in Figure 2, we demonstrated that light stimulation of ChR2 (inducing neuronal firing) and of eNpHR (silencing neuronal firing) expressing excitatory neurons in the primary somatosensory cortex (S1) is capable of modulating the sensory-evoked BOLD fMRI responses in rats (74,75).
Figure 2. Modulations in the BOLD fMRI responses in the rat’s primary somatosensory cortex (S1) induced by optogenetics stimulation.
a. Examples of BOLD fMRI activation z-maps (p<0.05) induced by sensory (contralateral forepaw) and ChR2 stimulation overlaid on the EPI images. The optic fiber was placed directly above the right S1. The time courses of BOLD fMRI responses across the different cortical laminae are shown. b. Light induced activation of eNpHR resulted in decreases in both the extent and the amplitude of BOLD fMRI responses during forepaw stimulation mainly in the upper cortical laminae. Z statistic activation maps (p<0.05) are overlaid on RARE anatomical images. Red bars represent forepaw stimulation. Images were acquired using a Bruker 9.4 T animal dedicated scanner.
The combinations of optogenetics with fMRI can also facilitate detailed investigation of brain connectivity and function of neuronal circuits associated with plasticity and brain pathologies. Light stimulation of ChR2 expressing excitatory neurons in M1 resulted in BOLD fMRI responses in downstream regions (thalamus) demonstrating their strong connectivity (44). In addition, anesthesia has been shown to dramatically decrease the optogenetics-induced BOLD fMRI connectivity between cortical and sub-cortical areas in mice (76).
The BOLD fMRI temporal and laminar characteristics of light stimulation of ChR2 expressing excitatory neurons in lamina V of S1 of mice were compared to conventional vibrissa (sensory) stimulation. Both stimulation modalities gave rise to identical BOLD fMRI and electrophysiological responses (77). To date, the groups that combined optogenetics with fMRI used different animal models, the light sensitive channels were expressed in different neuronal populations and the optogenetics stimulations took place in different sites. Thus, careful interpretation of the findings, in terms of the function of the neuronal circuitry and the neuronal basis of the BOLD fMRI signals, should be employed.
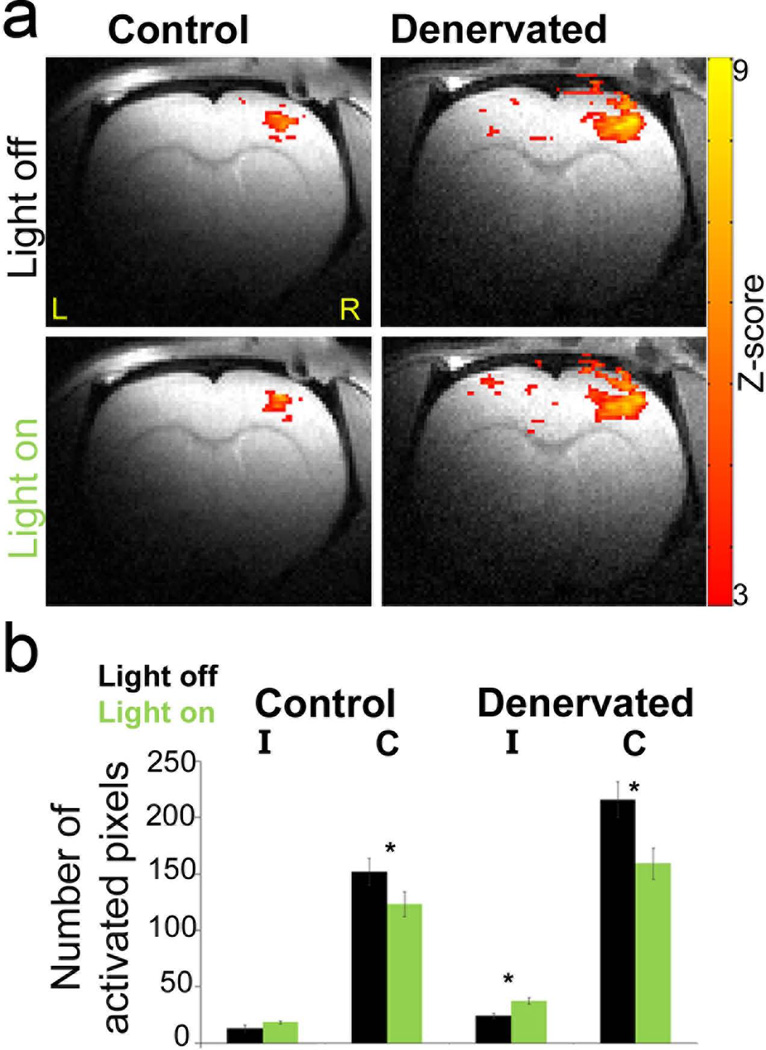
In our studies, we also used BOLD fMRI as a mean to monitor optogenetics-induced changes in post-injury plasticity in a rat model for peripheral nerve injury. Through optogenetics manipulation of cortical neurons in the hemisphere ipsilateral to the injured forepaw, we guided the cortical reorganization (35). As shown in Figure 3, we successfully decreased the inhibition in S1 ipsilateral to the injured forepaw, which in turn could facilitate recovery and rehabilitation following peripheral nerve injury.
Figure 3. BOLD fMRI shows increased responses within the primary somatosensory cortex (S1) contralateral to the injured forepaw during eNpHR stimulation.
a. In denervated rats, stimulation of eNpHR of the healthy (right) S1 induced increases in BOLD fMRI responses in the deprived S1, ipsilateral to intact forepaw stimulation. Z-maps (p<0.05) are overlaid on RARE anatomical images. b. Group average of the BOLD fMRI response spatial extent in S1 contralateral (C) and ipsilateral (I) to forepaw stimulation in control (n=5) and denervated (n=5) rats with or without eNpHR stimulation (*, p<0.05). Images were acquired using a Bruker 9.4 T animal dedicated scanner.
This work strengthens the notion that the combination of optogenetics and fMRI can provide an appropriate platform to resolve pre-synaptic vs. post-synaptic changes of neuronal activity and changes in the activity of large neuronal networks in the context of plasticity associated with development, learning and injury.
In summary, the application of novel genetic tools that can manipulate the MR contrast can open a new avenue for understanding complex biological and physiological systems.
Acknowledgments
GP is thankful to Bruker for their collaborative support on this work, and David Glover for assisting with manuscript preparation. This work was supported by NIH/NINDS 5R01NS072171.
References
- 1.Jackson DA, Symons RH, Berg P. Biochemical method for inserting new genetic information into DNA of Simian Virus 40: circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proc Natl Acad Sci U S A. 1972;69(10):2904–2909. doi: 10.1073/pnas.69.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koretsky AP, Traxler BA. The B isozyme of creatine kinase is active as a fusion protein in Escherichia coli: in vivo detection by 31P NMR. FEBS Lett. 1989;243(1):8–12. doi: 10.1016/0014-5793(89)81206-2. [DOI] [PubMed] [Google Scholar]
- 3.Koretsky AP, Brosnan MJ, Chen LH, Chen JD, Van Dyke T. NMR detection of creatine kinase expressed in liver of transgenic mice: determination of free ADP levels. Proc Natl Acad Sci U S A. 1990;87(8):3112–3116. doi: 10.1073/pnas.87.8.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walter G, Barton ER, Sweeney HL. Noninvasive measurement of gene expression in skeletal muscle. Proc Natl Acad Sci U S A. 2000;97(10):5151–5155. doi: 10.1073/pnas.97.10.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koretsky A, Lin Y-J, Schorle H, Jaenisch R. Genetic control of MRI contrast by expression of the transferrin receptor. Proc Intl Soc Mag Res. 1996;4:69. [Google Scholar]
- 6.Louie AY, Huber MM, Ahrens ET, Rothbacher U, Moats R, Jacobs RE, Fraser SE, Meade TJ. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol. 2000;18(3):321–325. doi: 10.1038/73780. [DOI] [PubMed] [Google Scholar]
- 7.Cohen B, Dafni H, Meir G, Harmelin A, Neeman M. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7(2):109–117. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Genove G, Demarco U, Xu H, Goins WF, Ahrens ET. A new transgene reporter for in vivo magnetic resonance imaging. Nat Med. 2005:450–454. doi: 10.1038/nm1208. [DOI] [PubMed] [Google Scholar]
- 9.Deans AE, Wadghiri YZ, Bernas LM, Yu X, Rutt BK, Turnbull DH. Cellular MRI contrast via coexpression of transferrin receptor and ferritin. Magn Reson Med. 2006;56(1):51–59. doi: 10.1002/mrm.20914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilad AA, McMahon MT, Walczak P, Winnard PT, Jr, Raman V, van Laarhoven HW, Skoglund CM, Bulte JW, van Zijl PC. Artificial reporter gene providing MRI contrast based on proton exchange. Nat Biotechnol. 2007;25(2):217–219. doi: 10.1038/nbt1277. [DOI] [PubMed] [Google Scholar]
- 11.Gilad AA, Winnard PT, Jr, van Zijl PC, Bulte JW. Developing MR reporter genes: promises and pitfalls. NMR Biomed. 2007;20(3):275–290. doi: 10.1002/nbm.1134. [DOI] [PubMed] [Google Scholar]
- 12.Gilad AA, Ziv K, McMahon MT, van Zijl PC, Neeman M, Bulte JW. MRI reporter genes. J Nucl Med. 2008;49(12):1905–1908. doi: 10.2967/jnumed.108.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui W, Liu L, Kodibagkar VD, Mason RP. S-Gal, a novel 1H MRI reporter for beta-galactosidase. Magn Reson Med. 2010;64(1):65–71. doi: 10.1002/mrm.22400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu G, Gilad AA. MRI of CEST-based reporter gene. Methods Mol Biol. 2011;771:733–746. doi: 10.1007/978-1-61779-219-9_36. [DOI] [PubMed] [Google Scholar]
- 15.Liu G, Liang Y, Bar-Shir A, Chan KW, Galpoththawela CS, Bernard SM, Tse T, Yadav NN, Walczak P, McMahon MT, Bulte JW, van Zijl PC, Gilad AA. Monitoring enzyme activity using a diamagnetic chemical exchange saturation transfer magnetic resonance imaging contrast agent. J Am Chem Soc. 2011;133(41):16326–16329. doi: 10.1021/ja204701x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapiro MG, Westmeyer GG, Romero PA, Szablowski JO, Kuster B, Shah A, Otey CR, Langer R, Arnold FH, Jasanoff A. Directed evolution of a magnetic resonance imaging contrast agent for noninvasive imaging of dopamine. Nat Biotechnol. 2010;28(3):264–270. doi: 10.1038/nbt.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapiro MG, Szablowski JO, Langer R, Jasanoff A. Protein nanoparticles engineered to sense kinase activity in MRI. J Am Chem Soc. 2009;131(7):2484–2486. doi: 10.1021/ja8086938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grabill C, Silva AC, Smith SS, Koretsky AP, Rouault TA. MRI detection of ferritin iron overload and associated neuronal pathology in iron regulatory protein-2 knockout mice. Brain Res. 2003;971(1):95–106. doi: 10.1016/s0006-8993(03)02366-7. [DOI] [PubMed] [Google Scholar]
- 19.Cohen B, Ziv K, Plaks V, Israely T, Kalchenko V, Harmelin A, Benjamin LE, Neeman M. MRI detection of transcriptional regulation of gene expression in transgenic mice. Nat Med. 2007;13(4):498–503. doi: 10.1038/nm1497. [DOI] [PubMed] [Google Scholar]
- 20.Ziv K, Meir G, Harmelin A, Shimoni E, Klein E, Neeman M. Ferritin as a reporter gene for MRI: chronic liver over expression of H-ferritin during dietary iron supplementation and aging. NMR Biomed. 2010;23(5):523–531. doi: 10.1002/nbm.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen AP, Hurd RE, Gu YP, Wilson DM, Cunningham CH. (13)C MR reporter probe system using dynamic nuclear polarization. NMR Biomed. 2011;24(5):514–520. doi: 10.1002/nbm.1618. [DOI] [PubMed] [Google Scholar]
- 22.Troughton JS, Greenfield MT, Greenwood JM, Dumas S, Wiethoff AJ, Wang J, Spiller M, McMurry TJ, Caravan P. Synthesis and evaluation of a high relaxivity manganese(II)-based MRI contrast agent. Inorg Chem. 2004;43(20):6313–6323. doi: 10.1021/ic049559g. [DOI] [PubMed] [Google Scholar]
- 23.Caravan P. Strategies for increasing the sensitivity of gadolinium based MRI contrast agents. Chem Soc Rev. 2006;35(6):512–523. doi: 10.1039/b510982p. [DOI] [PubMed] [Google Scholar]
- 24.Gottesfeld Z, Neeman M. Ferritin effect on the transverse relaxation of water: NMR microscopy at 9.4 T. Magn Reson Med. 1996;35(4):514–520. doi: 10.1002/mrm.1910350410. [DOI] [PubMed] [Google Scholar]
- 25.Iordanova B, Robison CS, Ahrens ET. Design and characterization of a chimeric ferritin with enhanced iron loading and transverse NMR relaxation rate. J Biol Inorg Chem. 2010;15(6):957–965. doi: 10.1007/s00775-010-0657-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weissleder R, Moore A, Mahmood U, Bhorade R, Benveniste H, Chiocca EA, Basilion JP. In vivo magnetic resonance imaging of transgene expression. Nat Med. 2000;6(3):351–355. doi: 10.1038/73219. [DOI] [PubMed] [Google Scholar]
- 27.Alfke H, Stoppler H, Nocken F, Heverhagen JT, Kleb B, Czubayko F, Klose KJ. In vitro MR imaging of regulated gene expression. Radiology. 2003;228(2):488–492. doi: 10.1148/radiol.2282012006. [DOI] [PubMed] [Google Scholar]
- 28.Zurkiya O, Chan AW, Hu X. MagA is sufficient for producing magnetic nanoparticles in mammalian cells, making it an MRI reporter. Magn Reson Med. 2008;59(6):1225–1231. doi: 10.1002/mrm.21606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogawa S, Lee TM. Magnetic resonance imaging of blood vessels at high fields: in vivo and in vitro measurements and image simulation. Magn Reson Med. 1990;16(1):9–18. doi: 10.1002/mrm.1910160103. [DOI] [PubMed] [Google Scholar]
- 30.Pelled G. MRI of neuronal plasticity in rodent models. Methods Mol Biol. 2011;711:567–578. doi: 10.1007/978-1-61737-992-5_29. [DOI] [PubMed] [Google Scholar]
- 31.Neeman M, Gilad AA, Dafni H, Cohen B. Molecular imaging of angiogenesis. J Magn Reson Imaging. 2007;25(1):1–12. doi: 10.1002/jmri.20774. [DOI] [PubMed] [Google Scholar]
- 32.Gross S, Gilead A, Scherz A, Neeman M, Salomon Y. Monitoring photodynamic therapy of solid tumors online by BOLD-contrast MRI. Nat Med. 2003;9(10):1327–1331. doi: 10.1038/nm940. [DOI] [PubMed] [Google Scholar]
- 33.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8(9):1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 34.Tye KM, Deisseroth K. Optogenetic investigation of neural circuits underlying brain disease in animal models. Nat Rev Neurosci. 2012;13(4):251–266. doi: 10.1038/nrn3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li N, Downey JE, Bar-Shir A, Gilad AA, Walczak P, Kim H, Joel SE, Pekar JJ, Thakor NV, Pelled G. Optogenetic-guided cortical plasticity after nerve injury. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1100815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, Wang L-P, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446(7136):633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 37.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458(7241):1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 38.Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O'Shea DJ, Prakash R, Gunaydin LA, Hyun M, Fenno LE, Gradinaru V, Yizhar O, Deisseroth K. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2012;9(2):159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris TJ, Green JJ, Fung PW, Langer R, Anderson DG, Bhatia SN. Tissue-specific gene delivery via nanoparticle coating. Biomaterials. 2010;31(5):998–1006. doi: 10.1016/j.biomaterials.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450(7168):420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai HC, Zhang F, Adamantidis A, Stuber GD, Bonci A, de Lecea L, Deisseroth K. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324(5930):1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459(7247):698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, de Lecea L, Deisseroth K. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc. 2010;5(3):439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee JH, Durand R, Gradinaru V, Zhang F, Goshen I, Kim DS, Fenno LE, Ramakrishnan C, Deisseroth K. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465(7299):788–792. doi: 10.1038/nature09108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dijkhuizen RM, Ren J, Mandeville JB, Wu O, Ozdag FM, Moskowitz MA, Rosen BR, Finklestein SP. Functional magnetic resonance imaging of reorganization in rat brain after stroke. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(22):12766–12771. doi: 10.1073/pnas.231235598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dijkhuizen RM, Singhal AB, Mandeville JB, Wu O, Halpern EF, Finklestein SP, Rosen BR, Lo EH. Correlation between brain reorganization, ischemic damage, and neurologic status after transient focal cerebral ischemia in rats: a functional magnetic resonance imaging study. J Neurosci. 2003;23(2):510–517. doi: 10.1523/JNEUROSCI.23-02-00510.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pelled G, Bergman H, Goelman G. Bilateral overactivation of the sensorimotor cortex in the unilateral rodent model of Parkinson's disease - a functional magnetic resonance imaging study. Eur J Neurosci. 2002;15(2):389–394. doi: 10.1046/j.0953-816x.2001.01866.x. [DOI] [PubMed] [Google Scholar]
- 48.Pawela CP, Biswal BB, Hudetz AG, Li R, Jones SR, Cho YR, Matloub HS, Hyde JS. Interhemispheric neuroplasticity following limb deafferentation detected by resting-state functional connectivity magnetic resonance imaging (fcMRI) and functional magnetic resonance imaging (fMRI) Neuroimage. 2010;49(3):2467–2478. doi: 10.1016/j.neuroimage.2009.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pelled G, Chuang KH, Dodd SJ, Koretsky AP. Functional MRI detection of bilateral cortical reorganization in the rodent brain following peripheral nerve deafferentation. Neuroimage. 2007;37(1):262–273. doi: 10.1016/j.neuroimage.2007.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pelled G, Dodd SJ, Koretsky AP. Catheter confocal fluorescence imaging and functional magnetic resonance imaging of local and systems level recovery in the regenerating rodent sciatic nerve. Neuroimage. 2006;30(3):847–856. doi: 10.1016/j.neuroimage.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 51.Yu X, Wang S, Chen DY, Dodd S, Goloshevsky A, Koretsky AP. 3D mapping of somatotopic reorganization with small animal functional MRI. Neuroimage. 2010;49(2):1667–1676. doi: 10.1016/j.neuroimage.2009.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Attwell D, Iadecola C. The neural basis of functional brain imaging signals. Trends Neurosci. 2002;25(12):621–625. doi: 10.1016/s0166-2236(02)02264-6. [DOI] [PubMed] [Google Scholar]
- 53.Ekstrom A. How and when the fMRI BOLD signal relates to underlying neural activity: the danger in dissociation. Brain Res Rev. 2010;62(2):233–244. doi: 10.1016/j.brainresrev.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Enager P, Piilgaard H, Offenhauser N, Kocharyan A, Fernandes P, Hamel E, Lauritzen M. Pathway-specific variations in neurovascular and neurometabolic coupling in rat primary somatosensory cortex. J Cereb Blood Flow Metab. 2009;29(5):976–986. doi: 10.1038/jcbfm.2009.23. [DOI] [PubMed] [Google Scholar]
- 55.Logothetis NK. What we can do and what we cannot do with fMRI. Nature. 2008;453(7197):869–878. doi: 10.1038/nature06976. [DOI] [PubMed] [Google Scholar]
- 56.van Eijsden P, Hyder F, Rothman DL, Shulman RG. Neurophysiology of functional imaging. Neuroimage. 2009;45(4):1047–1054. doi: 10.1016/j.neuroimage.2008.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vanzetta I, Grinvald A. Coupling between neuronal activity and microcirculation: implications for functional brain imaging. HFSP J. 2008;2(2):79–98. doi: 10.2976/1.2889618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yen CC, Fukuda M, Kim SG. BOLD responses to different temporal frequency stimuli in the lateral geniculate nucleus and visual cortex: insights into the neural basis of fMRI. Neuroimage. 2011;58(1):82–90. doi: 10.1016/j.neuroimage.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iadecola C, Nedergaard M. Glial regulation of the cerebral microvasculature. Nat Neurosci. 2007;10(11):1369–1376. doi: 10.1038/nn2003. [DOI] [PubMed] [Google Scholar]
- 60.Koehler RC, Roman RJ, Harder DR. Astrocytes and the regulation of cerebral blood flow. Trends Neurosci. 2009;32(3):160–169. doi: 10.1016/j.tins.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 61.Lauritzen M. Reading vascular changes in brain imaging: is dendritic calcium the key? Nat Rev Neurosci. 2005;6(1):77–85. doi: 10.1038/nrn1589. [DOI] [PubMed] [Google Scholar]
- 62.Lecrux C, Hamel E. The neurovascular unit in brain function and disease. Acta Physiol (Oxf) 2011;203(1):47–59. doi: 10.1111/j.1748-1716.2011.02256.x. [DOI] [PubMed] [Google Scholar]
- 63.Kim T, Masamoto K, Fukuda M, Vazquez A, Kim SG. Frequency-dependent neural activity, CBF, and BOLD fMRI to somatosensory stimuli in isoflurane-anesthetized rats. Neuroimage. 2010;52(1):224–233. doi: 10.1016/j.neuroimage.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature. 2001;412(6843):150. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- 65.Mukamel R, Gelbard H, Arieli A, Hasson U, Fried I, Malach R. Coupling between neuronal firing, field potentials, and FMRI in human auditory cortex. Science. 2005;309(5736):951–954. doi: 10.1126/science.1110913. [DOI] [PubMed] [Google Scholar]
- 66.Shmuel A, Augath M, Oeltermann A, Logothetis NK. Negative functional MRI response correlates with decreases in neuronal activity in monkey visual area V1. Nat Neurosci. 2006;9(4):569–577. doi: 10.1038/nn1675. [DOI] [PubMed] [Google Scholar]
- 67.Viswanathan A, Freeman RD. Neurometabolic coupling in cerebral cortex reflects synaptic more than spiking activity. Nat Neurosci. 2007;10(10):1308–1312. doi: 10.1038/nn1977. [DOI] [PubMed] [Google Scholar]
- 68.Caesar K, Thomsen K, Lauritzen M. Dissociation of spikes, synaptic activity, and activity-dependent increments in rat cerebellar blood flow by tonic synaptic inhibition. Proc Natl Acad Sci U S A. 2003;100(26):16000–16005. doi: 10.1073/pnas.2635195100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Devor A, Tian P, Nishimura N, Teng IC, Hillman EM, Narayanan SN, Ulbert I, Boas DA, Kleinfeld D, Dale AM. Suppressed neuronal activity and concurrent arteriolar vasoconstriction may explain negative blood oxygenation level-dependent signal. J Neurosci. 2007;27(16):4452–4459. doi: 10.1523/JNEUROSCI.0134-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pelled G, Bergstrom DA, Tierney PL, Conroy RS, Chuang KH, Yu D, Leopold DA, Walters JR, Koretsky AP. Ipsilateral cortical fMRI responses after peripheral nerve damage in rats reflect increased interneuron activity. Proc Natl Acad Sci U S A. 2009;106(33):14114–14119. doi: 10.1073/pnas.0903153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Logothetis NK, Augath M, Murayama Y, Rauch A, Sultan F, Goense J, Oeltermann A, Merkle H. The effects of electrical microstimulation on cortical signal propagation. Nat Neurosci. 2010;13(10):1283–1291. doi: 10.1038/nn.2631. [DOI] [PubMed] [Google Scholar]
- 72.Tolias AS, Sultan F, Augath M, Oeltermann A, Tehovnik EJ, Schiller PH, Logothetis NK. Mapping cortical activity elicited with electrical microstimulation using FMRI in the macaque. Neuron. 2005;48(6):901–911. doi: 10.1016/j.neuron.2005.11.034. [DOI] [PubMed] [Google Scholar]
- 73.Pan WJ, Thompson G, Magnuson M, Majeed W, Jaeger D, Keilholz S. Simultaneous FMRI and electrophysiology in the rodent brain. J Vis Exp. 2010;(42) doi: 10.3791/1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Downey JE, Li N, Gilad AA, Walczak P, Thakor NV, Pelled G. The laminar specific neuronal responses to forepaw and optogenetics stimulations. In Proceedings of the ISMRM. 2011;19 [Google Scholar]
- 75.Downey JE, Walczak P, Joel SE, Gilad AA, McMahon MT, Kim H, Pekar JJ, Thakor NV, Pelled G. Light-Induced Activation of Light-Sensitive Pumps Modulates FMRI Responses. In Proceedings of the ISMRM. 2010;18 [Google Scholar]
- 76.Desai M, Kahn I, Knoblich U, Bernstein J, Atallah H, Yang A, Kopell N, Buckner RL, Graybiel AM, Moore CI, Boyden ES. Mapping brain networks in awake mice using combined optical neural control and fMRI. J Neurophysiol. 2011;105(3):1393–1405. doi: 10.1152/jn.00828.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kahn I, Desai M, Knoblich U, Bernstein J, Henninger M, Graybiel AM, Boyden ES, Buckner RL, Moore CI. Characterization of the functional MRI response temporal linearity via optical control of neocortical pyramidal neurons. J Neurosci. 2011;31(42):15086–15091. doi: 10.1523/JNEUROSCI.0007-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]