Abstract
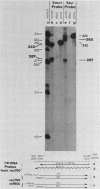
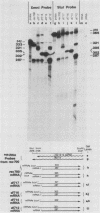
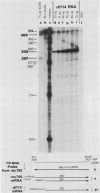
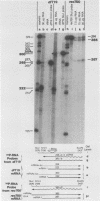
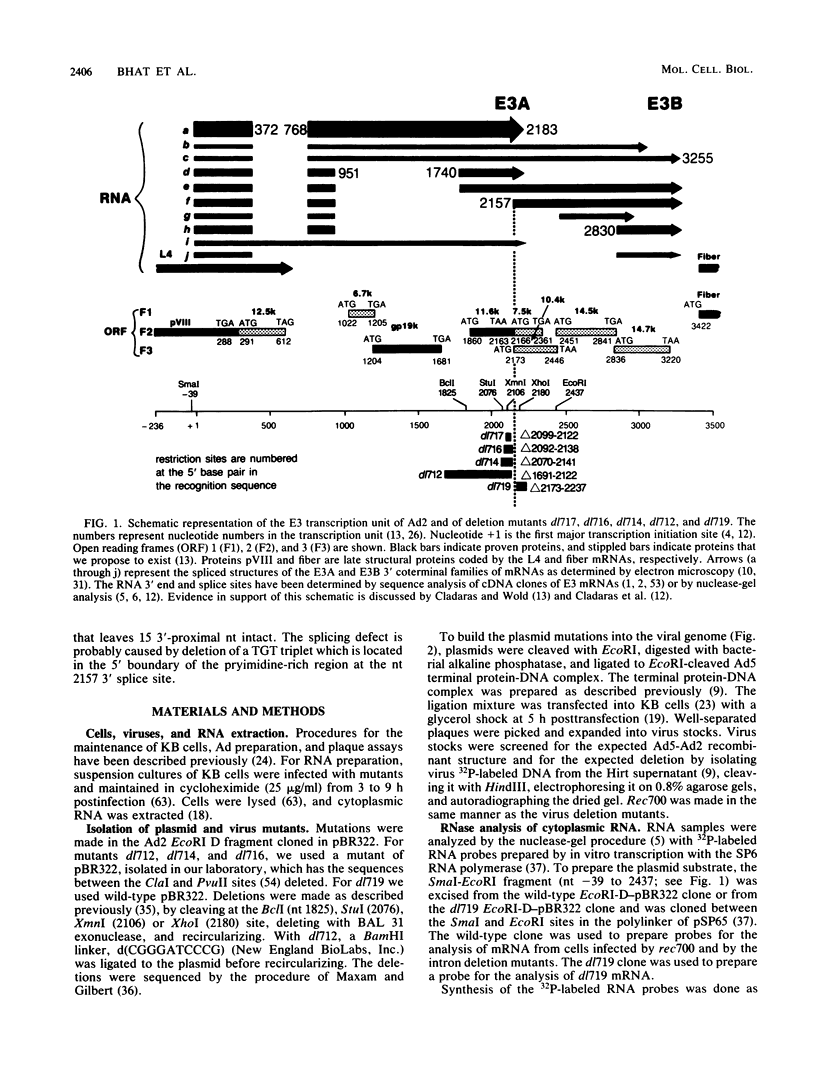
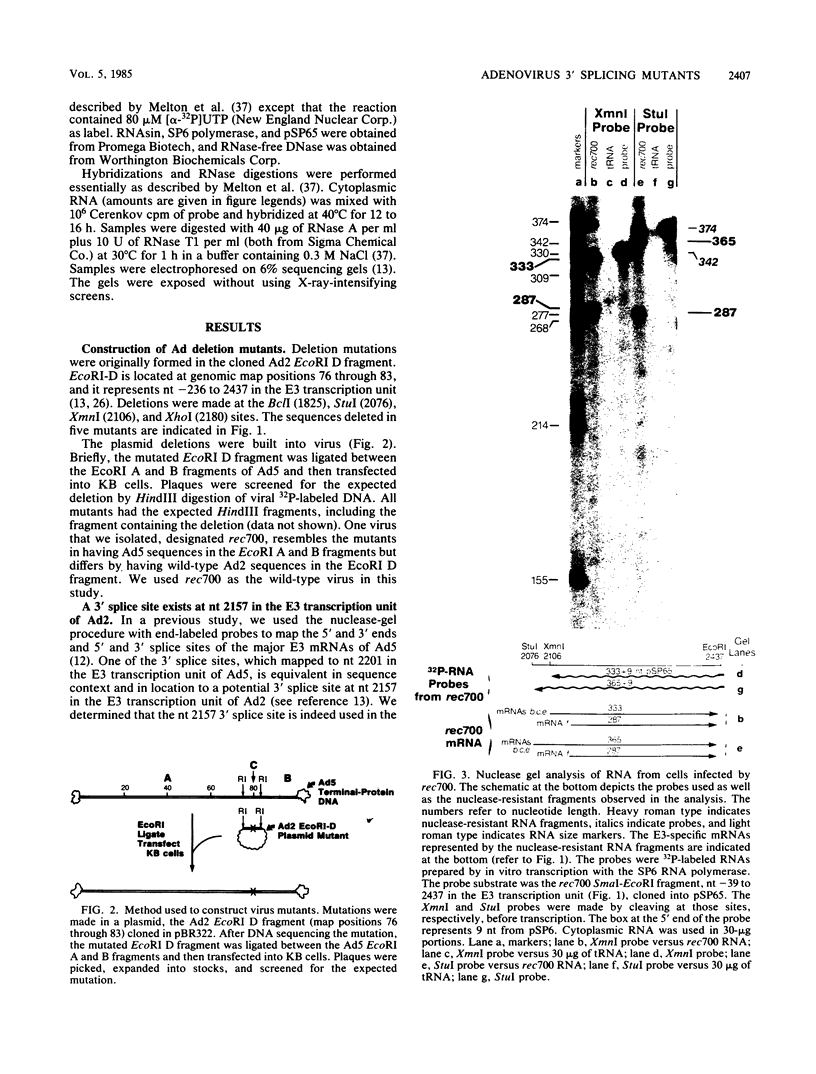
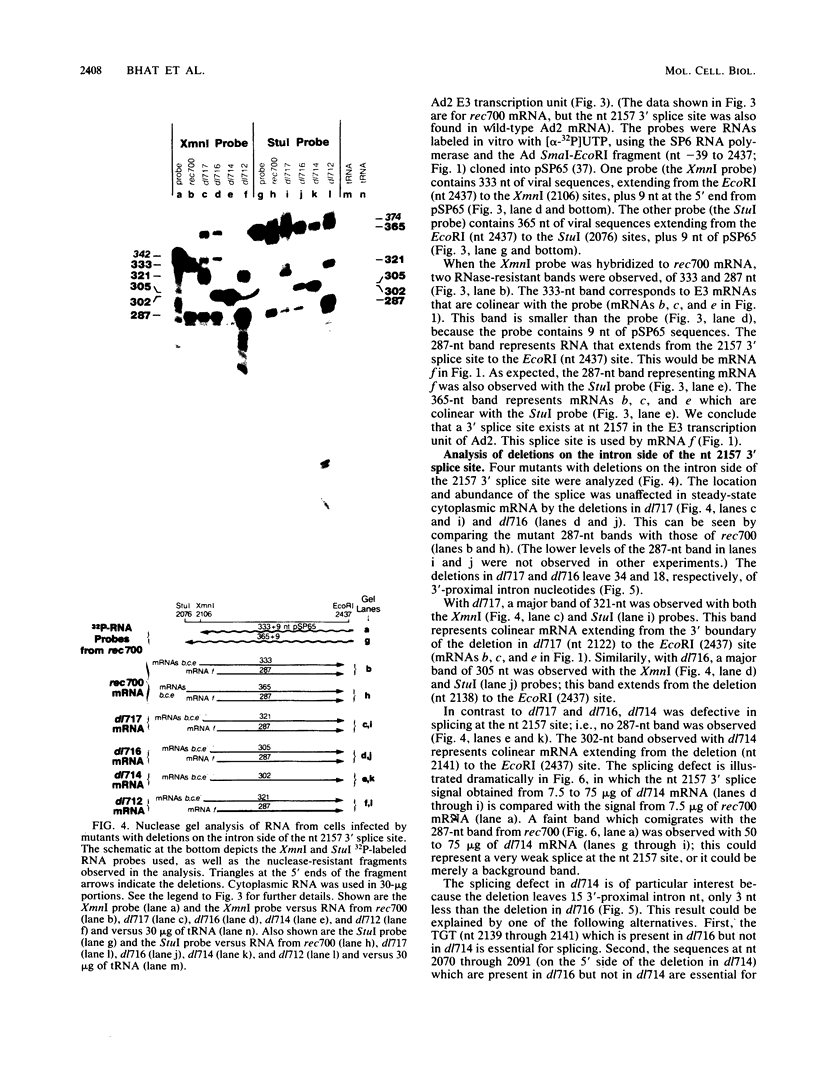
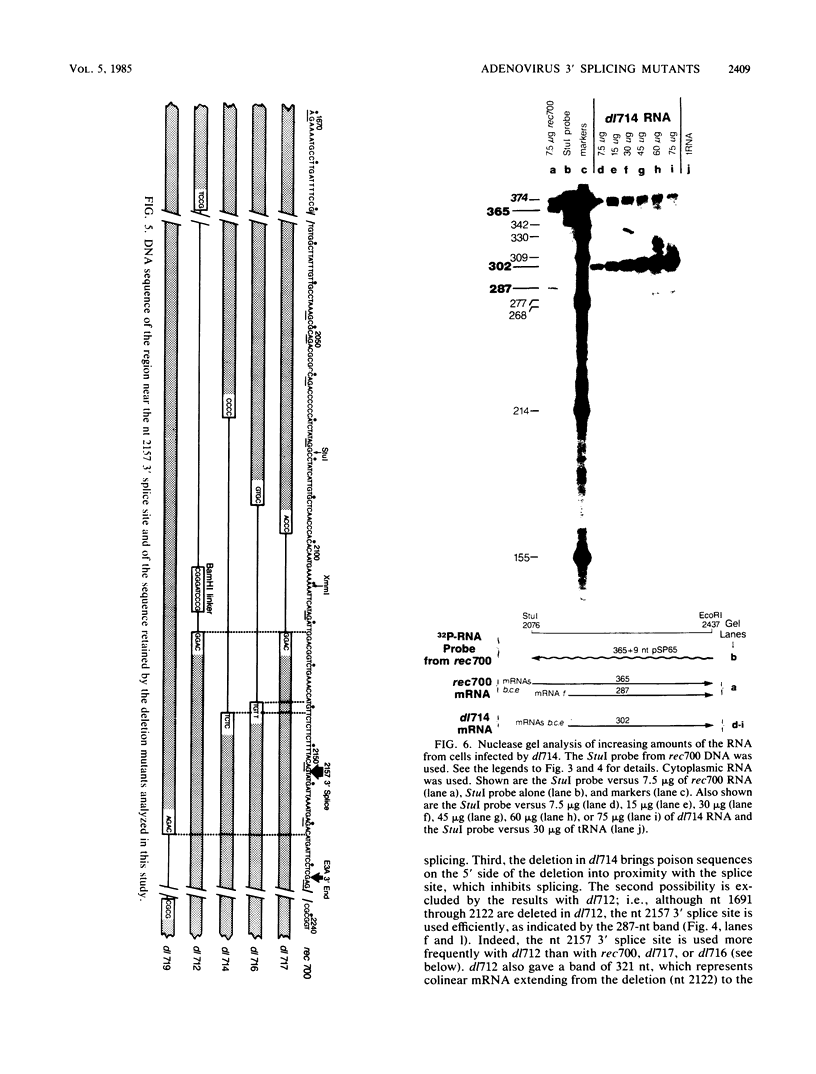
Five viable virus mutants were constructed with deletions near a 3' splice site located at nucleotide 2157 in the E3 transcription unit of adenovirus 2. The mutants were examined for splicing activity at the 2157 3' splice site in vivo by nuclease-gel analysis of steady-state cytoplasmic mRNA. Splicing was not prevented by an exon deletion (dl719) that leaves 16 5'-proximal exon nucleotides intact or by intron deletions that leave 34 (dl717, dl712) or 18 (dl716) 3'-proximal intron nucleotides intact. The sequences deleted in one of these intron mutants (dl716) include the putative branchpoint site used in lariat formation during splicing. Thus, a surrogate branchpoint site apparently can be used for splicing. Another intron mutant (dl714) has a deletion that leaves 15 3'-proximal intron nucleotides intact; remarkably, this deletion virtually abolished splicing, even though the deletion is only 3 nucleotides closer to the splice site than is the deletion in dl716 which splices normally. The three nucleotides deleted in dl714 that are retained by dl716 are the sequence TGT. The TGT sequence is located on the 5' boundary of the pyrimidine-rich region upstream of the nucleotide 2157 3' splice site. Such pyrimidine-rich regions are ubiquitous at 3' splice sites. Most likely, the TGT is required for splicing at the nucleotide 2157 3' splice site. The TGT may be important because of its specific sequence or because it forms the 5' boundary of the pyrimidine-rich region.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed C. M., Chanda R., Stow N., Zain B. S. The sequence of 3'-termini of mRNAs from early region III of adenovirus 2. Gene. 1982 Oct;19(3):297–301. doi: 10.1016/0378-1119(82)90019-1. [DOI] [PubMed] [Google Scholar]
- Antonarakis S. E., Irkin S. H., Cheng T. C., Scott A. F., Sexton J. P., Trusko S. P., Charache S., Kazazian H. H., Jr beta-Thalassemia in American Blacks: novel mutations in the "TATA" box and an acceptor splice site. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1154–1158. doi: 10.1073/pnas.81.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C. C., Ziff E. B. Promoters and heterogeneous 5' termini of the messenger RNAs of adenovirus serotype 2. J Mol Biol. 1981 Jun 25;149(2):189–221. doi: 10.1016/0022-2836(81)90298-9. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Structure of the adenovirus 2 early mRNAs. Cell. 1978 Jul;14(3):695–711. doi: 10.1016/0092-8674(78)90252-0. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Benoist C., O'Hare K., Gannon F., Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busslinger M., Moschonas N., Flavell R. A. Beta + thalassemia: aberrant splicing results from a single point mutation in an intron. Cell. 1981 Dec;27(2 Pt 1):289–298. doi: 10.1016/0092-8674(81)90412-8. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G., Chinnadurai S., Brusca J. Physical mapping of a large-plaque mutation of adenovirus type 2. J Virol. 1979 Nov;32(2):623–628. doi: 10.1128/jvi.32.2.623-628.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow L. T., Broker T. R., Lewis J. B. Complex splicing patterns of RNAs from the early regions of adenovirus-2. J Mol Biol. 1979 Oct 25;134(2):265–303. doi: 10.1016/0022-2836(79)90036-6. [DOI] [PubMed] [Google Scholar]
- Chu G., Sharp P. A. A gene chimaera of SV40 and mouse beta-globin is transcribed and properly spliced. Nature. 1981 Jan 29;289(5796):378–382. doi: 10.1038/289378a0. [DOI] [PubMed] [Google Scholar]
- Cladaras C., Bhat B., Wold W. S. Mapping the 5' ends, 3' ends, and splice sites of mRNAs from the early E3 transcription unit of adenovirus 5. Virology. 1985 Jan 15;140(1):44–54. doi: 10.1016/0042-6822(85)90444-1. [DOI] [PubMed] [Google Scholar]
- Cladaras C., Wold W. S. DNA sequence of the early E3 transcription unit of adenovirus 5. Virology. 1985 Jan 15;140(1):28–43. doi: 10.1016/0042-6822(85)90443-x. [DOI] [PubMed] [Google Scholar]
- Dobkin C., Pergolizzi R. G., Bahre P., Bank A. Abnormal splice in a mutant human beta-globin gene not at the site of a mutation. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1184–1188. doi: 10.1073/pnas.80.5.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Esumi H., Takahashi Y., Sato S., Nagase S., Sugimura T. A seven-base-pair deletion in an intron of the albumin gene of analbuminemic rats. Proc Natl Acad Sci U S A. 1983 Jan;80(1):95–99. doi: 10.1073/pnas.80.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Felber B. K., Orkin S. H., Hamer D. H. Abnormal RNA splicing causes one form of alpha thalassemia. Cell. 1982 Jul;29(3):895–902. doi: 10.1016/0092-8674(82)90451-2. [DOI] [PubMed] [Google Scholar]
- Frost E., Williams J. Mapping temperature-sensitive and host-range mutations of adenovirus type 5 by marker rescue. Virology. 1978 Nov;91(1):39–50. doi: 10.1016/0042-6822(78)90353-7. [DOI] [PubMed] [Google Scholar]
- Fukumaki Y., Ghosh P. K., Benz E. J., Jr, Reddy V. B., Lebowitz P., Forget B. G., Weissman S. M. Abnormally spliced messenger RNA in erythroid cells from patients with beta+ thalassemia and monkey cells expressing a cloned beta+-thalassemic gene. Cell. 1982 Mar;28(3):585–593. doi: 10.1016/0092-8674(82)90213-6. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Piatak M., Mertz J. E., Weissman S. M., Lebowitz P. Altered utilization of splice sites and 5' termini in late RNAs produced by leader region mutants of simian virus 40. J Virol. 1982 Nov;44(2):610–624. doi: 10.1128/jvi.44.2.610-624.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith M. E., Humphries R. K., Ley T., Cline A., Kantor J. A., Nienhuis A. W. "Silent" nucleotide substitution in a beta+-thalassemia globin gene activates splice site in coding sequence RNA. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2318–2322. doi: 10.1073/pnas.80.8.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Green M., Wold W. S. Human adenoviruses: growth, purification, and transfection assay. Methods Enzymol. 1979;58:425–435. doi: 10.1016/s0076-6879(79)58157-9. [DOI] [PubMed] [Google Scholar]
- Hamer D. H., Leder P. SV40 recombinants carrying a functional RNA splice junction and polyadenylation site from the chromosomal mouse beta maj globin gene. Cell. 1979 Jul;17(3):737–747. doi: 10.1016/0092-8674(79)90280-0. [DOI] [PubMed] [Google Scholar]
- Hwang L. S., Park J., Gilboa E. Role of intron-contained sequences in formation of moloney murine leukemia virus env mRNA. Mol Cell Biol. 1984 Nov;4(11):2289–2297. doi: 10.1128/mcb.4.11.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérissé J., Courtois G., Galibert F. Nucleotide sequence of the EcoRI D fragment of adenovirus 2 genome. Nucleic Acids Res. 1980 May 24;8(10):2173–2192. doi: 10.1093/nar/8.10.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. B., Noon W. A. Intron splicing: a conserved internal signal in introns of animal pre-mRNAs. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7417–7420. doi: 10.1073/pnas.81.23.7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Alwine J., Goldman N., Gruss P., Jay G. New chimeric splice junction in adenovirus type 2-simian virus 40 hybrid viral mRNA. J Virol. 1980 Oct;36(1):143–151. doi: 10.1128/jvi.36.1.143-151.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P., Dhar R., Lai C. J. Processing and expression of early SV40 mRNA: a role for RNA conformation in splicing. Cell. 1979 Sep;18(1):85–92. doi: 10.1016/0092-8674(79)90356-8. [DOI] [PubMed] [Google Scholar]
- Kitchingman G. R., Westphal H. The structure of adenovirus 2 early nuclear and cytoplasmic RNAs. J Mol Biol. 1980 Feb 15;137(1):23–48. doi: 10.1016/0022-2836(80)90155-2. [DOI] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Klinz F. J., Donath C., Gallwitz D. Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell. 1984 Mar;36(3):645–653. doi: 10.1016/0092-8674(84)90344-1. [DOI] [PubMed] [Google Scholar]
- Lee D. C., Roeder R. G., Wold W. S. DNA sequences affecting specific initiation of transcription in vitro from the EIII promoter of adenovirus 2. Proc Natl Acad Sci U S A. 1982 Jan;79(1):41–45. doi: 10.1073/pnas.79.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Berk A. J. Elimination of mRNA splicing by a point mutation outside the conserved GU at 5' splice sites. Nucleic Acids Res. 1984 May 11;12(9):3821–3827. doi: 10.1093/nar/12.9.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Courtois G., Eng C., Berk A. Complete transformation by adenovirus 2 requires both E1A proteins. Cell. 1984 Apr;36(4):951–961. doi: 10.1016/0092-8674(84)90045-x. [DOI] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Control of adenovirus E1B mRNA synthesis by a shift in the activities of RNA splice sites. Mol Cell Biol. 1984 May;4(5):966–972. doi: 10.1128/mcb.4.5.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C., Fisher E. F., Caruthers M. H., Berk A. J. Resolving the functions of overlapping viral genes by site-specific mutagenesis at a mRNA splice site. Nature. 1982 Feb 4;295(5848):380–384. doi: 10.1038/295380a0. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S. H., Goff S. C., Hechtman R. L. Mutation in an intervening sequence splice junction in man. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5041–5045. doi: 10.1073/pnas.78.8.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S. H., Kazazian H. H., Jr, Antonarakis S. E., Ostrer H., Goff S. C., Sexton J. P. Abnormal RNA processing due to the exon mutation of beta E-globin gene. Nature. 1982 Dec 23;300(5894):768–769. doi: 10.1038/300768a0. [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Sexton J. P., Goff S. C., Kazazian H. H., Jr Inactivation of an acceptor RNA splice site by a short deletion in beta-thalassemia. J Biol Chem. 1983 Jun 25;258(12):7249–7251. [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Piatak M., Ghosh P. K., Norkin L. C., Weissman S. M. Sequences locating the 5' ends of the major simian virus 40 late mRNA forms. J Virol. 1983 Nov;48(2):503–520. doi: 10.1128/jvi.48.2.503-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Rautmann G., Matthes H. W., Gait M. J., Breathnach R. Synthetic donor and acceptor splice sites function in an RNA polymerase B (II) transcription unit. EMBO J. 1984 Sep;3(9):2021–2028. doi: 10.1002/j.1460-2075.1984.tb02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. R., Pikielny C. W., Rosbash M. In vivo characterization of yeast mRNA processing intermediates. Cell. 1984 Dec;39(3 Pt 2):603–610. doi: 10.1016/0092-8674(84)90467-7. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Solnick D. An adenovirus mutant defective in splicing RNA from early region 1A. Nature. 1981 Jun 11;291(5815):508–510. doi: 10.1038/291508a0. [DOI] [PubMed] [Google Scholar]
- Stålhandske P., Persson H., Perricaudet M., Philipson L., Pettersson U. Structure of three spliced mRNAs from region E3 of adenovirus type 2. Gene. 1983 May-Jun;22(2-3):157–165. doi: 10.1016/0378-1119(83)90099-9. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Thimmappaya B., Shenk T. Nucleotide sequence analysis of viable deletion mutants lacking segments of the simian virus 40 genome coding for small t antigen. J Virol. 1979 Jun;30(3):668–673. doi: 10.1128/jvi.30.3.668-673.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R., Orkin S. H., Maniatis T. Specific transcription and RNA splicing defects in five cloned beta-thalassaemia genes. Nature. 1983 Apr 14;302(5909):591–596. doi: 10.1038/302591a0. [DOI] [PubMed] [Google Scholar]
- Volckaert G., Feunteun J., Crawford L. V., Berg P., Fiers W. Nucleotide sequence deletions within the coding region for small-t antigen of simian virus 40. J Virol. 1979 Jun;30(3):674–682. doi: 10.1128/jvi.30.3.674-682.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J. C., Edmonds M. Polyadenylylated nuclear RNA contains branches. Proc Natl Acad Sci U S A. 1983 Feb;80(4):950–954. doi: 10.1073/pnas.80.4.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring R. B., Ray J. A., Edwards S. W., Scazzocchio C., Davies R. W. The Tetrahymena rRNA intron self-splices in E. coli: in vivo evidence for the importance of key base-paired regions of RNA for RNA enzyme function. Cell. 1985 Feb;40(2):371–380. doi: 10.1016/0092-8674(85)90151-5. [DOI] [PubMed] [Google Scholar]
- White R. T., Berg P., Villarreal L. P. Simian virus 40-rabbit beta-globin recombinants lacking late mRNA splice sites express cytoplasmic RNAs with altered structures. J Virol. 1982 Apr;42(1):262–274. doi: 10.1128/jvi.42.1.262-274.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Wold W. S., Green M., Brackmann K. H., Cartas M. A., Devine C. Genome expression and mRNA maturation at late stages of productive adenovirus type 2 infection. J Virol. 1976 Nov;20(2):465–477. doi: 10.1128/jvi.20.2.465-477.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]