Abstract
DNA polymerase η (pol η) plays a critical role in suppressing mutations caused by the bypass of cis-syn cyclobutane pyrimidine dimers (CPD) that escape repair. There is evidence this is also the case for the oxidative lesion 7,8-dihydro-8-oxo-guanine (8-oxoG). Both of these lesions cause moderate to severe blockage of synthesis when encountered by replicative polymerases, while pol η displays little no to pausing during translesion synthesis. However, since lesion bypass does not remove damaged DNA from the genome and can possibly be accompanied by errors in synthesis during bypass, the process is often called ‘damage tolerance’ to delineate it from classical DNA repair pathways. The fidelity of lesion bypass is therefore of importance when determining how pol η suppresses mutations after DNA damage. As pol η has been implicated in numerous in vivo pathways other than lesion bypass, we wanted to better understand the molecular mechanisms involved in the relatively low-fidelity synthesis displayed by pol η. To that end, we have created a set of mutant pol η proteins each containing a single amino acid substitution in the active site and closely surrounding regions. We determined overall DNA synthesis ability as well as the efficiency and fidelity of bypass of thymine-thymine CPD (T-T CPD) and 8-oxoG containing DNA templates. Our results show that several amino acids are critical for normal polymerase function, with changes in overall activity and fidelity being observed. Of the mutants that retain polymerase activity, we demonstrate that amino acids Q38, Y52, and R61 play key roles in determining polymerase fidelity, with substation of alanine causing both increases and decreases in fidelity. Remarkably, the Q38A mutant displays increased fidelity during synthesis opposite 8-oxoG but decreased fidelity during synthesis opposite a T-T CPD.
Keywords: Mutagenesis, DNA damage, translesion synthesis, DNA polymerase eta, 8-oxoG, thymine-thymine CPD
1. Introduction
DNA polymerase η, a member of the Y-family, plays a critical role in bypassing DNA lesions, most notablycis-syn cyclobutane pyrimidine dimers (CPD) created by exposure of DNA to ultraviolet light. It synthesizes past lesions that block replicative polymerases and would otherwise halt replication fork progression. This action serves to lower the mutagenic potential they represent, and cells lacking functional pol η display markedly higher mutation rates after UV light exposure [1–4]. Despite this observation, the fidelity of pol η during the bypass event is far from perfect or even high fidelity in the classic sense of the word [5–7]. Replicative polymerases are capable of copying 105–106 nucleotides without making an error [8–10], while the fidelity of translesion synthesis (TLS) by polymeraseη can be 3–4 orders of magnitude lower. Despite early characterizations of TLS by pol η as being “error free” [11–14], during thymine-thymine CPD (T-T CPD) bypass the enzyme produces errors in the range of 1 in 30 [6], which is also the average error rate when copying large stretches of undamaged DNA [15, 16]. The fidelity of human pol η when bypassing 8-oxoG is even lower, with both steady state kinetic assays [7, 17] and those requiring complete bypass in the presence of all four deoxynucleotides [5] showing that dATP and dCTP are inserted at roughly equal frequencies. On an absolute scale this is very poor fidelity, although there is evidence that errors generated during bypass can be detected and removed by the exonuclease activities of the replicative polymerases [18]. It is often stated that 8-oxoG is not a blocking lesion, although there are multiple reports that demonstrate it can impede the normal synthesis of replicative polymerases [5, 19–21]. Additionally, while multiple polymerases have been shown to be able to bypass 8-oxoG under some conditions [17, 22, 23], pol η does so with greater than 100% efficiency compared to an undamaged G in the same context [5]. We hypothesize that the ability of pol η to efficiently synthesize past the lesion makes it one of the preferred polymerase to do so in vivo, acknowledging other polymerases may also play a role [17, 24]. In this scenario, the low-fidelity of bypass is a required trade-off to prevent the accumulation of single stranded regions of unreplicated DNA that are even more problematic. The combination of high-efficiency but low-fidelity lesion bypass by pol η therefore represents a balance that is assumed to lead to an overall positive outcome for the cell.
The TLS properties of pol η are conserved across a wide range of species. For instance, deep sea worms have pol η homologs that allow T-T CPD bypass, despite presumably never being exposed to UV light [25]. While the properties of pol η from different species are generally in agreement, there are a few differences. For instance, the human, mouse and budding yeast (S. cerevisiae) forms of pol η display similar properties when it comes to T-T CPD bypass and their fidelity when copying undamaged DNA [6, 11, 12, 14–16, 26, 27]. With respect to 8-oxoG bypass, however, there is a marked difference. DNA pol η from yeast displays the same high efficiency of bypass with bypass fidelity in the same range as a T-T CPD [5, 28, 29]. As noted above, while human and mouse pol η are able to efficiently bypass 8-oxoG, their fidelity when doing so is remarkably low, in the range of 1 in 2, or 50% [5, 7, 17]. Despite these in vitro findings there is evidence that in vivo pol η suppresses mutagenesis by this lesion [30]. Some possible explanations for these seemingly disparate results are extrinsic proofreading of errors, as has been shown for T-T CPD bypass, and modulation of the fidelity by association with replication accessory proteins [17, 18]. Interestingly, while the latter mechanism has been demonstrated by one assay for human pol η, it has been ruled out by a different assay for yeast pol η [5].
Early crystal structures of Y-family polymerases suggested that a very open, solvent accessible active site was one of the means by which the unique properties of the enzymes were achieved [31–35]. More recent crystal structures have shown for both human and yeast polη that the active site can in fact fit two template bases [36, 37]. This provides an explanation for how the chemically linked CPD can be so readily bypassed, as can lesions created by the crosslinking chemotherapy agent cisplatin [38–43]. Interestingly, the structures of DNA pol η in complex with undamaged DNA show two bases in the active site as well. This is in line with the observed low fidelity when copying undamaged DNA [6, 13, 15, 16, 26], and may help to explain how pol η is able to bypass other DNA lesions such as 8-oxoG and thymine glycol [7, 29, 40]. While the crystal structure of yeast pol η bound to 8-oxoG containing DNA is informative for the preferred dCTP incorporation by that enzyme [44], there is currently no structure available of human pol η in complex with 8-oxoG to shed light on the remarkably low fidelity bypass it exhibits [5, 7]. However, the available crystal structures of the human enzyme show which amino acid residues interact with the DNA and incoming nucleotide triphosphate and have allowed us to select several candidate residues for detailed biochemical analysis.
As pol η has been implicated in processes other than TLS [45–50], we sought to understand the molecular determinants that govern human pol η function. To investigate the link between lesion bypass fidelity and lesion bypass ability, we have generated several mutant forms of pol η, each containing a single amino acid substitution in the active site and surrounding regions of the catalytic core of human pol η (Figure 1). These include three single nucleotide polymorphisms (SNP’s) identified in the NCBI dbSNP database [51] as well as several picked from the crystal structures and alignments of pol η sequences [52]. Each mutant was purified and tested for overall DNA polymerase activity as well as lesion bypass ability and fidelity.
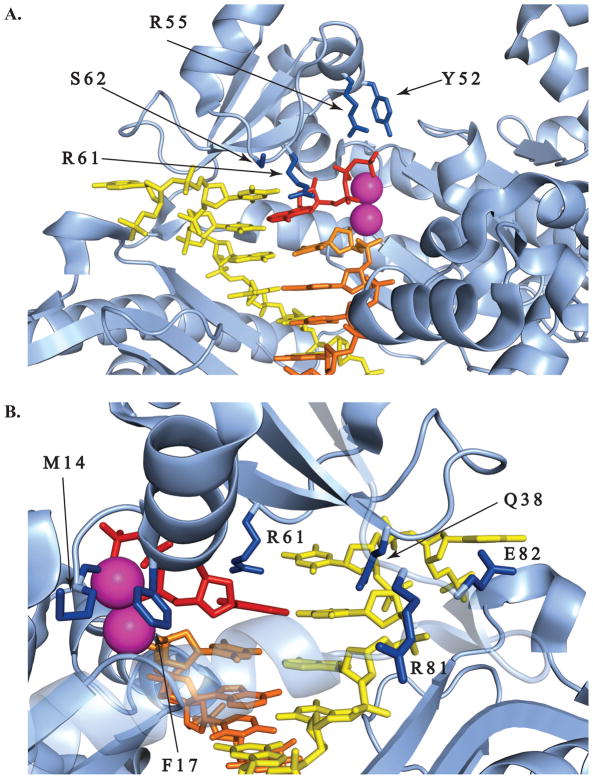
Figure 1.
Location of amino acids altered in this study within the active site of the protein. DNA template is shown in yellow, primer strand in orange, incoming nucleotide in red and metal ions as magenta spheres. Amino acids of interest are shown in dark blue. A. Y52, R55, R61 and S62 residues. B. M14, F17, Q38, R61, R81, E82 residues. The light blue transparent helix is ‘in front’ of the DNA (from the viewpoint of this image) but was lightened so as not to obscure the view. Images were created with PyMOL Molecular Graphics System (version 1.3, Schrodinger LLC) using coordinates in PDB: 3MR2 [36].
2. Materials and Methods
2.1 Reagents and materials
Oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA) and Midland Certified Reagent Co. (Midland, TX). Phosphoramidite precursors for the damaged nucleotides were purchased (by Midland Certified Reagent Co.) from Glen Research (Sterling, VA). Nucleotides and restriction enzymes were obtained from New England Biolabs (Ipswich, MA). All polymerase and exonuclease reactions were performed in 40 mM Tris pH 8.0, 250 μg/ml BSA, 10 mM DTT, 10 mM MgCl2, 60 mM KCl, and 1.25% glycerol. Polymerase reactions were supplemented with 1 mM or 0.1 mM final concentration of each dNTP for the forward gap filling or oligonucleotide based assays, respectively. All cell lines, bacteriophage and reagents for lesion bypass and forward mutation assays have been previously described [53, 54]
2.2 Expression vector and protein purification
The pET21b-XPV vector was obtained through the generosity of the laboratory of Dr. Thomas Kunkel (NIEHS, Research Triangle Park, NC). This vector codes for the first 511 amino acids of human pol η containing a C-terminal 6X histidine tag after a 2 amino acid linker (519 amino acids total; ~56 kD; vector sequence available on request). Amino acid changes were introduced using the Stratagene QuikChange II XL site-directed mutagenesis kit using conditions recommended by the manufacturer. Primers used for mutagenesis reactions are given in Supplementary Table 1. Changes were verified and no additional mutations in the ORF were confirmed by sequencing performed by Genewiz (South Plainfield, NJ). The expression vector (coding for either wild type or the single amino acid changes) was introduced into BL21 (DE3) cells by electroporation, grown overnight at 37°C on LB media containing 100 μg/ml ampicillin, and then a single colony was used to inoculate 10 ml LB broth with 100 μg/ml ampicillin and grown to saturation at 30 °C/250 RPM. Saturated 10 ml culture was used to inoculate 1 L LB broth (no ampicillin) that was incubated at 30 °C/250 RPM until the OD595 reached 0.4–0.6. IPTG was added to 0.5 mM final concentration and the culture was incubated at 15°C/250 RPM for an additional 18 hours. Cells were harvested by centrifugation, the pellet washed twice with PBS, and the cell pellet was resuspended in a small volume (~5 ml) of PBS that was dripped into liquid N2, creating ~3–5 mm diameter drops that were stored at −80 °C. Cells were lysed using a SPEX Sample Prep 6870 Freezer/Mill. Cell drops were cooled for 10 minutes in liquid N2 and then subjected to 6 cycles of: 1 minute grinding (10 impacts per second) and 1 minute cooling. The resulting lysed cell powder was thawed and resuspended in 20–40 ml ice cold Buffer A (25 mM sodium phosphate pH 7.4, 10% glycerol, 300 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol) supplemented with 0.2 mM PMSF and Roche Complete Protease inhibitor tablets (1 tablet per 50 ml total volume). The crude cell lysate was sonicated (Branson 250 sonifier; output-2, duty cycle 50%) 6–10 times for 30 seconds with 30 seconds on ice in between cycles. Cell lysate was centrifuged at 20,000g for 30 minutes at 4 °C. The clarified extract was loaded onto a 5 ml HiTrap Chelating HP column (GE Healthcare; charged with NiSO4 and then equilibrated in Buffer A). The column was washed with 10 column volumes (CV) Buffer A, then with 5 CV Buffer B (25 mM sodium phosphate pH 7.4, 10% glycerol, 100 mM NaCl, 5 mM β-mercaptoethanol) supplemented with 10 mM imidazole. Bound protein was eluted using a 20 CV linear gradient of Buffer B containing from 10 to 600 mM imidazole. Fractions containing protein (as judged from A280 readings) were analyzed by SDS-PAGE and fractions with the expected protein size were pooled and concentrated using an Amicon Ultra -15 (30K MWCO) centrifugal filter (Millipore), then diluted 1:10 with Buffer C (25mM Tris-Cl pH 7.4, 10% glycerol, 1 mM DTT) containing 100 mM NaCl. The resulting solution was filtered through a 0.2 μm filter and loaded onto Mono S 5/50 GL column (1ml bed volume) equilibrated with Buffer C (100 mM NaCl). The column was washed with 10 CV Buffer C containing 100 mM NaCl, then eluted with a 30 CV linear gradient of Buffer C containing from 100 to 600 mM NaCl. Protein fractions identified by A280 were analyzed by SDS-PAGE. Selected fractions containing highly pure polymerase were aliquoted and frozen in liquid N2 and stored at −80 °C until further use.
2.3 Polymerase and lesion bypass assay
Determination of polymerase activity and bypass efficiency were performed essentially as described previously [6, 55] using the template 5′-TCGGTACCGGGTTAxCCTTTGGAGTCGACCTGC-3′ (where x represents either undamaged G or 8-oxo-G and the underlined TT are either undamaged or a cis-syn cyclobutane thymine-thymine dimer), and primers 5′-Cy5-GCAGGTCGACTCCAAAG-3′ (G/8-oxoG reactions) or 5′-Cy5-GCAGGTCGACTCCAAAGGC-3′ (T-T CPD reactions). Substrate was created by mixing primer (5 μM final concentration) with 1.1X molar equivalent template in 25 mM Tris pH 8.0 and 100 mM NaCl, heating to 95 °C for 5 minutes and then cooling to room temperature over 3 hours, protected from light. Assays using these “running start” substrates were performed using substrate to enzyme ratios and time points (indicated in figure legends and text) empirically determined sufficient to keep termination probabilities constant over time. Reaction products were resolved by 10% dPAGE and imaged with a Storm 865 imager. Analysis of band intensity was done using Image Quant TL software (GE Healthcare) and the termination probabilities, bypass amounts, and primer utilization were quantitated as described in [5, 6, 55]. The overall activity on undamaged DNA was calculated from the primer utilization amounts and normalized for amount of polymerase in the reaction and time. Lesion bypass fidelity assays were performed as previously described as was calculation of mutant frequencies and error rates after sequencing [5, 54, 55]. Substrates for the lesion bypass fidelity assays were created as above using the template sequence 5′-CCAGCTCGGTACCGGGTTAxCCTTTGGAGTCGACCTGCAGAAATT-3′ for 8-oxoG reactions (x is either G or 8-oxoG) or 5′-AGGAAACAGCTATGACCATGATTACGAATTCCAGCTCGGTACCGGGxxAGCCTTTGGAGTCGACCTGCAGAAATT-3′ for T-T CPD experiments (x represents either undamaged TT or a cis-syn T-T CPD). Regions paired with primer are underlined.
2.4 Forward mutation assay
The fidelity of the purified polymerases when copying undamaged DNA was measured utilizing the well-characterized 407 base gap filling forward mutation assay [53]. Reactions contained between 50 and 500 fold excess polymerase over substrate and were incubated at 37°C for 1 hour. Between 1200 and 4800 plaques resulting from each gap filling reaction were counted for plaque color phenotype and from an unselected subset of the mutants, DNA was amplified using TempliPhi (GE Healthcare; following manufacturers recommended procedures) and the filled region analyzed for changes after sequencing. Mutation frequencies and error rates were calculated as described previously [53].
3. Results
3.1 Active site mutations of pol η
Candidate amino acids for change were determined from previously published reports and sequence alignments of Y-family polymerases [36, 56–59]. Single nucleotide polymorphisms in the candidate region were determined from the NCBI SNP database [51]. The group of mutants created and expressed is presented in Table 1 and their location within the structure of the protein is shown in Figure 1. Predicted function of SNP’s is based on the published structure of human pol η [36]. Other amino acids in these regions play roles in coordination of a magnesium ion required for catalytic activity, contact with the template bases both at the site of nucleotide insertion and slightly upstream, and alignment of the incoming nucleotide [36]. Purification of E. coli expressed protein was achieved for wild type and 10 individual amino acid substitution mutants and was consistently greater than 95% pure, as judged from SYPRO Red stained gels. Two attempts at purification of the F17L mutant resulted in inadequate purified protein yields, presumably from incorrectly folded protein as the expected overexpressed bands was observed in crude cell extracts. As would be expected from preparations of the naturally exonuclease deficient pol η [15], all preparations were confirmed to be free of detectable mismatched primer:terminus exonuclease activity (A:G mispair; data not shown).
Table 1.
Activity and lesion bypass fidelity for single amino acid mutants of human pol η
| Mutant | Function/Role* | Activity** | Dark Blue plaques (%)*** | ||
|---|---|---|---|---|---|
| Undamaged | T-T CPD | 8-oxoG | |||
| Wild Type | 170 ± 38 | 7.1% | 2.3% | 29% | |
| M14V | Active site magnesium coordination**** | 500 ± 140 (3.0X) | 5.3% | 2.2% | 27% |
| F17L | Lid of steric gate | NA | NA | NA | NA |
| Q38A | Van der Waals interaction with template base | 350 ± 32 (2.1X) | 6.4% | 7.0% | 14% |
| Q38V | Van der Waals interaction with template base | 16 ± 3 (0.1X) | 6.8% | 1.7% | 33% |
| Y52E | Interaction with incoming nucleotide | 120 ± 35 (0.7X) | 1.4% | 0.5% | 3.2% |
| R55A | Interaction with phosphate of incoming nucleotides | ND | NA | NA | NA |
| R61A | Hydrogen bonds with incoming nucleotide | 170 ± 20 (1.0X) | 2.7% | 0.9% | 24% |
| S62A | Contact with 5′ base upstream of 1st template base | 32 ± 14 (0.2X) | 7.4% | 2.1% | 17% |
| S62G | Contact with 5′ base upstream of 1st template base | 130 ± 20 (0.8X) | 3.6% | 2.8% | 23% |
| R81C | Structure of protein | 420 ± 90 (2.5X) | 6.1% | 4.3% | 26% |
| E82D | Structure of protein | 230 ± 87 (1.4X) | 4.0% | 2.1% | 21% |
Predicted function based on published reports and crystal structures
Picomoles of substrate extended per minute per μg protein. Values in parentheses are relative to WT. NA, not applicable. ND, none detected.
Frequency of dark blue plaques observed in lesion bypass assay.
This interaction is with the peptide bond, not the side chain.
3.2 Polymerase activity
To evaluate whether the amino acid substitutions had any effect on the ability to synthesize DNA, we first performed polymerase assays under conditions of substrate excess and calculated the amount of substrate used per minute per μg protein (Table 1). In these assays, we empirically determined the substrate to enzyme ratio needed to utilize no more than ~30% of the total substrate within 8 minutes. These values were chosen as they consistently give ‘single hit’ conditions, as defined by the termination probability of the enzyme at any given position remaining constant over time [55]. The overall polymerase activity of the various preparations (picomole substrate used/min/μg protein) was calculated from the actual primer utilization values. Preps with activity less than 10% compared to wild type were purified a second time to ensure it was not an artifact of the purification process. The various mutants tested ranged from complete lack of detectable polymerase activity to having several fold more activity. Interestingly, all three of the naturally occurring isoforms of pol η tested (M14V, R81C, and E82D) displayed increased overall activity while only one of the other mutants (Q38A) tested did. Three mutants (Y52E, R61A, and S62G) displayed no change or slightly reduced activity, while Q38V and S62A displayed drastically reduced but still detectable activity. It is interesting that the Q38V and S62A forms had reduced activity while changes to different amino acids at these same residues had near normal or increased polymerase activity (Q38A and S62G; 80% and 210% of wild type, respectively), demonstrating that it is not just changing the wild type sequence but to which residue that needs to be considered. The R55A form, which coordinates the incoming dNTP phosphate group, was devoid of any detectable polymerase activity despite twice being purified in a soluble form. Only forms of pol η that were able to generate completely copied duplex substrates (lesion bypass assay) and able to completely fill the 407-base gapped plasmid DNA (forward mutation assay) were evaluated further. Therefore, the R55A and F17L forms are not included in subsequent analyses. While we did not perform pre-steady state kinetic analysis to determine the active fraction of each preparation, all proteins were expressed and purified under similar conditions and we assume that the differences in observed activity are intrinsic to the different amino acid sequence. Repeat preparations of both wild type and selected mutants show similar activities (within 10%; data not shown).
3.3 Lesion bypass efficiency
We analyzed all mutants that had requisite levels of polymerase activity in order to determine their ability to bypass either 8-oxo-G or a T-T CPD relative to an otherwise identical undamaged sequence. The goal was to determine if the changes in activity observed on undamaged DNA extended to damaged DNA or if there was a differential effect caused by the lesion containing substrates. Short times and high substrate to enzyme ratios (20:1 to 400:1) were used to ensure single substrate-polymerase interactions [54, 55]. All preparations showed the expected low processivity, with none of the mutants being able to insert more than 8–10 nucleotides during a single round of synthesis (Figure 2). This is consistent with several published reports of pol η and other Y-family polymerase activity [6, 12, 15, 60, 61]. Only Q38A (71% efficiency) and Y52E (100% efficiency) display somewhat reduced ability to bypass 8-oxoG compared to wild type (150% efficiency). All other mutants tested displayed the preferred copying of damaged DNA compared to undamaged DNA that wild type polymerase shows (i.e. bypass efficiency values of greater than 100%) [5, 6, 27]. We note that even the lowest observed 8-oxoG bypass efficiency seen here is still 2–3 fold higher than S. cerevisiae replicative polymerase δ[5] and pol ε [19]. When measuring T-T CPD bypass efficiency, we also observed more efficient bypass of damaged DNA compared to undamaged (i.e. values greater than 100%) for most mutant forms of the protein, although the magnitude of the effect does seem to be diminished somewhat compared to wild type (Figure 3B). The only exceptions were Q38A and Y52E (91% and 46%, respectively). Again, these values are still much higher than the values for replicative polymerases [18, 62].
Figure 2.
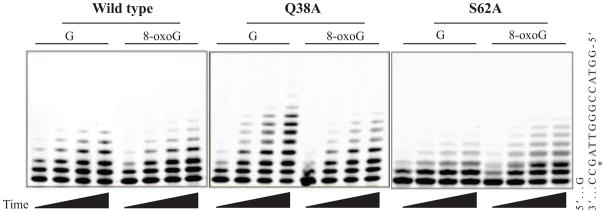
Denaturing polyacrylamide gel electrophoresis based separation of lesion bypass efficiency assay reaction products. Shown are 3 selected forms of the polymerase at time points ranging from 2–8 minutes. After analysis of fluorescently labeled primer strands using ImageQuant TL (GE Life Sciences), reactions were confirmed to be under single interaction conditions, demonstrating constant termination probabilities over time. Primer utilization and bypass amounts were calculated as previously described [54, 55].
Figure 3. Lesion bypass efficiency by pol η mutants.
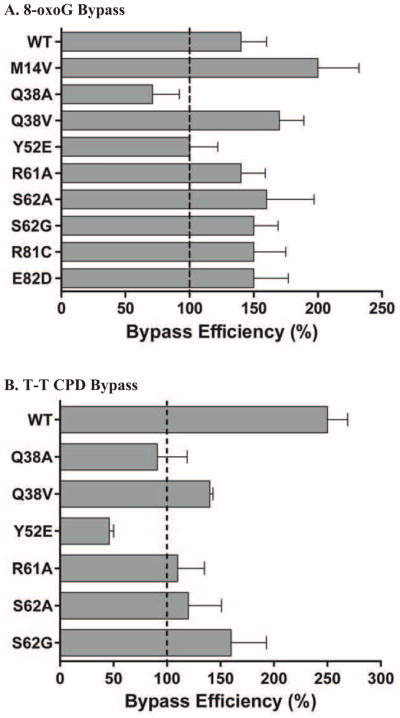
Lesion bypass efficiency assays were performed as described in Materials and Methods. Bypass of either undamaged or damaged template DNA was measured under conditions of single interactions between polymerase and substrate. Substrate:polymerase ratios used ranged from 20:1 (Q38V) to 400:1 (M14V) to account for the differences in overall activity of the various forms. Bypass efficiency is defined as the ratio of damaged to undamaged bypass, i.e. values greater than 100% reflect better bypass of the damaged base. The average bypass efficiency of 4 time points (2, 4, 6, 8 minutes) each from 2 reactions are graphed with the standard deviation shown in the error bars. A. Bypass efficiency of 8-oxoG containing templates. B. Bypass efficiency of T-T CPD containing templates.
3.4 Lesion bypass fidelity
We next investigated the bypass fidelity of these mutant forms of human pol η during synthesis past two common lesions: a cis-syn cyclobutane thymine-thymine dimer and 7,8-dihydro-8-oxo-guanine. The assay used employs a partially duplex oligonucleotide with sequence that matches a part of the LacZaopen reading frame but containing an amber stop codon, within which the lesion is located. After recovery of the synthesized strand, the newly copied DNA is annealed to gapped M13mp18 DNA and transfected into E. coli. Inaccurate bypass of either lesion generates a sequence that gives a dark blue M13 plaque phenotype, while accurate bypass causes a readily distinguishable light blue plaque [54, 55]. The frequency of dark blue plaques is therefore an indication of lesion bypass fidelity and detailed spectrum information is obtained from sequencing of DNA from mutant plaques. As can be seen in Table 1, of the mutants tested, there were few drastic differences in the overall fidelity of either undamaged or damaged DNA. With the exceptions of Y52E for all three templates and Q38A for the T-T CPD containing template, differences in dark blue plaque frequency were less than 3-fold compared to wild type. This indicates that overall none of these changes cause a major change in the fidelity of pol η. The Y52E mutant has previously been shown to have moderately higher fidelity than the wild type enzyme during synthesis when copying undamaged DNA [57] but here we show this extends to two different damaged templates as well. This is the first report of this mutant for bypass fidelity opposite 8-oxoG, where we observed an almost 10-fold drop in the dark blue plaque frequency compared to wild type protein (29% for wild type and 3.2% for Y52E). This frequency is very similar to that given by the yeast pol η enzyme in this same assay [5].
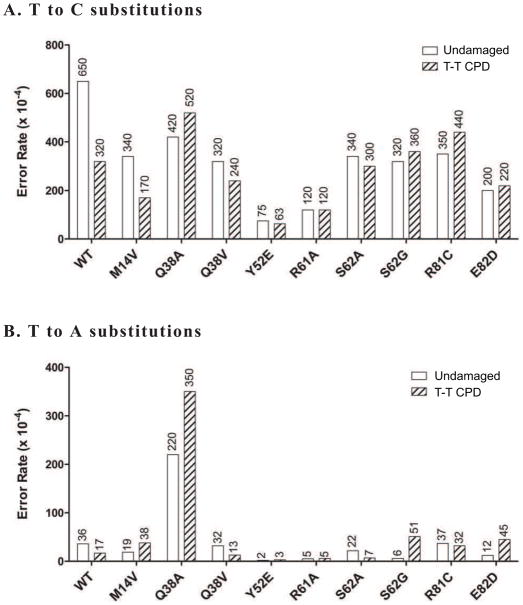
A more thorough analysis of the bypass fidelity is achieved by sequencing of mutant plaque DNA. This allows for a detailed analysis of the spectrum of changes that gives insight into exactly what bases are being inserted during bypass. From this analysis (Figures 4 and 5), we see that the increase (2.2% to 7.0% dark blue plaque frequency) by the Q38A form is caused almost solely by an increase in T misinsertions at the 3′T, causing the T→A error rate to increase ~20 fold compared to wild type (17 versus 350 × 10−4, WT and Q38A, respectively) (Figure 4B). The T→C error rates are the same as wild type (320 versus 520 × 10−4 for WT and Q38A, respectively) (Figure 4A). This same error occurs with increased frequency when copying undamaged DNA as well. None of the other mutants gave such a drastic increase in error rate during T-T CPD bypass. Both the Y52E and R61A mutants show more modest decreases in error rate during T-T CPD bypass for both T→A and T→C changes.
Figure 4. Error rate when copying a cis-syn cyclobutane thymine-thymine dimer by pol η mutants.
Values given are calculated as described in the Materials and Methods and represent errors per 10,000 bases copied. Bypass fidelity reactions used a 2:1 substrate:polymerase ratio and were incubated at 37 °C for 30 minutes. Values come from sequencing (for each form) 83–135 dark blue plaques for undamaged DNA and 38–52 dark blue plaques for T-T CPD reactions, with each plaque representing a unique bypass event. A. Values for T→C errors (dGTP misinsertion) opposite the 3′T of either undamaged (white bars) or damaged (hatched bars) template DNA. B. Values for T→A errors (dTTP misinsertion) opposite the 3′T of either undamaged (white bars) or damaged (hatched bars) template DNA. In both panels the values for wild type pol η are consistent with values generated using full length pol η in the same assay [6].
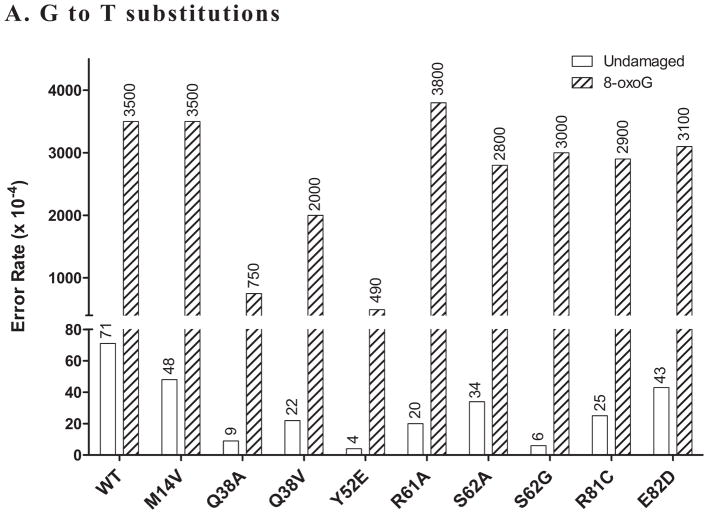
Figure 5. Error rate when copying an 8-oxoG lesion by pol η mutants.
Values given are calculated as described in the Materials and Methods and represent errors per 10,000 bases copied. Bypass fidelity reactions used a 2:1 substrate:polymerase ratio and were incubated at 37 °C for 30 minutes. Values come from sequencing (for each form) 83–135 dark blue plaques for undamaged DNA and 14–48 dark blue plaques for 8-oxoG reactions, with each plaque representing a unique bypass event. The undamaged samples are the same as those described in Figure 4. A. Values for G→T errors (dATP misinsertion) opposite the G/8-oxoG base of either undamaged (white bars) or damaged (hatched bars) template DNA. The Y-axis is broken to allow plotting of both undamaged and damaged rates, despite orders of magnitude difference. Only the G→T change is shown as it is by far the most frequent error when copying 8-oxoG (data not shown and [5]). The values for wild type pol η are consistent with values generated using full length pol η in the same assay [5].
When copying template G bases, misinsertion of dATP causes a G→T mutation. Even though this specific error is much less frequent than the most common pol η error, T→C (71 vs 650 × 10−4, G→T and T→C, respectively for WT protein), the Q38A (9 × 10−4), Y52E (4 × 10−4), R61A (20 × 10−4), and S62G (6 × 10−4) mutants all show a decrease in the error rate of at least 3-fold compared to wild type (Figure 5). However, of these only the Y52E (490 × 10−4) and Q38A (750 × 10−4) proteins also show a decrease in error rate when copying 8-oxoG (wild type rate of 3500 × 10−4; 7.1 and 4.6 fold decreases, respectively). This supports the idea that Y52E is in general a higher fidelity enzyme [57] while suggesting that the R61A and S62G changes have more limited effects. It is remarkable that Q38A displays better fidelity than wild type when copying both undamaged G and 8-oxoG, because of the lower fidelity it displays when copying either undamaged or damaged template T. To our knowledge, this is the first report of a single amino acid change in pol η (or any polymerase) that causes a lower fidelity phenotype when copying one lesion (T-T CPD) and a higher fidelity phenotype when copying another lesion (8-oxoG). It is also interesting that the Q38V protein displays neither of these attributes, being essentially like WT for both T-T CPD and 8-oxoG bypass. It should be noted here that none of the SNP’s tested (M14V, R81C and E82D) displayed any difference compared to wild type for lesion bypass fidelity of either lesion.
3.5 Forward Gap Fidelity
To further investigate the changes in fidelity we observed, a more robust forward fidelity assay was performed. In this assay, a gap of 407 bases is copied by the purified polymerase, allowing determination of a diverse group of changes including all 12 base:base mismatches, insertions, deletions, and other complex mutations [53]. These experiments confirmed and extended the results of the lesion bypass assay (Table 2). The overall single base substitution error rate, the most common of all errors detected, was the lowest for Y52E and R61A forms (~3 fold reduction compared to WT), similar to the lesion bypass assay. We use a 3 fold change as the minimum required to be considered different. Most samples were less drastically affected and displayed fold changes between 0.5 and 1.1 compared to wild type. That is, not significantly different when all possible errors are grouped together. The spectrum of errors from the various forms did, however, generate some interesting results.
Table 2.
Base substitution and insertion/deletion error rates of pol η mutants copying undamaged DNA in a gapped plasmid
| Mutant | Error rate (× 10−4) * | ||||
|---|---|---|---|---|---|
| Single base substitution | Single base deletion | Single base insertion | Complex** | Tandem base substitution | |
| WT | 230 | 25 | 3 | 35 | 13 |
| M14V | 170 (0.7) | 9 (0.4) | 5 (1.8) | 22 (0.6) | 9 (0.7) |
| Q38A | 260 (1.1) | 18 (0.7) | 16 (5.3) | 100 (2.8) | 43 (3.3) |
| Q38V | 220 (1.0) | 12 (0.5) | 9 (3.1) | 46 (1.3) | 10 (0.8) |
| Y52E | 73 (0.3) | 10 (0.4) | ≤1 (0.5) | 3 (0.1) | 1 (0.1) |
| R61A | 74 (0.3) | 7 (0.3) | 4 (1.4) | 7 (0.2) | 1 (0.1) |
| S62A | 170 (0.7) | 11 (0.4) | 5 (1.8) | 16 (0.5) | 8 (0.6) |
| S62G | 170 (0.7) | 14 (0.6) | 3 (1.0) | 20 (0.6) | 8 (0.6) |
| R81C | 180 (0.8) | 21 (0.9) | 7 (2.3) | 36 (1.0) | 9 (0.7) |
| E82D | 120 (0.5) | 13 (0.5) | 8 (2.6) | 20 (0.6) | 5 (0.4) |
Values given correspond to errors per 10,000 bases copied.
Complex errors are defined as multiple changes occurring within 3 bases, such as substitution-deletions and substitution-insertions. The tandem base substitutions error rates shown are a subset of the Complex errors reported. Values in parentheses for mutants are relative to WT rate.
The Q38A protein was more likely to give single base insertions (5.3 fold increase) and complex errors, including tandem base substitutions (3.3 fold increase). This did not extend to single base deletions however. In fact, all of the mutant proteins were less likely to generate single base deletions compared to wild type (0.3 to 0.9 fold decrease), while all but the Y52E protein seemed more likely to generate single base insertions (up to 5.3 fold increase). Again, these changes are relatively small. Interestingly, we never observed a single base insertion in mutant plaques from the Y52E form (28 samples, 11,396 bases analyzed). This is consistent with the larger decrease in complex mutations observed by this form (0.1 fold compared to WT). Further breaking down the single base substitution error rates (Supplementary Table 2) shows that each of the different mutant forms of pol η has a distinct means of achieving the overall fidelity it does. For example, the higher fidelity Y52E form displays reduced error rates at template purines and only modest changes (if any) at template pyrimidines. This is also the case for the R61A protein. Interestingly, the Q38A protein displayed an increase in error rate for two specific changes: C→A and T→A, both of which are caused by dTTP misinsertion (10.4 and 8.2 fold increases compared to WT, respectively). The latter is the same error the Q38A protein made in the lesion bypass reversion assay.
4. Discussion
We have generated multiple single amino acid substitution mutants of human DNA polymerase η in and around the active site in an attempt to better understand the molecular mechanism by which the unique properties of this polymerase are achieved. Specifically, we are interested in the high efficiency but moderate-to-low fidelity bypass of two common lesions, 8-oxoG and a T-T CPD. By studying both properties with these mutant forms of the polymerase we were able to identify several amino acid residues that affect the overall function of the polymerase and some that have very specifics effects on fidelity. We used a truncated version of pol η (encoding the N-terminal 511 amino acids of the enzyme) as a model for the polymerase catalytic activity. This form of the protein can be generated in highly pure form and in relatively large quantities using an E. coli expression system and is amenable to creation of site specific mutants. While lacking the C-terminal regions of the polymerase required in vivo for ubiquitination and other critical protein-protein interactions [1, 2], an even shorter form of pol η has been used to describe the crystal structure of the catalytic residues and in that report they argue the truncated form is a suitable model of polymerase catalytic activity [36]. We also provide evidence for this, as the wild type of our truncated protein displays very similar biochemical properties to the full length enzyme for both lesion bypass efficiency and fidelity. To our knowledge, the regulatory and protein interaction domains of the full length protein have not been shown to alter the nucleotide incorporation properties of the enzyme. Indeed, using yeast proteins we have previously shown that the fidelity and efficiency of both T-T CPD and 8-oxoG are unchanged by the presence of replication accessory proteins [5, 27].
One hallmark of pol η is the ability to preferentially copy damaged DNA over undamaged DNA. We feel this property is of critical importance and at times overlooked in its role in suppressing mutagenesis during TLS. Of the amino acid residues investigated, there was not a correlation between lesion bypass efficiency and fidelity. One change (Y52E) caused an increase in overall polymerase fidelity that was accompanied by a decrease in lesion bypass efficiency opposite both 8-oxoG and a T-T CPD, although the observed efficiency is still greater than that of the replicative polymerases [5, 18]. One potential consequence of this observation would be an increased chance of polymerase stalling opposite a lesion during bypass, leading to an increased chance of polymerase switching or extrinsic proofreading. But another form (Q38A) also displayed decreased bypass efficiency of both lesions that was accompanied by a decrease in fidelity opposite one of them (T-T CPD) and an increase in fidelity opposite the other (8-oxoG). Other changes (R61A and S62A) caused varying degrees of fidelity changes when copying either undamaged or damaged DNA but displayed little to no changes in bypass efficiency. From these results we conclude that the bypass efficiency of pol η opposite these two lesions is not entirely dependent on what nucleotide is inserted during the synthesis step. There also did not seem to be a correlation between the overall activity of the enzyme and either fidelity or bypass efficiency. Of note is the fact that none of the three SNP’s tested (M14V, R81C and E82D) had any major effect on the polymerase properties. While this is a negative finding, it is important to keep in mind this applies only the outcomes described here. It is possible these SNPs affect the protein in other ways that are undetectable in these assays. Analyzing the in vivo effects of these SNPs as well as selected other mutants might allow for a more thorough understanding of the connection between bypass efficiency, fidelity and cell fate after DNA damage has occurred.
We found that one of our mutants, R55A, was completely devoid of DNA polymerase activity. This arginine is conserved in Y-family enzymes in archaebacteria, eubacteria, yeast, plants, flies, worms, mice and in multiple human Y-family enzymes [52, 58]. This supports the notion it is a critical residue for polymerase function. Based on the recent crystal structures of human pol η [36] and modeling of earlier yeast pol η structures [57], it appears the positively charged arginine interacts with the negatively charged phosphate of the incoming nucleotide, stabilizing it in the active site. Replacing this residue with an uncharged alanine would be expected therefore to affect polymerization. Evidence for this is contained in a previously reported random mutation screen of human pol η that selected for still active protein [58]. In that experiment, only a single mutant was recovered that contained a mutation at this residue (from arginine to lysine). Presumably this mutant was still active because the change did not remove the positive charge. It will be interesting to assay the properties of a R55L form of pol η as well as other Y-family proteins to test this idea.
The finding that Y52E displays better than wild type fidelity confirms and expands previous work [57]. This residue is also involved in coordinating the incoming nucleotide, so changing from tyrosine to glutamic acid is predicted to destabilize the incoming nucleotide. A mismatched nucleotide requires even more stabilization so it is not surprising that Y52E displays higher fidelity. Our data is the first to demonstrate this quantitatively for lesion bypass as well (Figures 4 and 5) and supports the idea that pol η copies damaged and undamaged DNA in the same way. These results suggest that the Y52E form of pol η is a general antimutator, copying both damaged and undamaged substrates with higher fidelity. The R61A change has recently been shown (using steady-state kinetics of single nucleotide insertion) to reduce overall polymerase activity as well as misinsertion of dGTP opposite undamaged template T [36]. We see the same effect for both undamaged and the 3′T of a T-T CPD, and also show that it has decreased (relative to wild type) misinsertion of dCTP opposite both undamaged and damaged T’s (Figure 4). In fact, we see relatively large (at least 5 fold) decreases in the misinsertion of at least one dNTP opposite all four template bases when the R61A protein copies large stretches of undamaged DNA (Supplementary Table 2). This arginine residue is conserved in pol η from multiple species [58]. We do not see a similar increase in fidelity when this mutant copies 8-oxoG (Figure 5). Human pol η has yet to be crystallized in complex with 8-oxoG, but it is suspected that the misincorporation of dATP opposite this lesion occurs via Hoogsteen pairing as has been observed for T7 DNA pol and pol β [21, 63]. We note here that since R61 normally serves to prevent Hoogsteen pairing [36], our results suggest that perhaps the mechanism of pol η copying past 8-oxoG is in fact different than other higher fidelity polymerases. This idea has also been put forth for yeast pol η, which has been crystallized bound to 8-oxoG templates [44]. Of interest here is the observation that in yeast pol η, when a dATP residue is modeled opposite 8-oxoG, there are no steric conflicts, but that in order to be in ideal position for catalysis, the dATP residue would need to “move slightly inward’” [44]. In yeast pol η, R73 (equivalent to R61 in human) sits on top of the incoming dCTP residue during 8-oxoG bypass. The adjacent M74 (yeast) amino acid is larger than the adjacent S62 (human). It’s possible that the smaller residue allows more room for the dATP to move inward, explaining the large increase in dATP insertion by human pol η opposite 8-oxoG compared to the yeast enzyme [5, 7, 28, 29]. Changing R61 to alanine or S62 to alanine or glycine, as we have done here, would allow even more room and hence explain why no increase in fidelity was observed with these mutants.
Perhaps the most interesting finding was the fact that the Q38A mutant displayed both increased and decreased fidelity, dependent on the template being copied. While it displayed an increase in fidelity compared to wild type polymerase opposite both G and 8-oxoG (8 and 5 fold, respectively), its fidelity when copying the undamaged or T-T CPD sequence was the same as wild type for dGTP misinsertion and worse for dTTP misinsertion (6 and 20 fold lower fidelity for undamaged TT and T-T CPD, respectively). The homologous Q55 in yeast pol η stabilizes 8-oxoG in the active site, perhaps explaining the observation that of all the mutants we tested, Q38A displayed the lowest bypass efficiency for 8-oxoG. This same form of pol η was recently shown to display increased stalling when bypassing a T-T CPD [36] and we see a similar result here (Figure 3B). We suggest that this may be due to the increased misinsertion of dTTP during bypass (Figure 4B). The data presented here shows it also has decreased fidelity, at least for a T→A change. These results indicate that the link between bypass efficiency and fidelity is complicated and possibly lesion specific. The Q38A and R61A results suggest that the ability to bypass either 8-oxoG or a T-T CPD may occur by different molecular interactions between the protein and DNA. This could explain the extraordinarily low fidelity of wild type pol η for 8-oxoG bypass and may mean that other factors (e.g. accessory proteins, extrinsic proofreading) play a larger role in determining the mutagenicity of TLS on this lesion after bypass. An in vivo analysis of selected mutants is needed to determine if similar changes in the mutation rates in cells after genotoxic exposure occurs in a manner consistent with these in vitro experiments.
Supplementary Material
Highlights.
We analyze the properties of several polymerase η mutants.
We find several amino acid residues critical for fidelity.
Three identified SNP’s have no detectable affect on polymerase function
Lesion bypass efficiency and fidelity are not closely linked
Acknowledgments
We thank Dr. Robert Smart and Dr. Thomas Kunkel for helpful conversations and materials. We thank the reviewers for numerous comments and helpful suggestions to increase the quality of this manuscript.
Funding information: This work was supported by the National Institutes of Health [R01 ES016942 to S.M.] and [T32 ES007046]. Partial support of S.T. was by the College of Agriculture and Life Sciences at North Carolina State University. Funding for open access charge: National Institutes of Health.
Abbreviations
- pol η
DNA polymerase η
- CPD
cis-syn cyclobutane pyrimidine dimers
- 8-oxoG
7,8-dihydro-8-oxo-guanine
- T-T CPD
thymine-thymine cyclobutane pyrimidine dimer
- TLS
translesion synthesis
- SNP
single nucleotide polymorphisms
Footnotes
Conflict of interest statement: The authors declare no conflict of interest.
Supplementary data: Supplementary data are available at Mutation Research online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cellular and Molecular Life Sciences. 2009;66:2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nature reviews. Molecular cell biology. 2012;13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maher VM, Ouellette LM, Curren RD, McCormick JJ. Caffeine enhancement of the cytotoxic and mutagenic effect of ultraviolet irradiation in a xeroderma pigmentosum variant strain of human cells. Biochem Biophys Res Commun. 1976;71:228–234. doi: 10.1016/0006-291x(76)90272-2. [DOI] [PubMed] [Google Scholar]
- 4.Maher VM, Ouellette LM, Curren RD, McCormick JJ. Frequency of ultraviolet light-induced mutations is higher in xeroderma pigmentosum variant cells than in normal human cells. Nature. 1976;261:593–595. doi: 10.1038/261593a0. [DOI] [PubMed] [Google Scholar]
- 5.McCulloch SD, Kokoska RJ, Garg P, Burgers PM, Kunkel TA. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases δ and η. Nucleic Acids Res. 2009;37:2830–2840. doi: 10.1093/nar/gkp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cis-syn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Yuan F, Wu X, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Error-prone lesion bypass by human DNA polymerase η. Nucleic Acids Res. 2000;28:4717–4724. doi: 10.1093/nar/28.23.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 9.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 10.McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol η. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 12.Washington MT, Johnson RE, Prakash S, Prakash L. Fidelity and processivity of Saccharomyces cerevisiae DNA polymerase η. J Biol Chem. 1999;274:36835–36838. doi: 10.1074/jbc.274.52.36835. [DOI] [PubMed] [Google Scholar]
- 13.Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase η. J Biol Chem. 2000;275:7447–7450. doi: 10.1074/jbc.275.11.7447. [DOI] [PubMed] [Google Scholar]
- 14.Washington MT, Johnson RE, Prakash S, Prakash L. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc Natl Acad Sci USA. 2000;97:3094–3099. doi: 10.1073/pnas.050491997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-η. Nature. 2000;404:1011–1013. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda T, Bebenek K, Masutani C, Rogozin IB, Hanaoka F, Kunkel TA. Error rate and specificity of human and murine DNA polymerase η. J Mol Biol. 2001;312:335–346. doi: 10.1006/jmbi.2001.4937. [DOI] [PubMed] [Google Scholar]
- 17.Maga G, Villani G, Crespan E, Wimmer U, Ferrari E, Bertocci B, Hubscher U. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature. 2007;447:606–608. doi: 10.1038/nature05843. [DOI] [PubMed] [Google Scholar]
- 18.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 2004;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 2008;36:5660–5667. doi: 10.1093/nar/gkn555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Einolf HJ, Guengerich FP. Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase δ. Steady-state and pre-steady-state kinetic analysis. J Biol Chem. 2001;276:3764–3771. doi: 10.1074/jbc.M006696200. [DOI] [PubMed] [Google Scholar]
- 21.Brieba LG, Eichman BF, Kokoska RJ, Doublie S, Kunkel TA, Ellenberger T. Structural basis for the dual coding potential of 8-oxoguanosine by a high-fidelity DNA polymerase. EMBO J. 2004;23:3452–3461. doi: 10.1038/sj.emboj.7600354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibutani S, Bodepudi V, Johnson F, Grollman AP. Translesional synthesis on DNA templates containing 8-oxo-7,8-dihydrodeoxyadenosine. Biochemistry. 1993;32:4615–4621. doi: 10.1021/bi00068a019. [DOI] [PubMed] [Google Scholar]
- 23.Vaisman A, Woodgate R. Unique misinsertion specificity of pol τ may decrease the mutagenic potential of deaminated cytosines. EMBO J. 2001;20:6520–6529. doi: 10.1093/emboj/20.22.6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markkanen E, Castrec B, Villani G, Hubscher U. A switch between DNA polymerases delta and lambda promotes error-free bypass of 8-oxo-G lesions. Proc Natl Acad Sci U S A. 2012;109:20401–20406. doi: 10.1073/pnas.1211532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashiwagi S, Kuraoka I, Fujiwara Y, Hitomi K, Cheng QJ, Fuss JO, Shin DS, Masutani C, Tainer JA, Hanaoka F, Iwai S. Characterization of a Y-Family DNA Polymerase eta from the Eukaryotic Thermophile Alvinella pompejana. J Nucleic Acids. 2010;2010 doi: 10.4061/2010/701472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Washington MT, Johnson RE, Prakash L, Prakash S. Accuracy of lesion bypass by yeast and human DNA polymerase η. Proc Natl Acad Sci USA. 2001;98:8355–8360. doi: 10.1073/pnas.121007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCulloch SD, Wood A, Garg P, Burgers PM, Kunkel TA. Effects of accessory proteins on the bypass of a cis-syn thymine-thymine dimer by Saccharomyces cerevisiae DNA polymerase eta. Biochemistry. 2007;46:8888–8896. doi: 10.1021/bi700234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nat Genet. 2000;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- 29.Yuan F, Zhang Y, Rajpal DK, Wu X, Guo D, Wang M, Taylor JS, Wang Z. Specificity of DNA lesion bypass by the yeast DNA polymerase η. J Biol Chem. 2000;275:8233–8239. doi: 10.1074/jbc.275.11.8233. [DOI] [PubMed] [Google Scholar]
- 30.Lee DH, Pfeifer GP. Translesion synthesis of 7,8-dihydro-8-oxo-2′-deoxyguanosine by DNA polymerase η in vivo. Mutat Res. 2008;641:19–26. doi: 10.1016/j.mrfmmm.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 32.Uljon SN, Johnson RE, Edwards TA, Prakash S, Prakash L, Aggarwal AK. Crystal structure of the catalytic core of human DNA polymerase kappa. Structure. 2004;12:1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Ling H, Boudsocq F, Plosky BS, Woodgate R, Yang W. Replication of a cis-syn thymine dimer at atomic resolution. Nature. 2003;424:1083–1087. doi: 10.1038/nature01919. [DOI] [PubMed] [Google Scholar]
- 34.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 35.Ling H, Boudsocq F, Woodgate R, Yang W. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 36.Biertümpfel C, Zhao Y, Kondo Y, Ramón-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase η. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase η. Nature. 2010;465:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bassett E, Vaisman A, Havener JM, Masutani C, Hanaoka F, Chaney SG. Efficiency of extension of mismatched primer termini across from cisplatin and oxaliplatin adducts by human DNA polymerases β and η in vitro. Biochemistry. 2003;42:14197–14206. doi: 10.1021/bi035359p. [DOI] [PubMed] [Google Scholar]
- 39.Bassett E, Vaisman A, Tropea KA, McCall CM, Masutani C, Hanaoka F, Chaney SG. Frameshifts and deletions during in vitro translesion synthesis past Pt-DNA adducts by DNA polymerases β and η. DNA Repair (Amst) 2002;1:1003–1016. doi: 10.1016/s1568-7864(02)00150-7. [DOI] [PubMed] [Google Scholar]
- 40.Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J. 2000;19:3100–3109. doi: 10.1093/emboj/19.12.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaisman A, Masutani C, Hanaoka F, Chaney SG. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase η. Biochemistry. 2000;39:4575–4580. doi: 10.1021/bi000130k. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y, Biertümpfel C, Gregory MT, Hua Y-J, Hanaoka F, Yang W. Structural basis of human DNA polymerase η-mediated chemoresistance to cisplatin. Proc Natl Acad Sci USA. 2012;109:7269–7274. doi: 10.1073/pnas.1202681109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ummat A, Rechkoblit O, Jain R, Roy Choudhury J, Johnson RE, Silverstein TD, Buku A, Lone S, Prakash L, Prakash S, Aggarwal AK. Structural basis for cisplatin DNA damage tolerance by human polymerase η during cancer chemotherapy. Nat Struct Mol Biol. 2012;19:628–632. doi: 10.1038/nsmb.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silverstein TD, Jain R, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural Basis for Error-free Replication of Oxidatively Damaged DNA by Yeast DNA Polymerase η. Structure. 2010;18:1463–1470. doi: 10.1016/j.str.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase η error spectrum. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 46.Zeng X, Winter DB, Kasmer C, Kraemer KH, Lehmann AR, Gearhart PJ. DNA polymerase η is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 47.Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, Takeda S. Dual roles for DNA polymerase η in homologous DNA recombination and translesion DNA synthesis. Mol Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 48.Kamath-Loeb AS, Lan L, Nakajima S, Yasui A, Loeb LA. Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc Natl Acad Sci USA. 2007;104:10394–10399. doi: 10.1073/pnas.0702513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rey L, Sidorova JM, Puget N, Boudsocq F, Biard DS, Monnat RJ, Jr, Cazaux C, Hoffmann JS. Human DNA polymerase η is required for common fragile site stability during unperturbed DNA replication. Mol Cell Biol. 2009;29:3344–3354. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bétous R, Rey L, Wang G, Pillaire M-J, Puget N, Selves J, Biard DSF, Shin-ya K, Vasquez KM, Cazaux C, Hoffmann J-S. Role of TLS DNA polymerases η and κ in processing naturally occurring structured DNA in human cells. Mol Carcinog. 2009;48:369–378. doi: 10.1002/mc.20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boudsocq F, Ling H, Yang W, Woodgate R. Structure-based interpretation of missense mutations in Y-family DNA polymerases and their implications for polymerase function and lesion bypass. DNA Repair (Amst) 2002;1:343–358. doi: 10.1016/s1568-7864(02)00019-8. [DOI] [PubMed] [Google Scholar]
- 53.Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- 54.McCulloch SD, Kunkel TA. Measuring the fidelity of translesion DNA synthesis. Methods Enzymol. 2006;408:341–355. doi: 10.1016/S0076-6879(06)08021-9. [DOI] [PubMed] [Google Scholar]
- 55.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase η and Sulfolobus solfataricus Dpo4. J Biol Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 56.Glick E, Anderson JP, Loeb LA. In vitro production and screening of DNA polymerase η mutants for catalytic diversity. Biotechniques. 2002;33:1136–1142. 1144. doi: 10.2144/02335dd08. [DOI] [PubMed] [Google Scholar]
- 57.Glick E, Chau JS, Vigna KL, McCulloch SD, Adman ET, Kunkel TA, Loeb LA. Amino acid substitutions at conserved tyrosine 52 alter fidelity and bypass efficiency of human DNA polymerase η. J Biol Chem. 2003;278:19341–19346. doi: 10.1074/jbc.M300686200. [DOI] [PubMed] [Google Scholar]
- 58.Glick E, Vigna KL, Loeb LA. Mutations in human DNA polymerase η motif II alter bypass of DNA lesions. EMBO J. 2001;20:7303–7312. doi: 10.1093/emboj/20.24.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Niimi A, Limsirichaikul S, Yoshida S, Iwai S, Masutani C, Hanaoka F, Kool ET, Nishiyama Y, Suzuki M. Palm Mutants in DNA Polymerases α and η Alter DNA Replication Fidelity and Translesion Activity. Mol Cell Biol. 2004;24:2734–2746. doi: 10.1128/MCB.24.7.2734-2746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, Ohmori H, Kunkel TA. Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J Biol Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 61.Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol Cell. 2001;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- 62.McCulloch SD, Kunkel TA. Multiple solutions to inefficient lesion bypass by T7 DNA polymerase. DNA Repair (Amst) 2006;5:1373–1383. doi: 10.1016/j.dnarep.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH. Structure of DNA polymerase beta with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure. 2003;11:121–127. doi: 10.1016/s0969-2126(02)00930-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.