Abstract
Atomic force microscopy (AFM) is widely used in the biological sciences. Despite 25 years of technical developments, two popular modes of bioAFM, imaging and single molecule force spectroscopy, remain hindered by relatively poor force precision and stability. Recently, we achieved both sub-pN force precision and stability under biologically useful conditions (in liquid at room temperature). Importantly, this sub-pN level of performance is routinely accessible using a commercial cantilever on a commercial instrument. The two critical results are that (i) force precision and stability were limited by the gold coating on the cantilevers, and (ii) smaller yet stiffer cantilevers did not lead to better force precision on time scales longer than 25 ms. These new findings complement our previous work that addressed tip-sample stability. In this review, we detail the methods needed to achieve this sub-pN force stability and demonstrate improvements in force spectroscopy and imaging when using uncoated cantilevers. With this improved cantilever performance, the widespread use of nonspecific biomolecular attachments becomes a limiting factor in high-precision studies. Thus, we conclude by briefly reviewing site-specific covalent-immobilization protocols for linking a biomolecule to the substrate and to the AFM tip.
Keywords: Atomic force microscopy, scanning probe microscopy, single molecule force spectroscopy, drift, overstretching DNA, cantilever, imaging
1. Introduction
Drift, in position and force, is a long-standing problem that limits the application of atomic force microscopy (AFM) in biology [1, 2]. Poor long-term force stability hinders AFM-based single-molecule force spectroscopy experiments (SMFS), particularly those occurring over longer (>1 s) time frames. For example, state-of-the-art AFMs can be used to study the equilibrium folding and unfolding of proteins only over a few seconds [3], rather than the hundreds of seconds achieved with optical traps [4]. In addition, lack of force control hinders AFM imaging — the force set point during long scans often needs to be manually updated [1]. To quantify the scale of this problem, a force precision and stability of 5–10 pN is typical for commercial instruments, with custom instruments achieving 2 pN [3]. A related problem is the extended periods of time, often hours or even overnight [3], required for the AFM to “settle” after loading an AFM tip. Hence, routine and timely sub-picoNewton (pN) force precision and stability would accelerate a wide range of AFM-based biophysical assays, particularly if it could be achieved with commercial cantilevers on commercial instruments.
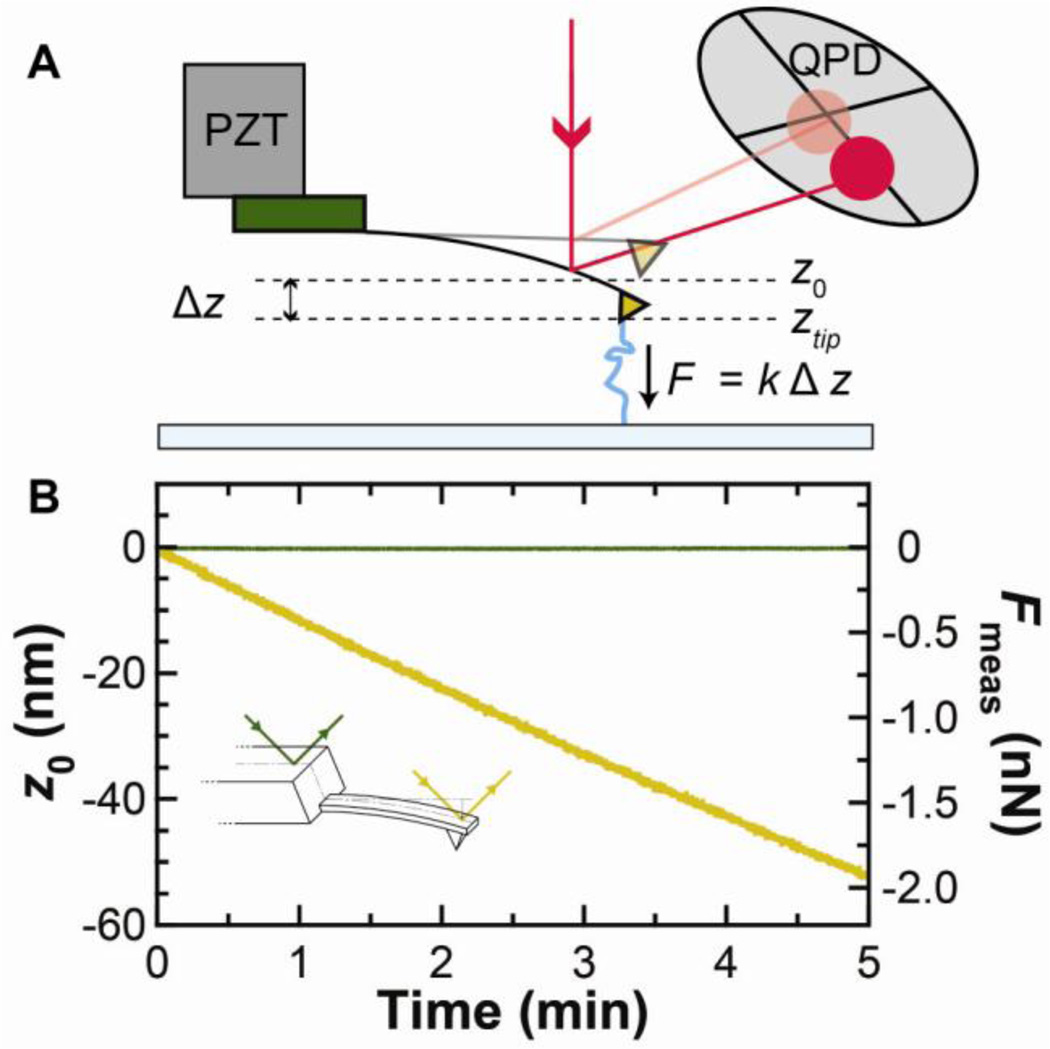
Recently, we found that the cantilever itself is the major source of force drift [5]. To understand this result, we need to review an underlying assumption in how force (F) is measured in AFM (Fig. 1A). It is assumed that changes in tip deflection (Δz) arise only from changes in the applied force. The force is then determined using F = −kΔz = −k(ztip−z0) where k is the cantilever stiffness and ztip the instantaneous deflection of the cantilever. Implicit in this assumption is that the zero-force position of the cantilever (z0) does not depend on time. Contrary to this expectation, a simple test showed that z0 is not constant, but drifts significantly for a popular class of soft silicon nitride cantilevers (BioLevers, Olympus) [5]. Specifically, the cantilever deflection laser measured an 800-fold higher drift rate when focused onto the cantilever than onto the base of the chip on which the cantilever was mounted (Fig. 1B, inset). This test unambiguously shows that the cantilever is the primary source of force drift (Fig. 1B), rather than external opto-mechanical sources (i.e., laser pointing noise). As we will discuss in detail below, the primary cause of this cantilever drift is its gold coating (Fig. 2).
Fig. 1.
Source of force drift in an atomic force microscope (AFM). (A) Schematic of a force spectroscopy experiment. The AFM tip is attached to a molecule and retracted from the surface. Force is determined by the deflection (ztip−z0), as measured on a quadrant photodiode (QPD). (B) With no molecule attached, the zero force position z0 (gold) of a cantilever (short BioLever) was measured as a function of time 2 h after wetting. A similar record (green) was measured after repositioning the detection laser onto the chip holding the cantilever. These measurements were scaled using the cantilever’s sensitivity (S = 0.043 V/nm) and stiffness (k = 37 pN/nm). This comparison demonstrates that the primary source of force drift is the cantilever. PZT: piezoelectric stage. This figure is reprinted from [5] with permission from the American Chemical Society.
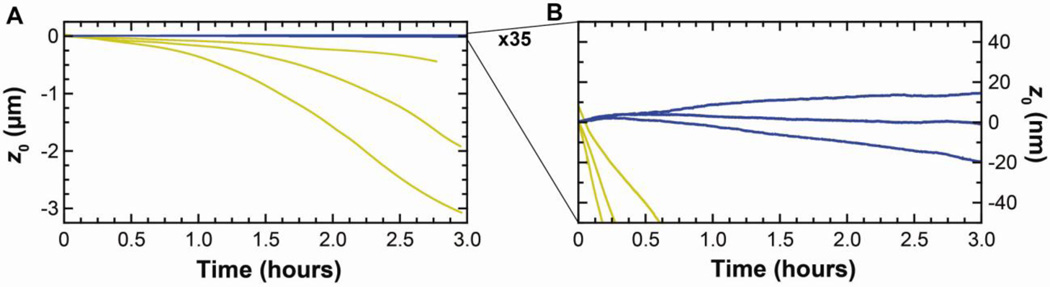
Fig. 2.
Drift in the zero force position (z0) on gold-coated (gold) versus uncoated (blue) BioLever Mini cantilevers. (A) Three-hour-long traces of z0 show that gold-coated BioLever Mini cantilevers drift significantly more than uncoated ones. (B) Scale expanded by a factor of 35 shows minimal residual drift on uncoated BioLever Mini cantilevers.
Gold coatings are added to cantilevers to enhance their reflectivity and are traditionally seen as critical to improving the signal-to-noise ratio in AFM experiments [6]. Coating a cantilever only on its back side leads to a substantial thermally induced force drift because of the bimetallic effect [7, 8]. Hence, many cantilevers, including the ones we studied, are coated on both sides to minimize such temperature-induced drift. Even with cantilevers coated on both sides, drift due to the gold coating has been previously reported [9, 10]. The novel result from our work is not that the gold is associated with drift [7–11]. Rather, the key insight arises from a pair of results that are contrary to the conventional wisdom in AFM: (i) removing a cantilever’s gold coating does not sacrifice the signal-to-noise ratio over relevant bandwidths (0.001–10,000 Hz) and (ii) smaller cantilevers do not always lead to better force precision. These results led to the unexpected insight that uncoated long BioLevers outperform uncoated BioLever Minis on time scales longer than 25 ms.
In this paper, we first discuss the current state of high-precision AFM, focusing on recent work on calmodulin that highlights the need for greater precision and stability in AFM. We then discuss our recent demonstration that removing the gold coating from cantilevers significantly increases the force precision and force stability [5]. We extend this result with related new results on the use of uncoated cantilevers in bio-imaging. With these advancements, the force stability during a typical AFM-based SMFS experiment is now limited by the use of nonspecific biomolecular attachments. We thus conclude by briefly reviewing site-specific covalent-immobilization protocols for linking biomolecules to the substrate and to the AFM tip.
1.1 Recent advances in sub-pN force resolution
The rates of folding and unfolding of biomolecules under constant force are sensitive to sub-pN changes in the applied load [12, 13]. Hence, both sub-pN force precision and stability are critical to studying macromolecular folding at equilibrium conditions. As a result, most AFM-based force spectroscopy experiments have focused on non-equilibrium stretching protocols where the molecule under study is stretched at a relatively high rate (50–5,000 nm/s). Such fast stretching protocols minimize the effect of positional and mechanical drift. If the whole experiment is over before there is significant drift, then the drift does not affect the results. In contrast, state-of-the-art optical traps can study equilibrium folding and unfolding because sub-pN force precision and stability are readily accessible. Clearly, the range of experiments that could be performed with AFM as well as their experimental precision would be enhanced if AFM could achieve a comparable level of instrumental performance.
Sub-pN force precision in a limited bandwidth has been observed during AFM-based SMFS experiments using lock-in amplification [14]. In this study, a 5-nm oscillation was applied to the tip at 20 Hz. These improvements allowed the folding pathway of an immunoglobulin to be more carefully examined. Overall, lock-in detection improves force precision in a specified bandwidth but can complicate interpretation.
A more general approach is to minimize drift and improve precision of the instrument. For example, the Rief group developed a custom-built, low-drift AFM. The stability of this instrument enabled them to investigate the conformational fluctuations of the calcium-sensing protein calmodulin at the single-molecule level [3] with high force precision (~2 pN) due, in part, to their extraordinarily slow pulling velocity (1 nm/s). At this stretching rate, they observed equilibrium hopping between two folding sub-states over 1–2 s. They went on to show how the kinetics of this hopping depended on the Ca2+ concentration. This example shows how a unique low-drift AFM facilitated partial reconstruction of calmodulin’s folding/unfolding kinetic pathway.
Two years later, the same group elucidated the full kinetic pathway for calmodulin folding and unfolding using an ultrastable dual-beam optical trap [4]. The motivation for changing measurement platform was better force stability and precision. The researchers were now able to achieve 300-s long records of folding and unfolding at a fixed extension (v = 0). These records revealed a complex folding pathway containing six states, including transiently populated folding intermediates. More generally, this paper highlights the ability of optical traps to reveal and quantitatively measure the full energy landscape of a single protein.
Given this success, it is reasonable to question whether AFM should continue to be a popular tool for SMFS. One critical advantage of AFM is that highly stable instruments are commercially available, broadening the range of users. Moreover, AFM is inherently a surface-based technique, allowing for a broader range of systems to be studied than in a dual-beam optical-trap assay. For example, the small surface area of an AFM tip facilitates extracting an individual membrane protein from a lipid bilayer for folding and unfolding studies [15, 16]. Furthermore, AFMs combine force spectroscopy with sub-nanometer resolution imaging [16]. On a more technical level, dual-beam traps require relatively long handles (~300–600 nm), while AFMs can use much shorter constructs (20–40 nm), dramatically decreasing compliance issues. A final advantage is AFM’s ability to access higher forces without significant radiation damage. Hence, there is a bright future for AFM-based force spectroscopy if its strengths could be combined with the force precision and stability of state-of-the-art optical traps.
Recently, we showed that removing the gold coating from cantilevers significantly improved the force precision and the force stability of AFM in liquid. Specifically, we achieved a 0.5-pN force precision over a broad bandwidth (0.01–10 Hz) [5]. Importantly, this precision is typical and timely. A majority (60%) of the cantilevers tested showed a 0.54 ± 0.02 pN (N = 15) integrated force noise in just 30 min after mounting. Moreover, the uncoated cantilever with the worst integrated force noise (1.6 pN) outperformed the quietest gold-coated cantilever (2.6 pN), when both cantilevers were allowed to settle for 3 h. This improvement in precision does not require a custom cantilever or a custom AFM. Rather, we achieved this state-of-the-art sensitivity using popular commercial cantilevers (BioLevers) on a commercial AFM (Cypher, Asylum Research). We emphasize that the measured bandwidth covers a 100-s time window, potentially allowing for equilibrium studies similar to what has been achieved in optical traps.
2. Sub-picoNewton force stability and precision for biological applications of AFM
2.1 Description of method and analysis
In this section, we first describe the process for removing the gold coating from commercial cantilevers (BioLevers). Next, we detail the acquisition and analysis of several hour-long drift traces. We then conclude by discussing sample preparation for DNA-stretching and bacteriorhodopsin-imaging experiments. Much of the following description has been adapted from our recent publication [5].
Although BioLever cantilevers without gold are not commercially distributed, it is straightforward to chemically remove the gold and chromium coatings. Gold was etched for ~30 s in either aqua regia (nitric acid and hydrochloric acid, 1:3 by volume) or in a commercial solution (Transene type TFA) at room temperature. The cantilevers were then rinsed in deionized water, and the chromium was subsequently etched using a commercial chromium etchant (Transene Type 1020) for another ~30 s at room temperature. The cantilevers were again rinsed in deionized water and dipped in isopropanol to prevent them from sticking to the chip. Finally, the chips were blotted dry onto filter paper. A detailed protocol is provided in supplementary material. For studies of MLCT cantilevers (Bruker) that use titanium rather than chromium underneath the gold layer, the titanium was etched using a commercial solution (Transene TFTN) at 75 °C for 60 s.
Since removing the gold coating dramatically decreased the sensitivity (V/nm) of the cantilever, we needed to compute performance metrics in the relevant units (nm and pN) rather than raw voltage data. As is standard in AFM, the sensitivity was determined by touching the mica substrate with the cantilever, and the spring constant was determined using the thermal noise method [17]. We then measured z0 for 3 h. These long records were automatically acquired using an algorithm implemented in Macrobuilder, part of the Asylum Research software (Macrobuilder script available upon request). Specifically, to keep the laser spot in the linear region of the detector, we touched the surface and re-zeroed the quadrant photodiode (QPD) every 5 min. Note that this periodic contact with the surface (i.e., acquisition of a force curve) caused discontinuities in the trace (Fig. 3A) (see below). All drift experiments were conducted with freshly cleaved mica in buffer (150 mM KCl, 10 mM Tris, pH 7.8). The duration of the experiment was limited by sample evaporation. Temperature of the AFM head was regulated to ± 0.2 °C using an automated control unit.
Fig. 3.
Data acquisition and analysis. (A) Representative raw drift trace of a gold-coated long BioLever. The discontinuities in the trace represent re-zeroing of the QPD every 5 min. (B) Continuous trace corresponding to the raw drift trace shown in (A) was generated after applying our simple splicing algorithm. This figure is extracted from [5] with permission from the American Chemical Society.
To analyze the resulting traces, we developed a simple splicing algorithm to create continuous traces (Fig. 3B). This algorithm (i) finds the discontinuities by differentiating the trace and looking for peaks, (ii) determines the value of the trace before and after the discontinuity by averaging 20 points on each side of it, (iii) takes the difference between these values, and (iv) shifts the portion of the trace after the discontinuity by this difference. Individual points, used as delimiters, were also removed. In an alternative “manual” splicing technique for particularly troublesome traces, the user placed cursors before and after the touchoff event. Then, the data between the cursors was removed, and the trace aligned as with the automatic algorithm.
Removing the gold coating decreased the cantilever’s reflectivity, so it was important to determine if this decreased sensitivity led to decreased force precision. To do so, we measured five consecutive 100-s records of z0 at 50 kHz bandwidth at the end of the aforementioned ~3-h record. The built-in software with our commercial AFM normally does not provide for accurate quantification of the positional power spectral density (PSD) over this broad bandwidth (0.01–25,000 Hz), due to high pass filters at ~1 kHz. However, these filters can be bypassed using the “cross-link panel.” Force PSDs were then calculated by scaling the positional PSD by the cantilever stiffness. We then integrated the area under the PSD and obtained the integrated force noise curve. As another metric of force noise, Allan variance σ was determined using the formula in Eq. 1 [18]
| (1) |
where τ is the time interval, and xi is the mean value of the data over the ith time interval. The Allan variance at a given time interval therefore represents the difference between the means of neighboring sections, averaged over all the sections.
We demonstrated the improved force precision of uncoated cantilevers by stretching DNA, a common biophysical assay [19, 20]. For these experiments, the DNA was adsorbed nonspecifically to the surface. Briefly, freshly cleaved mica was first pretreated with 40 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 6.8) and 5 mM NiCl2 buffer for 1 h, and then incubated with 100 ng/µL of an unlabeled double-stranded (ds) DNA construct (2082 or 5966 base pairs long) for 12–24 hrs inside a humidity chamber. Force curves were then acquired by pushing the cantilever onto the surface at a contact force of 120–300 pN for 1 s and retracting at 400 nm/s. This protocol is a modification of the protocol provided by Sophia Hohlbauch (Asylum Research) and reference [20].
Stability during imaging was demonstrated by imaging bacteriorhodopsin (BR). Patches of BR (~2 ng) were deposited onto a freshly cleaved mica surface in adsorption buffer (300 mM KCl, 10 mM Tris, pH 7.8) for 10 min. The mica was subsequently rinsed with imaging buffer (150 mM KCl, 10 mM Tris, pH 7.8). The BR was imaged in contact mode, a technique that can achieve molecular resolution images of membrane proteins [15, 16, 21].
2.2 Uncoated cantilevers drift less
As discussed in the introduction, the gold coating on the cantilever was the dominant cause of long-term drift in the zero force position (z0), even for cantilevers coated on both sides. More quantitatively, gold-coated cantilevers (long BioLever; k ≈ 6 pN/nm; 100-µm long) drifted, on average, 900 nm (rms, N = 10), while uncoated cantilevers drifted by only 70 nm (rms, N = 10) over 3 h [5]. Hence, removing the gold reduced the drift by more than a factor of 10. We also showed, but did not quantify, similar results for the short BioLever and BioLever Mini (k ≈ 60 pN/nm; 38 µm long). In this present paper, BioLever Mini cantilevers were explored in more depth, since they are commonly used in high-resolution imaging of biomolecules in liquid [22–24]. Interestingly, these stiffer cantilevers showed a >75-fold decrease in drift when their gold coating was removed (Fig. 2; 1030 vs 13.5 nm rms, N = 3 for each case). Additionally, these gold-coated BioLever Minis exhibit a drift profile that tends to bend downwards, indicative of the tip moving toward the surface. Moreover, even after 3 h, the drift rate was not tending towards zero. This data shows that simply waiting for the experiment to “settle” is an inefficient strategy for achieving high-precision results.
2.3 Loss in sensitivity does not affect force precision
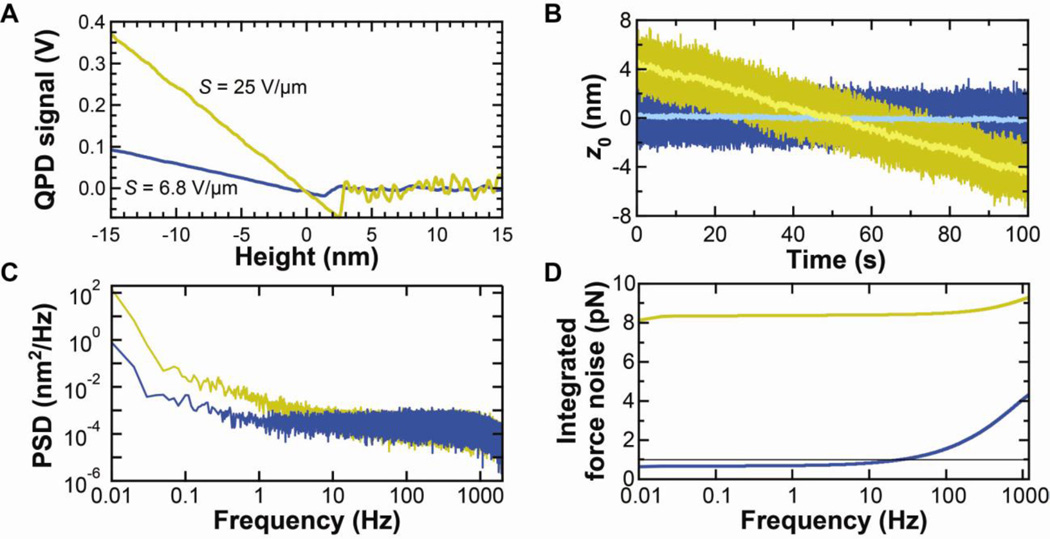
Traditionally, gold coating of AFM cantilevers is assumed to improve the signal-to-noise ratio by improving reflectivity. In contrast, our recent research shows that there is no sacrifice in positional or force precision for time scales longer than 0.1 ms when using the soft cantilevers (k = 6–100 pN/nm). As expected, uncoated cantilevers are less reflective than coated ones, and that decrease in reflectivity led to reduced optical-lever-arm sensitivity (V/nm), hereafter referred to as sensitivity (Fig. 4A). However, reflectivity or sensitivity is not the critical performance metric. The key question is: does this reduced sensitivity affect positional precision?
Fig. 4.
Effect of gold removal on sensitivity and precision. (A) Sensitivity of gold-coated (gold) and uncoated (blue) cantilevers. (B) Time records of zero-force position (z0) for gold-coated (k = 6.8 pN/nm) and uncoated (k = 7.1 pN/nm) cantilevers. High-bandwidth data (2.5 kHz) are shown in dark colors and smoothed data (10 Hz) in light colors. Note that, while the gold-coated cantilever has an approximately linear drift, there can be significant short-term fluctuations. (C) Averaged power spectral density of five consecutive 100-s records such as those shown in (B). (D) Integrated force noise from the records shown in (C). Data taken on the commercial AFM. This figure is reprinted from [5] with permission from the American Chemical Society.
Reduced light incident on the photodetector could lead to reduced positional precision at higher frequencies. We determined the positional precision of long BioLevers as a function of frequency by computing the power spectral density (PSD) of z0 from five consecutive 100-s long records. These records were taken 3 h after mounting. As expected, the gold-coated cantilevers showed significantly more drift than exhibited by the uncoated cantilevers (Fig. 4B). A comparison of the PSDs of these positional records revealed two interesting features (Fig. 4C). First, at lower frequencies, uncoated cantilevers outperformed coated cantilevers over three decades of bandwidth (0.01–10 Hz). Second, at higher frequencies, the positional PSD of the coated and uncoated cantilevers were the same, indicating that reduced reflectivity does not limit positional (or force) precision. The fact that a loss in sensitivity (signal) does not reduce the signal-to-noise ratio implies that the detection system is not the limiting factor, a result that may not generalize to older AFMs with older detection systems.
It also important to point out that these soft cantilevers do not have a resonance when near the surface. Rather, their motion is over damped (Q < 1) with a PSD similar to optical traps. The resulting Lorentzian-shaped PSD is flat up to a characteristic rolloff frequency (f0) and then decreases as 1/f2 (Fig. 4C, blue). The inverse of the rolloff frequency denotes the timescale at which statistically independent data can be collected [25].
2.4 Sub-pN force stability over extended periods
A common metric used in reporting the force precision in AFM is to specify the force spectral sensitivity (pN/√Hz) at a particular frequency. Another commonly reported metric is the force precision. For AFM, this metric is often calculated by integrating the thermal motion within a narrow bandwidth around the cantilever’s fundamental resonant peak. However, the folding and unfolding of biological molecules are sensitive to the total applied force. Thus, force stability over the full duration of the experiment is crucial. Otherwise, the dynamics of the system under study will be different between the start and end of the record. Two different metrics are useful in quantifying force stability: integrated force noise and Allan variance [18]. Both metrics investigate the force noise over a specified bandwidth or period. The integrated-force-noise analysis starts at a low frequency and calculates the noise over larger bandwidths incorporating higher frequencies (Fig. 4D). The Allan variance calculates the average force noise between pairs of successive points separated by a specified time (Fig. 5A). One advantage of this analysis over the integrated noise is that the Allan variance is not weighted by the low-frequency instrumental noise.
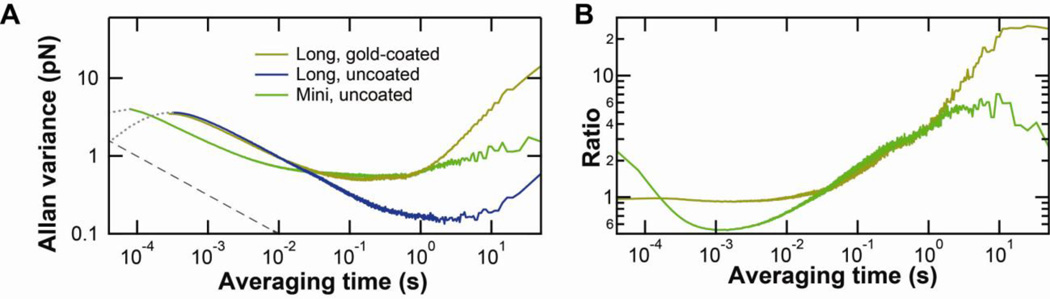
Fig. 5.
Allan variance analysis on soft AFM cantilevers. (A) Allan variance on BioLever Mini (green); long, gold-coated BioLever (gold); and long, uncoated BioLever (blue). The grey dashed line is a reference with a slope consistent with averaging Brownian motion. (B) Ratio of Allan variance of BioLever Mini (green) and long, gold-coated BioLever (gold) to that of the long, uncoated BioLever. At each frequency, the ratio represents how many times larger the Allan variance is for that cantilever relative to the uncoated long BioLever. At values less than 1, the cantilever in question outperforms the long, uncoated cantilever. We note that, at shorter time scales, there is a region in which the BioLever Mini outperforms the long BioLever by 50%. Yet, the long BioLever outperforms the Biolever Mini by factors of 4–7 on time scales longer than 1 s. Finally, at very shortest times, when the motion of the cantilever is correlated, the Allan variance yields misleading results on force precision and this region of the trace is deemphasized using a dotted gray line. The gold–coated long BioLever data, which is similar to the uncoated data, is not plotted in this region for clarity.
Using the integrated-force metric, we found that uncoated cantilevers achieved a sub-pN force stability over a broad bandwidth (Δf = 0.01–10 Hz) (Fig. 4D). This force stability was a ~10-fold improvement over that of gold-coated cantilevers. This difference is dominated by the drift in z0. So, the magnitude of the difference will depend on the duration of the experiment. We choose a 100-s period to quantify how well AFMs could perform extended studies of protein folding similar to state-of-the-art optical-trapping-based studies [4].
The Allan variance is useful for asking a different question [26]: to what degree can Brownian motion be averaged to trade off temporal resolution for improved force precision before instrumental noise starts to dominate? At the shortest times, the Allan variance initially increases suggesting worse performance at longer times. This is an artifact due to correlated motion of the cantilever on times scales shorter than 1/f0. As such, we have de-emphasized this portion of the record (Fig. 5A, grey dotted line). The peak in the Allan variance occurs at ~1/f0. On longer time scales, the Brownian motion is averaged, and the Allan variance decreases. At some point, instrumental noise starts to dominate and averaging over longer periods no longer improves force precision. To investigate this crossover point in AFM, we analyzed three different cantilevers, an uncoated BioLever Mini and both a gold-coated and an uncoated long BioLever. All three cantilevers showed the general trends outlined above.
Which cantilever yields the best force precision? The answer depends on the duration of the experiment. If there is no instrumental noise, then the best force precision would be achieved with shorter cantilevers with a reduced drag coefficient (β). This result is based on the fluctuation-dissipation theorem, which yields a force precision of where kBT is the thermal energy, and Δf is the bandwidth of the experiment. For times <0.01 s, the BioLever Mini shows better performance than the long BioLever, which is 2.6-fold longer and therefore has greater drag. However, on time scales longer than 0.01 s, the performance of the BioLever Mini degrades due to instrumental noise associated with cantilever detection. This limitation adds a fixed amount of positional noise so its effect on softer cantilevers is not as pronounced [5]. For experiments lasting 10 s, the uncoated long BioLever has a ~7-fold better performance than an uncoated BioLever Mini despite its longer length and ~20-fold better performance than a gold-coated long BioLever. To highlight this comparison, we plot these ratios of Allan variance over the full temporal range of the experiment (Fig. 5B). Interestingly, and somewhat surprisingly, the long BioLever outperformed the BioLever Mini on time scales longer than 25 ms. Thus, smaller cantilevers are not always better. The optimum cantilever depends on the individual experiment, in general, and the duration of the experiment, in particular.
Overall, integrated force and Allan variance analyses both demonstrate that uncoated cantilevers offer superior performance over gold-coated cantilevers for the cantilevers tested. After the gold coating is removed, the cantilever detection system is the most significant source of low-frequency instrumental noise. Our data on two different optical-lever-arm systems — a state-of-the-art commercial AFM and our custom-built ultrastable AFM [2] — showed essentially the same level of instrumental noise in the positional PSD (data not shown). One method to address this low-frequency noise, albeit at added complexity, is to use interferometric detection systems [27, 28].
2.5 Routine and timely sub-pN precision
Often a few selected traces or portions of traces are reported when describing stability. What is needed is both routine and timely access to state-of-the-art force stability. Such routine and timely access would improve the throughput of high-precision AFM-based biophysical measurements. And, it is exactly what removing the gold coating from the cantilevers achieved: a majority (9 out of 15) of the uncoated cantilevers that we tested had an integrated force noise below 1 pN (0.54 ± 0.02 pN) just 30 min after mounting [5]. On average, the uncoated cantilever had an integrated force noise more than 10 times quieter than the coated cantilevers at 3 h (0.8 vs 9.4 pN; N = 10). Moreover, the uncoated cantilever with the worst integrated force noise (1.6 pN) outperformed the quietest gold-coated cantilever (2.6 pN). Perhaps most importantly, this level of performance was achieved on a commercial AFM using a commercial cantilever. Thus, we expect the results presented here to immediately impact a wide range of AFM-based experiments in biophysics and nanotechnology.
2.6 Sub-pN precision and stability while stretching DNA
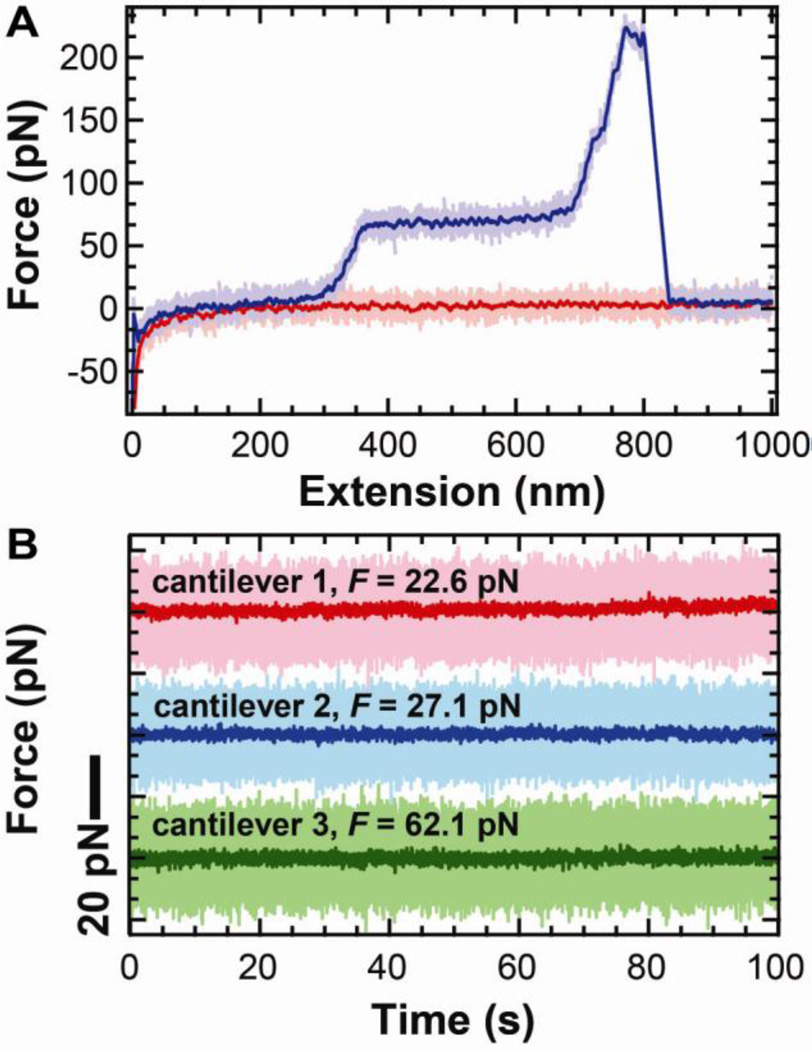
To demonstrate this improved instrumental performance for biophysics, we stretched DNA, a common single-molecule assay [19, 20, 29, 30]. The resulting force-extension curve (Fig. 6A) is very quiet by AFM standards and shows the canonical overstretching transition at ~65 pN [31, 32]. Further, we quantified the force precision by stretching a DNA molecule >30 nm from the surface and holding it at a constant extension for ~100 s. Analysis from three different cantilevers and DNA molecules shows that the force stability and precision were excellent, as demonstrated by full-bandwidth records smoothed to 10 Hz (effective data rate = 20 pts/s) (Fig. 6B). As will be discussed below (§3), the main limitation to achieving this level of performance is the non-specific attachment of the DNA to the tip and surface.
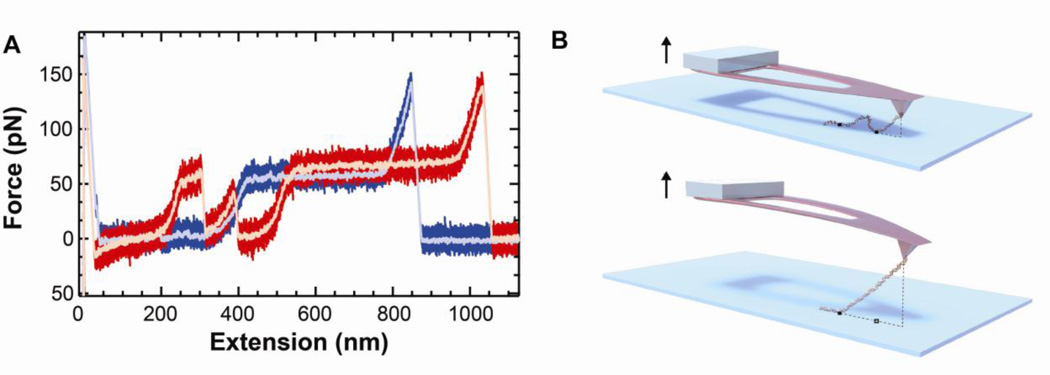
Fig. 6.
Stretching DNA with uncoated cantilevers exhibits sub-pN force stability. (A) Force-extension curve of DNA at a 400 nm/s pulling velocity. The high-bandwidth data (2.5 kHz) for approach (light pink) and retraction curves (light purple) were smoothed (100 Hz, purple and red). (b) Force-versus-time trace smoothed to 10 Hz while holding DNA at constant extension at 22.6 pN (pink), 27.1 pN (blue), and 62.1 pN (green). High-bandwidth data are shown in light colors with traces displaced for clarity. This figure is reprinted from [5] with permission from the American Chemical Society.
2.7 Force stability facilitates imaging
The connection between force stability and data quality in SMFS experiments is immediately clear; however, force stability is also critical for imaging applications. Molecules can change conformation depending on the force [15], and the imaging conditions can be degraded if the force drifts outside of an acceptable range. Although software-based techniques have been implemented [1], an AFM operator typically adjusts the force set point manually during imaging. Force drift is a more significant concern for contact mode imaging; AC modes of imaging are less susceptible to force drift.
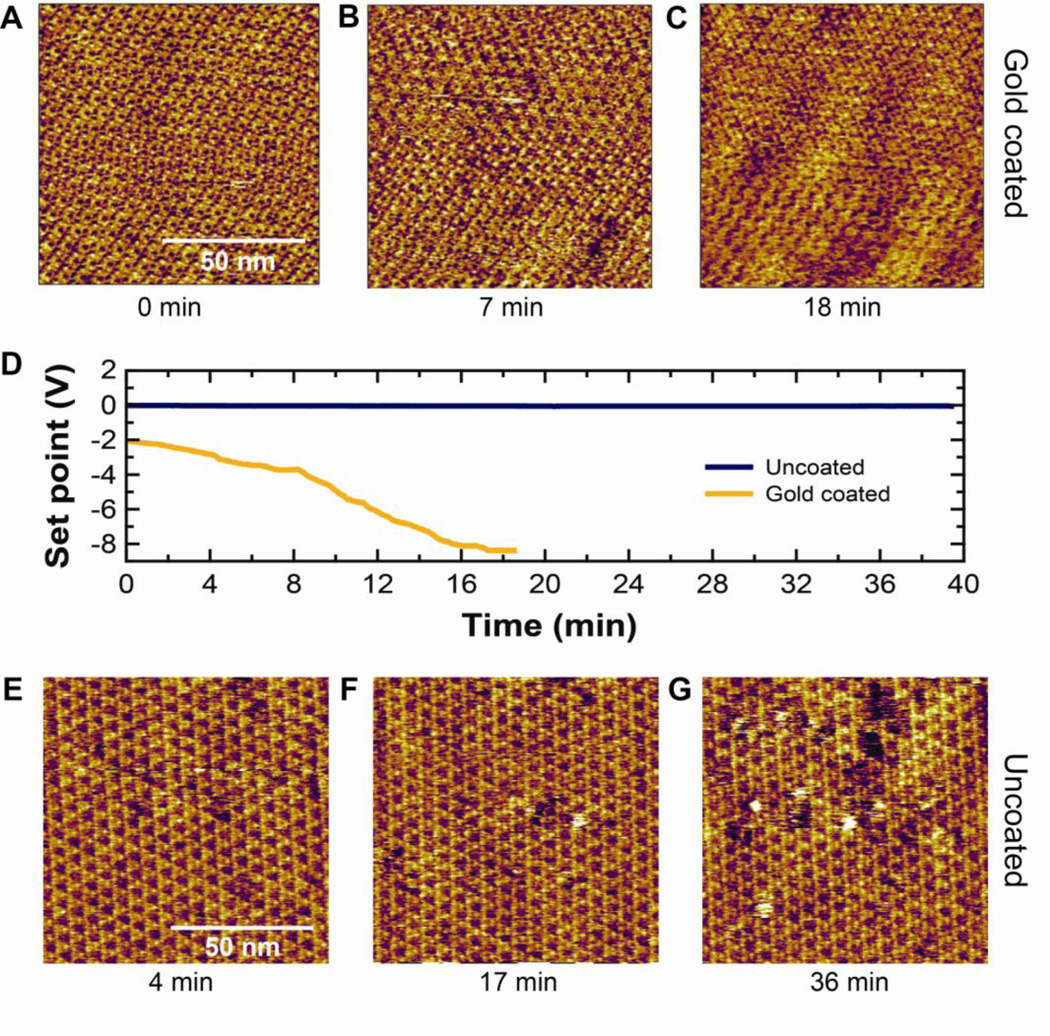
To demonstrate the utility of uncoated gold cantilevers for imaging with molecular resolution, we imaged bacteriorhodopsin, a model membrane protein that is widely studied by AFM. Images were acquired with the measured force being held constant by varying the position of the base of the cantilever (zPZT). If zPZT were changed to compensate for vertical mechanical drift, we would expect no change in actual applied force. However, if zPZT were to change to compensate for changes in z0, then the applied force to the BR would change. Our "constant force" imaging (Fig. 7A–C) using a gold-coated BioLever Mini demonstrates the problem of force drift in imaging. To maintain images of the BR lattice, we had to adjust the set point substantially during imaging (Fig. 7D) even though the system had “settled” or equilibrated for ~5 h. In contrast, when we imaged BR with an uncoated BioLever Mini (Fig. 7 E–G), the set point needed no such substantial change (Fig. 7D, blue) indicating that there was minimal force drift. We note that molecular-resolution images with uncoated cantilevers can be achieved ~30 min after mounting. Thus, the lack of force drift when using uncoated cantilevers facilitates taking molecular-resolution images. These improvements in instrumentation are synergistic with software-based force-drift compensation [1]. Together, they could make automated high-resolution AFM imaging more robust.
Fig. 7.
Time-lapse imaging of a 100 × 100 nm area (~4 Å/pixel) of a BR patch using a gold coated and uncoated BioLever Mini. (A–C) Images acquired at times 0, 7 and 18 min using a gold-coated BioLever Mini. (D) Plot showing the set point during imaging using gold-coated (gold) and uncoated (blue) BioLever Minis, respectively. (E–G) Images acquired at times 4, 17, and 36 min using an uncoated BioLever Mini.
2.8 Minimizing interference artifacts in the AFM deflection signal
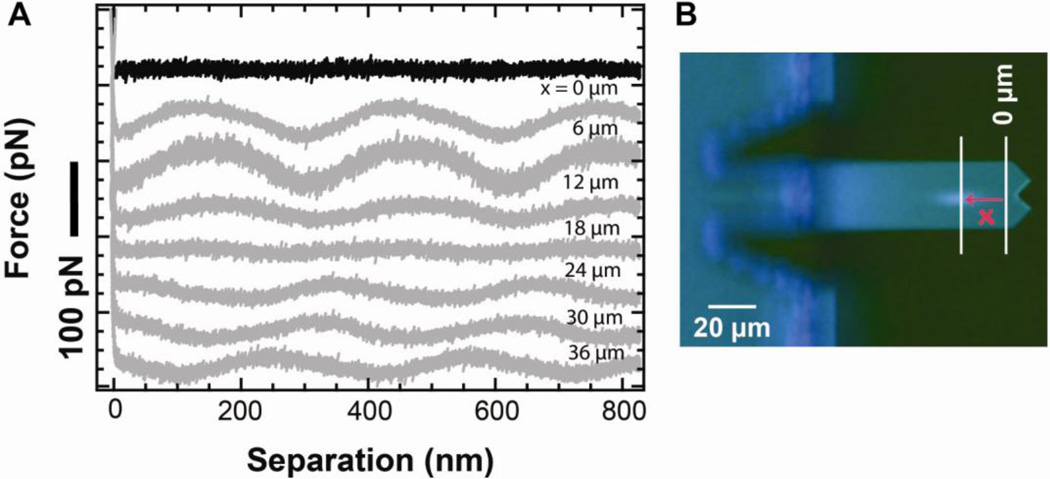
Force measurements in AFM originate from voltage measurements from a photodetector. Larger cantilever deflections lead to larger voltage changes. Touching the cantilever off the surface in a standard force-extension curve yields both the sensitivity of the optical lever arm system and a voltage offset associated with z0, the zero force position of the cantilever. It is common to assume that this voltage does not depend on height of the tip over the surface. We can make this assumption partly because researchers have made significant progress in reducing artifactual, height-dependent modulations in the deflection signal [33]. These modulations are caused, in part, by unwanted interference between light reflected from the sample surface and light reflected from the cantilever. With a modern commercial AFM, such fringes are absent in 95% of our force-extension records when using uncoated cantilevers on a mica surface (Fig. 8A). Mica is the most common surface for bioAFM, but gold-coated surfaces are also widely used in nanoscience [34, 35]. Such gold-coated surfaces are reflective and therefore contribute to unwanted fringes when using uncoated cantilevers [Fig. 8A, grey (x = 0)]. Interestingly, we found the magnitude of the fringes depended on the laser spot position along the long axis of the cantilever (Fig. 8B). Moreover, proper placement of the laser spot could essentially eliminate this unwanted effect. We note that the cantilever sensitivity did not change significantly with the laser-spot position in the range presented (Fig. S1); the measured stiffness did vary by 1–15%, but without a systematic trend as has been seen in earlier work [36].
Fig. 8.
Substrate composition and laser spot dependence of interference artifacts in the optical lever arm deflection signal. (A) Force-extension curves taken on mica (black) and on gold (grey) for different positions of the laser spot on an uncoated long BioLever. Only the approach portion of the force curve is shown for clarity. (B) Optical microscope image of a long uncoated BioLever showing the laser spot position and defining x as the distance of the center of the laser spot from the reference point (x = 0 µm).
2.9 Mechanism of gold-induced drift
In the preceding sections, we have demonstrated how removing gold improves AFM performance, but it is reasonable to ask the question: why does the gold coating cause cantilever drift? The most obvious answer is thermal drift; a residual bi-metallic effect remains despite the cantilever being coated on both sides. If that were the case, one would expect the drift rate to be correlated with temperature. However, we have previously shown that the drift rate does not depend on the temperature of the AFM head [5], contradicting this hypothesis. It is possible that the head temperature is enough different than that of the tip, several millimeters away, that a thermal effect is still present. A second possibility is that the optical-lever-arm laser affects the gold coating, causing it to crack or deform. However, we did not find that the drift rate depends on the laser power [5], so we do not believe the laser light is the primary cause. Another possibility is stress caused by the bending of the cantilevers upon wetting (Fig. 9A). These soft, 170-nm thick cantilevers typically fold completely flat against the chip due to the meniscus force. Damage from this process can be seen in a cantilever after it was unfolded using an alcohol dip (Fig. 9B). We speculate that this damage is different for a gold-coated than an uncoated cantilever and therefore causes gold-coated cantilevers to drift more. Finally, it has been shown that the viscoelasticity of gold causes noise at frequencies >1 Hz in air [10]. It is likely that this effect is also present in our data in fluid. This raises the interesting possibility that less viscoelastic metals might reduce the drift while remaining reflective. Whatever the mechanism(s) of gold-induced drift, the key result is that the gold coating can be removed without reducing the signal-to-noise ratio.
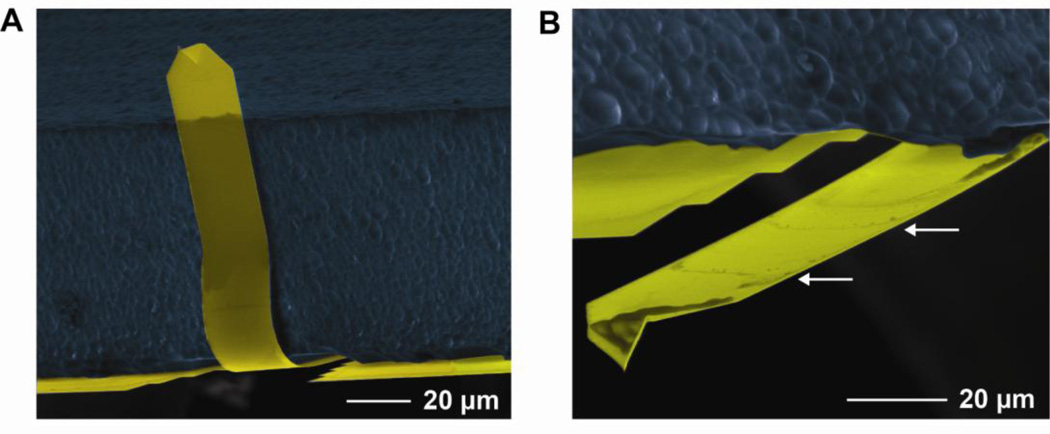
Fig. 9.
Soft cantilevers tend to buckle with wetting. (A) Scanning electron micrograph (SEM) of a long BioLever stuck to the chip after wetting. (B) SEM of the same individual cantilever after unfolding. Damage can be seen in the unfolded image (white arrows). Figure 9A is reprinted from [5] with permission from the American Chemical Society.
An interesting additional question is: do cantilevers that have been gold-coated on both sides drift less than cantilevers coated only on one side? To explore this question, we studied another soft cantilever commonly used in SMFS, the MLCT (k = 10 pN/nm; Bruker) [37–40]. This cantilever is coated only on one side and has a stiffness comparable to the long BioLever. The gold-coated MLCTs had approximately the same positional drift over 3 h as the long BioLevers [980 nm (N = 3) vs 900 nm (N = 10)]. This comparison suggests that temperature instability of the AFM head is not the primary cause of gold-induced drift, since the MLCT is coated only on one side, whereas the long BioLever is coated on both sides. We also confirmed that removing the gold on MLCT led to a reduced force drift. However, this reduction was not as large as that seen in the BioLevers (Fig. S2) and led to an average integrated force noise of 2.9 pN (Δf = 0.01–10 Hz; N = 3).
Under ideal conditions, one might expect cantilevers that have never been coated to perform better, since standard etching procedures are expected to leave a monolayer or two of the residual metal on top of the silicon nitride [41]. We note that MLCT-UC, described as uncoated in the product literature, have previously been coated with gold and titanium and have had these coatings removed. We suggest that users confirm that all metal has been removed both by visual inspection and an accompanying reduction in the reflected light off of the cantilever. More generally, we hope that the results presented here on the detrimental effects of gold coating of AFM cantilevers motivates the commercial distribution of cantilevers that have never been coated in gold.
3. Stability of biomolecular attachment
Is the force stability in our DNA records limited by the instrumental performance? At the moment, this is not the case for a majority of our records. The main limitation in our DNA pulling assay is the non-specific attachment technique used to couple the DNA to both the tip and the surface. A common feature in assays based on non-specific attachment is records containing drops in F (Fig. 10A, red) consistent with changes in the attachment pattern of the DNA to the tip and the surface (Fig. 10B). Such records are distinct from canonical DNA stretching curves (Fig. 10A, blue) and, therefore, records of this class are often excluded from analysis using objective criteria [16, 42]. Nonetheless, this physical adsorption (or physisorption) [10, 43] leads to two well-known issues that hinder precise AFM-based SMFS studies [3, 44–46]: (i) the site of the biomolecular attachment to the tip and the surface is not known, and (ii) the strength of these attachments is variable. The first issue complicates analysis because nominally identical molecules, such as the DNA shown in Fig. 10A, show different contour lengths. The latter issue of unpredictable bond strength limits long-duration records, precluding, for instance, studying protein folding over extended periods. The solution to these interrelated problems is a protocol that enables site-specific covalent attachment of the biomolecule to the substrate and to the AFM tip. Fortunately, there has been significant effort in the last decade to address exactly this issue, including a number of papers and reviews [47–50]. In this section, we discuss a few recent advances of covalent anchoring that have relevance for AFM-based SMFS.
Fig. 10.
DNA stretching experiment demonstrating one pitfall of non-specific attachment. (A) Two records of DNA elasticity. One record (blue) shows the canonical DNA elasticity, including the overstretching transition. The other curve (red) shows unwanted force ruptures caused by instability of the attachment point. The pink and blue curves are data smoothed to 100 Hz. (B) Schematic showing one likely cause for these artifacts. Because the molecule is non-specifically adsorbed, it can be attached to the surface at multiple points. At first, one attachment point is bearing the force, but it lets go, and the molecule is then being pulled from a second attachment point, changing the effective contour length of the molecule.
Site-specific labeling generally requires heterobifunctional crosslinkers. Such a bifunctional crosslinker provides one chemical mechanism to attach to the protein and another to anchor to glass, mica, or oxidized silicon (which is chemically similar to glass). Often these cross linkers are small molecules (§3.1) but increasingly protein-based cross-linkers (§3.2) are being used. A key advantage of protein-based crosslinkers is that they are genetically encoded into proteins, similar to green fluorescent proteins. However, in SFMS studies, these protein-based tags will also unfold, introducing additional folding events that could be conflated with events due to the system under study. Thus, there is no single best coupling protocol for all common biomolecules. Rather, users will need to balance advantages and disadvantages of each type of approach. Finally, we note that, in conjunction with attaching molecules to surfaces by a specific mechanism, one also has to eliminate physisorption of biomolecules to surfaces. Such suppression of non-specific sticking is typically achieved in single-molecule assays by coating the surface with poly(ethylene glycol)(PEG) [51].
3.1 Small molecule based coupling
To date, thiol-maleimide chemistry is one of the most commonly used conjugation techniques for anchoring biomolecules to surfaces [52]. The protein under study needs to contain sulfhydryl (free thiol) groups, which means that it should have one solvent-exposed cysteine for the anchoring to be site specific. To achieve such selectivity, it is not uncommon to make “cys-lite” proteins where all but one of the solvent exposed cysteines has been mutated away [53]. The power of this chemistry is that it is selective, precise, and stable. Typically, these cysteines are used to anchor proteins to gold-coated surfaces using a gold-thiol bond. Yet, for precision SMFS, we want to avoid such gold-based coupling. Hence the use of a gold-maleimide bond allows for bioconjugation to silanized glass, mica, and silicon-nitride surfaces. Thiol-based anchoring chemistry extends beyond proteins to nucleic acids. Commercial solid-state synthesis of DNA and RNA makes the introduction of a terminal thiol group easy and inexpensive. As a result, AFM-based SMFS on non-gold surfaces have been used for a wide variety of molecules, including DNA [29, 30], RNA [54], and recombinant proteins [46]. Moreover, Zimmerman, et. al., provides a detailed protocol for anchoring molecules to silanized surfaces, including uncoated silicon-nitride cantilevers [50]. The heterobifunctionality is provided by a poly(ethylene glycol) (PEG) crosslinker with an N-hydroxy succinimide (NHS) group on one end and a maleimide group on the other. The NHS group reacts with aminosilane-functionalized glass [3-aminopropyldimethylethoxysilane (APDMES)]. Low nonspecific binding and a large number of reactive sites for biomolecule coupling have been achieved using this protocol.
Another promising small-molecule immobilization approach that is compatible with thiolcontaining biological samples uses silatrane chemistry [49]. Lyubchenko and colleagues developed this chemistry to minimize the problem of non-specific adhesion at small distances, which is common with short linkers. Short linkers have a number of desirable benefits [55], but non-specific adhesion between the tip and the surface can mask the system under study. These researchers developed a nanoscale tetrahedral-shaped tripodal silatrane (T-silatrane) as a linker. T-silatrane consists of four arms, three of which enable immobilization of the construct on the surface, while its fourth arm, with a maleimide group, allows for covalent coupling with the cysteine of the peptide [38, 56]. They simplified the process by developing a one-step surface-preparation protocol, instead of the more common two-step protocol (silanization of the glass and then coupling a derivatized PEG to the functionalized glass). This chemistry has been used in dynamic force spectroscopy experiments of alpha-synuclein aggregation [38, 56], though it is not yet commercially available.
These coupling protocols have shown real advantages in molecular recognition force spectroscopy, a class of experiments in which one molecule is coupled to a surface and a second molecule to the tip. In this situation, the same thiol-based coupling chemistry can be used for each surface (tip and substrate). A more general need is to couple a single molecule between the tip and the surface. This general class of experiments requires a second orthogonal coupling chemistry. One excellent candidate to provide a fast and highly efficient reaction is click chemistry, a coupling between an azide and an alkyne [57]. Click chemistry is now being used in optical-trapping [58] and single-molecule fluorescence [59] assays. Thus, we expect that recent advances in protein labeling for related single-molecule assays can be leveraged to aid AFM-based experiments.
3.2 Protein-based tags
The primary advantage of protein-based tags, over small molecule crosslinkers, is that they can be genetically encoded. There are two general types of protein tags: small peptides and protein domains. Small peptides may anchor the protein either directly to a surface or bind to a partner protein. For instance, a small tag has been fused to RecBCD, a helicase, to create a genetically encoded biotin tag [60]. Biotin forms a strong non-covalent bond with streptavidin. The biotin-streptavidin bond is widely used in optical-trapping studies [25], and supports a force of ~25 pN for hundreds of seconds, sufficient for equilibrium studies of protein [4, 61] and riboswitch [62] folding. More recently, proteins with a small added peptide tag were efficiently converted to contain an aldehyde group [63]. The aldehyde can then react with a hydrazide-labeled reagent with near 100% efficiency [63]. Finally, small peptides can be used to site specifically anchor the target protein to surfaces through a very strong bond to a protein. A recent example is the SpyCatcher:SpyTag system that forms a covalent amide bond. The mean rupture force of this bond is 20 times greater than that of a streptavidin-biotin bond [64].
Instead of fusing a small peptide with the target protein, anchoring can also be achieved by fusing the protein of interest to another protein domain. These domains can then be used to anchor to a small ligand that is covalently attached to a PEG-coated surface. Two systems that have shown success in AFM are the SNAP and HALO tags [44, 65]. HALO tags are reported to have less nonspecific protein-substrate and tip-substrate interactions [65]. Reduced non-specific interactions should facilitate visualizing unfolding events occurring close to the surface. Additionally, optical-trapping studies applied ~10 pN over 100s of seconds using a HALO tag [66], suggesting that its structure may be stable enough to allow for equilibrium investigation of the folding and unfolding of some proteins without the HALO tag unfolding. Overall, the power of these protein-based anchoring techniques is that they provide orthogonal chemistries to cysteine-based anchoring.
4. Conclusion and outlook
Almost three decades since the invention of the AFM, technical improvements are still expanding the range of biological systems and assays that AFM can study. The most notable improvement is scan speed with the advent of video rate imaging [67]. On the other end of the speed spectrum are precision studies of the equilibrium folding and unfolding of proteins. To facilitate this research, our group has focused on improving the stability and precision of AFM [2, 5]. Initially, we focused on tip-sample stability, developing an optically stabilized AFM with a residual drift rate of 0.005 mn/min at ambient conditions [2]. Yet, for force spectroscopy applications, tip-sample stability was only half of the problem: force drift also occurred. Routine and timely access to sub-pN force resolution and stability was achieved by the simple removal of gold from a popular class of cantilevers [5]. The first key insight was that gold coating was not needed to make precise force measurements over the relevant bandwidths. Thus, removing the gold coating led to significantly enhanced force stability without any loss in force precision. This simple procedure only takes a ~1 min chemical etch and does not require custom cantilevers or a custom AFM. The second key result was that instrumental drift in the cantilever detection system degrades the performance of BioLever Mini on surprisingly short time scales (>0.01 s). This result led to uncoated long BioLevers outperforming uncoated BioLever Minis on time scales longer than 25 ms. Thus, smaller cantilevers do not always give lower force noise; the duration of the experiments is an important parameter to consider. We anticipate that these results can be applied to a wide range of AFM-based assays for biomolecular imaging and biophysics. Combining sub-pN AFM force spectroscopy with recently reported covalent, site-specific biomolecular immobilization techniques will pave the way to precise investigation of dynamical processes over hundreds to thousands of seconds, a new regime for AFM-based studies.
Supplementary Material
Acknowledgment
The authors thank Violet Roskens for preparing the DNA and BR samples used in the stretching and imaging experiment, respectively; Linda Randall for providing the halobacterium and protocols for preparing the BR samples; Jason Cleveland and Deron Walters for fruitful discussions; Brad Baxley for illustrating Fig. 10B; and Michael Johnson for sharing his Allan variance code. This work was supported by an NIH Molecular Biophysics Training Scholarship (ABC, T32 GM-065103), the National Science Foundation [DBI-0923544, Phys-1125844], and NIST. Mention of commercial products is for information only; it does not imply NIST’s recommendation or endorsement. TTP is a staff member of NIST’s Quantum Physics Division.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Casuso I, Scheuring S. Automated setpoint adjustment for biological contact mode atomic force microscopy imaging. Nanotechnology. 2010;21:035104. doi: 10.1088/0957-4484/21/3/035104. [DOI] [PubMed] [Google Scholar]
- 2.King GM, Carter AR, Churnside AB, Eberle LS, Perkins TT. Ultrastable Atomic Force Microscopy: Atomic-Scale Stability and Registration in Ambient Conditions. Nano Lett. 2009;9:1451–1456. doi: 10.1021/nl803298q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Junker JP, Ziegler F, Rief M. Ligand-Dependent Equilibrium Fluctuations of Single Calmodulin Molecules. Science. 2009;323:633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 4.Stigler J, Ziegler F, Gieseke A, Gebhardt JCM, Rief M. The Complex Folding Network of Single Calmodulin Molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 5.Churnside AB, Sullan RMA, Nguyen DM, Case SO, Bull MS, King GM, Perkins TT. Routine and Timely Sub-picoNewton Force Stability and Precision for Biological Applications of Atomic Force Microscopy. Nano Lett. 2012;12:3557–3561. doi: 10.1021/nl301166w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bippes CA, Muller DJ. High-resolution atomic force microscopy and spectroscopy of native membrane proteins. Rep. Prog. Phys. 2011;74:086601. [Google Scholar]
- 7.Radmacher M, Cleveland JP, Hansma PK. Improvement of Thermally-Induced Bending of Cantilevers Used for Atomic-Force Microscopy. Scanning. 1995;17:117–121. [Google Scholar]
- 8.Wenzler LA, Moyes GL, Beebe TP. Improvements to atomic force microscopy cantilevers for increased stability. Rev. Sci. Instrum. 1996;67:4191–4197. [Google Scholar]
- 9.Labuda A, Bates JR, Grutter PH. The noise of coated cantilevers. Nanotechnology. 2012;23:025503. doi: 10.1088/0957-4484/23/2/025503. [DOI] [PubMed] [Google Scholar]
- 10.Paolino P, Bellon L. Frequency dependence of viscous and viscoelastic dissipation in coated micro-cantilevers from noise measurement. Nanotechnology. 2009;20:405705. doi: 10.1088/0957-4484/20/40/405705. [DOI] [PubMed] [Google Scholar]
- 11.Summary Notes - Force spectroscopy measurements and processing. JPK Instruments. 2009 [Google Scholar]
- 12.Liphardt J, Onoa B, Smith SB, Tinoco I, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 13.Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. U.S.A. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlierf M, Berkemeier F, Rief M. Direct observation of active protein folding using lock-in force spectroscopy. Biophys. J. 2007;93:3989–3998. doi: 10.1529/biophysj.107.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller DJ, Buldt G, Engel A. Force-Induced Conformational Change of Bacteriorhodopsin. J. Mol. Biol. 1995;249:239–243. doi: 10.1006/jmbi.1995.0292. [DOI] [PubMed] [Google Scholar]
- 16.Oesterhelt F, Oesterhelt D, Pfeiffer M, Engel A, Gaub HE, Muller DJ. Unfolding pathways of individual bacteriorhodopsins. Science. 2000;288:143–146. doi: 10.1126/science.288.5463.143. [DOI] [PubMed] [Google Scholar]
- 17.Hutter JL, Bechhoefer J. Calibration of Atomic-Force Microscope Tips. Rev. Sci. Instrum. 1993;64:3342–3342. [Google Scholar]
- 18.Sullivan DB, Allan DW, Howe DA, Walls EL, editors. Characterization of Clocks and Oscillators. Washington: U.S. Government Printing Office; 1990. [Google Scholar]
- 19.Noy A, Vezenov DV, Kayyem JF, Meade TJ, Lieber CM. Stretching and breaking duplex DNA by chemical force microscopy. Chem. Biol. 1997;4:519–527. doi: 10.1016/s1074-5521(97)90324-0. [DOI] [PubMed] [Google Scholar]
- 20.Rief M, Clausen-Schaumann H, Gaub HE. Sequence-dependent mechanics of single DNA molecules. Nat. Struct. Biol. 1999;6:346–349. doi: 10.1038/7582. [DOI] [PubMed] [Google Scholar]
- 21.Butt HJ, Downing KH, Hansma PK. Imaging the Membrane-Protein Bacteriorhodopsin with the Atomic Force Microscope. Biophys. J. 1990;58:1473–1480. doi: 10.1016/S0006-3495(90)82492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greif D, Wesner D, Regtmeier J, Anselmetti D. High resolution imaging of surface patterns of single bacterial cells. Ultramicroscopy. 2010;110:1290–1296. doi: 10.1016/j.ultramic.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Vegh AG, Nagy K, Balint Z, Kerenyi A, Rakhely G, Varo G, Szegletes Z. Effect of Antimicrobial Peptide-Amide: Indolicidin on Biological Membranes. J. Biomed. Biotechnol. 2011;2011:670589. doi: 10.1155/2011/670589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto D, Nagura N, Omote S, Taniguchi M, Ando T. Streptavidin 2D Crystal Substrates for Visualizing Biomolecular Processes by Atomic Force Microscopy. Biophys. J. 2009;97:2358–2367. doi: 10.1016/j.bpj.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perkins TT. Optical traps for single molecule biophysics: a primer. Laser Photonics Rev. 2009;3:203–220. [Google Scholar]
- 26.Czerwinski F, Richardson AC, Oddershede LB. Quantifying Noise in Optical Tweezers by Allan Variance. Opt. Express. 2009;17:13255–13269. doi: 10.1364/oe.17.013255. [DOI] [PubMed] [Google Scholar]
- 27.Hoogenboom BW, Frederix PLTM, Yang JL, Martin S, Pellmont Y, Steinacher M, Zach S, Langenbach E, Heimbeck HJ. A Fabry-Perot interferometer for micrometer-sized cantilevers. Appl. Phys. Lett. 2005;86:074101. [Google Scholar]
- 28.Kumanchik L, Schmitz TL, Pratt JR. An interferometric platform for studying AFM probe deflection. Precis. Eng. 2011;35:248–257. [Google Scholar]
- 29.Kuhner F, Morfill J, Neher RA, Blank K, Gaub HE. Force-induced DNA slippage. Biophys. J. 2007;92:2491–2497. doi: 10.1529/biophysj.106.095836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morfill J, Kuhner F, Blank K, Lugmaier RA, Sedlmair J, Gaub HE. B-S transition in short oligonucleotides. Biophys. J. 2007;93:2400–2409. doi: 10.1529/biophysj.107.106112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith SB, Cui Y, Bustamante C. Overstretching of B-DNA: The elastic response of individual double-stranded and single stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 32.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy JL, Chatenay D, Caron F. DNA: An Extensible Molecule. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 33.Hansma HG, Schaffer TE, Cleveland JP. Atomic force microscope for generating a small incident beam spot. European Patent EP1012862. 1998 [Google Scholar]
- 34.Liu MZ, Amro NA, Chow CS, Liu GY. Production of nanostructures of DNA on surfaces. Nano Lett. 2002;2:863–867. [Google Scholar]
- 35.Piner RD, Zhu J, Xu F, Hong SH, Mirkin CA. "Dip-pen" nanolithography. Science. 1999;283:661–663. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- 36.Proksch R, Schaffer TE, Cleveland JP, Callahan RC, Viani MB. Finite optical spot size and position corrections in thermal spring constant calibration. Nanotechnology. 2004;15:1344–1350. [Google Scholar]
- 37.Oroz J, Hervas R, Carrion-Vazquez M. Unequivocal single-molecule force spectroscopy of proteins by AFM using pFS vectors. Biophys. J. 2012;102:682–690. doi: 10.1016/j.bpj.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim BH, Palermo NY, Lovas S, Zaikova T, Keana JFW, Lyubchenko YL. Single- Molecule Atomic Force Microscopy Force Spectroscopy Study of A beta-40 Interactions. Biochemistry. 2011;50:5154–5162. doi: 10.1021/bi200147a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahl SW, Nash MA, Fried DB, Slutzki M, Barak Y, Bayer EA, Gaub HE. Single-molecule dissection of the high-affinity cohesin-dockerin complex. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20431–20436. doi: 10.1073/pnas.1211929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan C, Chen A, Kolb P, Moy VT. Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy. Biochemistry. 2000;39:10219–10223. doi: 10.1021/bi992715o. [DOI] [PubMed] [Google Scholar]
- 41.Popa AM, Niedermann P, Heinzelmann H, Hubbell JA, Pugin R. Fabrication of nanopore arrays and ultrathin silicon nitride membranes by block-copolymer-assisted lithography. Nanotechnology. 2009;20 doi: 10.1088/0957-4484/20/48/485303. [DOI] [PubMed] [Google Scholar]
- 42.Janovjak H, Kessler M, Oesterhelt D, Gaub H, Muller DJ. Unfolding pathways of native bacteriorhodopsin depend on temperature. EMBO J. 2003;22:5220–5229. doi: 10.1093/emboj/cdg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 44.Kufer SK, Dietz H, Albrecht C, Blank K, Kardinal A, Rief M, Gaub HE. Covalent immobilization of recombinant fusion proteins with hAGT for single molecule force spectroscopy. Eur. Biophys. J. Biophy. 2005;35:72–78. doi: 10.1007/s00249-005-0010-1. [DOI] [PubMed] [Google Scholar]
- 45.Kufer SK, Puchner EM, Gumpp H, Liedl T, Gaub HE. Single-molecule cut-and-paste surface assembly. Science. 2008;319:594–596. doi: 10.1126/science.1151424. [DOI] [PubMed] [Google Scholar]
- 46.Morfill J, Blank K, Zahnd C, Luginbuhl B, Kuhner F, Gottschalk KE, Pluckthun A, Gaub HE. Affinity-matured recombinant antibody fragments analyzed by single-molecule force Spectroscopy. Biophys. J. 2007;93:3583–3590. doi: 10.1529/biophysj.107.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Camarero JA. New developments for the site-specific attachment of protein to surfaces. Biophys. Rev. Lett. 2005;1:1–28. [Google Scholar]
- 48.Gu JH, Xiao ZD, Yam CM, Qin GT, Deluge M, Boutet S, Cai CZ. Attaching single biomolecules selectively to the apex of AFM tips for measuring specific interactions. Biophys. J. 2005;89:L31–L33. doi: 10.1529/biophysj.105.069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lyubchenko YL, Shlyakhtenko LS, Gall AA. Atomic force microscopy imaging and probing of DNA, proteins, and protein DNA complexes: silatrane surface chemistry. Methods Mol. Biol. 2009;543:337–351. doi: 10.1007/978-1-60327-015-1_21. [DOI] [PubMed] [Google Scholar]
- 50.Zimmermann JL, Nicolaus T, Neuert G, Blank K. Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nat. Protoc. 2010;5:975–985. doi: 10.1038/nprot.2010.49. [DOI] [PubMed] [Google Scholar]
- 51.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–641. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 52.Hermanson GT. Bioconjugate Techniques. 2nd Edition edn. London, UK: Academic Press; 2008. [Google Scholar]
- 53.Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M. et al, A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 54.Heus HA, Puchner EM, van Vugt-Jonker AJ, Zimmermann JL, Gaub HE. Atomic force microscope-based single-molecule force spectroscopy of RNA unfolding. Anal. Biochem. 2011;414:1–6. doi: 10.1016/j.ab.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 55.Riener CK, Kienberger F, Hahn CD, Buchinger GM, Egwim IOC, Haselgrubler T, Ebner A, Romanin C, Klampfl C, Lackner B, et al. Heterobifunctional crosslinkers for tethering single ligand molecules to scanning probes. Anal. Chim. Acta. 2003;497:101–114. [Google Scholar]
- 56.Yu JP, Malkova S, Lyubchenko YL. alpha-Synuclein Misfolding: Single Molecule AFM Force Spectroscopy Study. J. Mol. Biol. 2008;384:992–1001. doi: 10.1016/j.jmb.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 57.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 58.Yu ZB, Koirala D, Cui YX, Easterling LF, Zhao Y, Mao HB. Click Chemistry Assisted Single-Molecule Fingerprinting Reveals a 3D Biomolecular Folding Funnel. J. Am. Chem. Soc. 2012;134:12338–12341. doi: 10.1021/ja303218s. [DOI] [PubMed] [Google Scholar]
- 59.Milles S, Tyagi S, Banterle N, Koehler C, VanDelinder V, Plass T, Neal AP, Lemke EA. Click Strategies for Single-Molecule Protein Fluorescence. J. Am. Chem. Soc. 2012;134:5187–5195. doi: 10.1021/ja210587q. [DOI] [PubMed] [Google Scholar]
- 60.Dohoney KM, Gelles J. chi-Sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature. 2001;409:370–374. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 61.Yu H, Liu X, Neupane K, Gupta AN, Brigley AM, Solanki A, Sosova I, Woodside MT. Direct observation of multiple misfolding pathways in a single prion protein molecule. Proc. Natl. Acad. Sci. U.S.A. 2012;109:5283–5288. doi: 10.1073/pnas.1107736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi X, Jung Y, Lin LJ, Liu C, Wu C, Cann IK, Ha T. Quantitative fluorescence labeling of aldehyde-tagged proteins for single-molecule imaging. Nat. Methods. 2012;9:499–503. doi: 10.1038/nmeth.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. U.S.A. 2012;109:E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taniguchi Y, Kawakami M. Application of HaloTag Protein to Covalent Immobilization of Recombinant Proteins for Single Molecule Force Spectroscopy. Langmuir. 2010;26:10433–10436. doi: 10.1021/la101658a. [DOI] [PubMed] [Google Scholar]
- 66.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-Molecule Protein Unfolding and Translocation by an ATP-Fueled Proteolytic Machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ando T, Kodera N, Naito Y, Kinoshita T, Furuta K, Toyoshima YY. A high-speed atomic force microscope for studying biological macromolecules in action. ChemPhysChem. 2003;4:1196–1202. doi: 10.1002/cphc.200300795. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.