Abstract
The detailed mechanisms of how DNA that is assembled around a histone core can be accessed by DNA-binding proteins for transcription, replication, or repair, remain elusive nearly 40 years after Kornberg's nucleosome model was proposed. Uncovering the structural dynamics of nucleosomes is a crucial step in elucidating the mechanisms regulating genome accessibility. This requires the deconvolultion of multiple structural states within an ensemble. Recent advances in single molecule methods enable unprecedented efficiency in examining subpopulation dynamics. In this review, we summarize studies of nucleosome structure and dynamics from single molecule approaches and how they advance our understanding of the mechanisms that govern DNA transactions.
Keywords: nucleosome, single molecule, dynamics, fluorescence, force
Nucleosome structure and dynamics
The 3 billion base pairs (bp) of DNA that encode the human genome extend to a length of ~2 meters when stretched out. To store this enormous amount of material in a nucleus requires that the DNA be folded into highly condensed chromatin structures. This inevitably leads to problems in accessing DNA during transactions such as transcription, replication, and repair. At the heart of chromatin is its most basic repeat unit, the nucleosome. The composition of nucleosome core particles (NCPs) was first reported by Roger Kornberg nearly 40 years ago [1, 2]. Work by Kornberg and others revealed that the NCP is comprised of two copies of H2A, H2B, H3 and H4, which form the histone octamer core, wrapped with ~147bp of DNA in 1.65 superhelical turns [3–6]. Obtaining the X-ray structure of nucleosomes at an atomic resolution was a crucial milestone that provided a framework for understanding how DNA transactions occur in a chromatin context.
Major insights into the physical properties of nucleosomes have been provided by experiments using purified histones and both natural and selected DNA sequences from many laboratories [7–17]. In vitro and in vivo nucleosome and chromatin studies have identified ATP-dependent and independent mechanisms that cells use to modulate chromatin structure. These mechanisms include mechanical alteration of nucleosome structures and positions and covalent modifications of histones and/or DNA [18–21]. Although this large body of groundbreaking work illuminates the diversity of controls used by eukaryotes to regulate access to the genome, insight into the structure and dynamics of nucleosomes is required for a comprehensive understanding of how chromatin transactions are governed. One requirement in studying the dynamic nucleosome structure is the ability to monitor structural fluctuations and to detect short-lived states. Standard biochemical assays are based on ensemble-averaging (see glossary) techniques that can miss subpopulations representing transient states. Single molecule methods provide a means to study such problems, as they are highly efficient in probing subpopulation dynamics of complex systems.
In this review we summarize the findings from single molecule approaches to study the structure and dynamics of nucleosomes, and how these results impact our understanding of DNA transactions. The distance and time scale of nucleosomal motion, which modulate genomic transactions, are well suited for single molecule studies. The mechanical forces involved in nucleosome structural integrity and chromatin remodeling have also been investigated at single particle resolution with optical and magnetic traps (Figures 1–4, Table 1 and Box 1). Collectively, the field of chromatin biology has seen important advances in understanding the fundamental dynamics of nucleosome structures aided by single molecule studies.
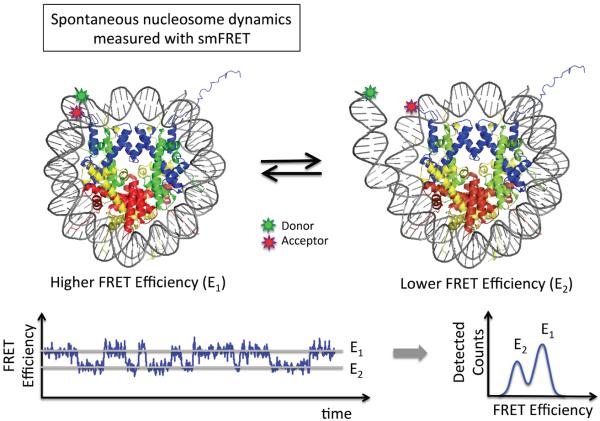
Figure 1. Single molecule fluorescence resonance energy transfer (FRET) studies on the structure and structural dynamics of nucleosomes.
FRET labels are attached to DNA and/or histones to elucidate the structure of nucleosomes and to follow the dynamic unwrapping and/or rewrapping of the nucleosomal DNA. The fluorophore positions in the example shown are from the fluorescence correlation spectroscopy (FCS) study by the Widom group [22]. The time trajectory of FRET efficiency or FCS measurement reveals dynamic motions of the nucleosomal DNA termini within the temporal resolution of the measurement. A simulated ideal FRET time trajectory of a surface immobilized single nucleosome particle is shown to illustrate the measurement scheme. The time-averaged FRET efficiency histogram reveals the closed (E1; high FRET) and opened (E2; low FRET) states of a nucleosome.
Table 1.
Methods for studying single molecule nucleosome structures and dynamics
| Method | Advantages | Limits | |
|---|---|---|---|
| Fluorescence | Fluorescence methods in general | Low level of system perturbation High temporal resolution Native environment |
No control over system dynamics Limited fluorophore labeling strategies Artifacts due to fluorophores |
| Single molecule FRET (smFRET) in general | No need for calibration for internal comparison (ratiometric measurement) High spatiotemporal resolution |
One-dimensional measurement | |
| smFRET with surface-immobilized nucleosomes | Efficient to study detailed dynamics of a complex process with more than 2 states Easy to deconvolve heterogeneous populations (i.e. easy to filter out contaminants and inactive species) Limit of temporal resolution: with wide-field imaging: a few milliseconds with one point detection: tens to hundreds of microseconds (limited by photon emission rate) |
Possible effects of surface interactions with nucleosomes | |
| smFRET and/or FCS with diffusing nucleosomes | No effects of surface interactions with nucleosomes FCS: Efficient to test if fast dynamics exist (tens to hundreds of microseconds) |
Impractical or impossible to study detailed dynamics of a complex process with more than 2 states Impractical or impossible to monitor dynamics with time scales longer than diffusion of nucleosomes |
|
| Force | Force methods in general | Direct control of system dynamics Direct measure of interactions Native environment |
Low level of control over reaction coordinate High level of system perturbation Limited force-handle labeling strategies |
| Optical tweezers | High force resolution (sub-pN) | Inefficient data collection (typically one molecule at a time) | |
| Magnetic tweezers | Efficient data collection in a wide-field imaging setup (multiple molecules at a time) | Higher level of noise (i.e. limited precision in controlling and/or measuring force at a rate higher than a few Hz) | |
| Scanning probe | Multi-dimensional imaging | Mostly under non-native conditions Typically very low or no temporal resolution |
|
Dynamic nucleosome structures and their role in DNA-protein transactions
Although it has been well accepted that DNA is packaged into chromatin structures since the 1970s, the mechanism(s) of how DNA-binding proteins target nucleosomal DNA remained unknown for 20 years. In 1995, Pollach and Widom first measured the equilibrium constants for spontaneous formation of `opened' and `closed' nucleosome conformations by assaying DNA accessibility with restriction enzymes [14]. This work demonstrated that the inherent dynamics of DNA-histone interactions play a fundamental role in how proteins can bind to target sequences located within nucleosomes. In 2005, the Widom lab determined the rates of spontaneous un/rewrapping of nucleosomal DNA using fluorescently labeled nucleosomes and fluorescence correlation spectroscopy (FCS) measurements [22]. The authors demonstrated that spontaneous unwrapping of DNA facilitated the binding of the LexA transcription factor near the termini of the nucleosomal DNA and the rewrapping of DNA limits the efficiency of LexA binding. Recently, the same group reported slower dynamics in internal regions of DNA [23]. Importantly, the kinetics of DNA-histone interactions can be markedly altered by cationic concentration [24]. Thus these data, which were obtained in the absence of salt, must be applied with caution in a cellular context, where the collective concentration of monovalent ions can be as high as ~150mM. Nonetheless, these reports provided the first direct physical evidence that spontaneous nucleosome structural dynamics modulate DNA accessibility. However, it was impossible to deconvolve intermediate un- or rewrapping states that might render valuable information on the regulation of DNA accessibility, as they were likely hidden in the ensemble-averaged behavior.
One approach to circumvent the limitations of ensemble-averaging is to study the dynamics of surface immobilized nucleosomes that contain a fluorophore pair, using fluorescence resonance energy transfer (FRET) measurements (Figure 1). The first reported studies using this strategy were from Tomschik et al. [25, 26]. The time scale of DNA un- and rewrapping, however, overlapped with those of fluorophore blinking. This confounds the interpretation because one cannot distinguish if the dynamic FRET signals are a result of blinking or because of FRET efficiency changes due to the un- and rewrapping of DNA. The Langowski group showed by FRET measurements that nucleosomes, which are freely diffusing in solution, have conformational heterogeneity, likely due to conformational dynamics [24]. They also reported that the conformational heterogeneity had a strong dependence on salt conditions and assigned the mixed conformational states to intermediate states during nucleosome disassembly [27]. The salt concentrations used in nucleosome studies need to be carefully considered because they can result in striking differences in nucleosome dynamics. Notably, in FCS studies, as NaCl concentration increased up to 100mM, so did the dynamics of nucleosomal DNA, particularly in the region of 1 to 100μsec [24]. On the other hand, van Noort and colleagues reported that, for surface immobilized nucleosomes, virtually all fully assembled nucleosomes (over 95%) remained stably wrapped with a high FRET efficiency that appeared unchanged whereas 3% showed FRET dynamics [28]. This work indicates that only a small minority of nucleosomes displayed repeated unwrapping and rewrapping events at 150mM NaCl. The kinetic rates differed at least by an order of magnitude from the rates reported in studies by Widom and colleagues in which salt was absent.. Considering that nucleosomes are composed of highly charged molecules, their dynamics are predicted to strongly depend on salt conditions [24, 29, 30]. Moreover, at physiological salt concentrations no significant dynamic motions of terminal DNA was observed on the millisecond time scale. We propose that the un/rewrapping dynamics of terminal DNA at a physiological salt concentration might be on the order of nanosecond to microsecond time scales. As there are only trivial energy barriers for the terminal DNA to unwrap from the histone core [31, 32], the time scale of this dynamics should be close to the time scale for diffusion within a few nanometers, which is in nanosecond to microsecond range. In summary, the difference in the kinetics reported by several groups must be at least in part due to the difference in the salt concentrations and, therefore, the results should be interpreted with care. Another source of the discrepancy might be the effect of surface attachment of nucleosomes and one approach to resolve this issue would be to compare intranucleosomal dynamics with varying linker lengths. Using varying salt conditions for studies of freely diffusing nucleosomes would help test how salt affects nucleosome dynamics and if the conditions are consistent with studies of surface immobilized nucleosomes. Together this would help identify any surface immobilization artifacts and to determine the detailed effects of ionic strength on nucleosome dynamics. Despite the complications, these studies tested and validated the hypothesis that a key step in processes requiring DNA binding involves the spontaneous excursions between wrapped and unwrapped DNA states.
Another approach to examine the structure and dynamics of individual nucleosomes is to characterize their mechanical properties based on force spectroscopic measurements using optical or magnetic traps and atomic force microscopy (AFM) [33–36]. In 2002, using an optical trap, the Wang group reported reversible assembly and disassembly of nucleosomes that takes place in multiple steps, suggesting multiple energy barriers along un/rewrapping reaction coordinates [33]. This study was followed by work from several groups [37–39] reporting intermediate structures that result from partial nucleosome disassembly, which indicated that there was a low energy barrier for detachment of DNA ends from the histone octamer. This result supports the observed spontaneous un- and rewrapping of DNA termini reported by the Widom group. Further, reports from the Bustamante and Bednar groups are in agreement with a two-step disassembly model, in which H2A-H2B dimers dissociate followed by H3–H4 tetramer dissociation. This is consistent with observations from fluorescence based confocal measurements of solution-borne nucleosomes and recent biochemical studies [24, 27, 29, 40–42]. Importantly, in the study by the Bednar group, DNA dissociation depended on the stability of nucleosomes in solution, which was modulated by varying the concentration of the chromatin template. This confirms that the experimental conditions, in particular ionic strength and nucleosome concentration, affect the stabilities of highly charged DNA and core histones, and consequently their spontaneous interactions. Therefore, it is important to design single molecule experiments with these results in mind, especially when studying factor-independent spontaneous dynamics of nucleosomes.
In the above studies, the resolution of measurements was too coarse to give detailed information on the energy barriers for nucleosomal DNA dynamics. Recently, a higher-resolution map of DNA-histone interactions was reported, based on the measurement of the mechanical force to unzip the DNA in a single nucleosome [31]. This study revealed a periodicity in the DNA-histone interactions that resembles the pause patterns of RNA polymerase, suggesting that the nucleosomal structure is the major energy barrier for RNA polymerase to overcome during transcription. Interestingly, this report revealed that these energy barriers are not uniform which may play a crucial role in governing the dynamics of nucleosomal DNA motion [31, 32]. The report also showed that the terminal regions of nucleosomal DNA have significantly weaker interactions with histones, in line with previous low-resolution force studies [38, 39] and the spontaneous unwrapping and rewrapping dynamics reported from fluorescence studies. These force spectroscopic studies provided the first direct implications of energy barriers posed by histone-DNA interactions during DNA transactions.
Another aspect of structural dynamics in chromatin relevant to the control of gene accessibility, in vivo, is the dynamic packing of nucleosomes in nucleosome arrays, which are assemblies of multiple nucleosomes connected to each other by DNA linkers. Studies that measured the equilibrium constant for site accessibility of nucleosomal DNA within a 17-mer nucleosome array and a trinucleosome array revealed an accessibility equilibrium similar to mononucleosomes [43, 44]. These studies were performed in a range of divalent ion (i.e. Mg2+) concentrations that promoted array compaction. Therefore, even in folded chromatin, nucleosome structures remain dynamic and allow for DNA binding proteins to access target sequences. Chromatin folds hierarchically at different levels in cells and consequently the un- and refolding behavior of short nucleosome arrays in vitro would be only one aspect of nucleosome packing in chromatin. However, intrinsic interactions between nucleosomes should still act as a physical basis for higher order chromatin folding. Therefore, these studies and further details on the thermodynamics and kinetics of the internucleosomal interactions in short nucleosome arrays are a good starting point in elucidating the mechanisms of nucleosome packing in regulating gene accessibility.
Single molecule force measurements suggest that there are numerous energy barriers in a single nucleosome that a polymerase must overcome to read through the genetic code. Various hypotheses have been proposed to explain this process. Early studies by the Felsenfeld and Bradbury labs elegantly showed evidence of a `pass-back' mechanism whereby the nucleosome is transiently displaced and passed back to the DNA as an RNA polymerase traverses the DNA [45–48]. In addition, their studies showed that a single H2A-H2B dimer was displaced, which might disrupt the structural integrity of the octamer core and consequently weaken the interactions between histones and DNA. In vivo studies have also provided support for such a partial disassembly mechanism. More recent single molecule studies by the Langowski group [24, 27] identified several partially disassembled nucleosome conformations, some of which were consistent with a loss in H2A-H2B dimers. However, results from high-resolution mapping of histone-DNA interactions suggest that RNA polymerase might be a molecular motor strong enough to overcome the energy barriers posed by histone-DNA interactions. In contrast, a Brownian ratchet model of RNA polymerase action has also been suggested based on the measurements with optical tweezers on the RNA polymerase pause dynamics [49]. Given that different regions of DNA (e.g. promoters vs. open reading frames) can have different extents of nucleosome positioning strength, both strategies (i.e. partial or full eviction of nucleosomes, and RNA polymerase acting as a strong motor protein) might be implemented during transcription. A recent report on the forced topology transitions of nucleosomal DNA with magnetic tweezers may also contribute to the elucidation of the relationship between nucleosome structures and RNA polymerase activity [35]. Further studies on various nucleosomes with sequences of different positioning strengths will help elucidate why some nucleosomes need to be partially or fully evicted before transcription proceeds and how nucleosomes in open reading frames might keep their structural integrity during transcription.
These single molecule studies tested and validated the hypothesis that rapid and dynamic changes observed in nucleosome structures allow for chromatin remodelers, polymerases and transcription factors to bind and function. Yet, there remain many unanswered questions such as what effects ionic strength, DNA sequence, linker DNA, linker histones, and modifications of DNA and histones, have on the structural dynamics of nucleosomes in both inter- and intra-nucleosomal contexts. In particular, the question of how covalent modifications of DNA and histones influence nucleosomal structure and dynamics is central to many long-standing problems in the field of chromatin biology. The following section summarizes efforts to address this question.
Effects of post-translational histone modifications and DNA methylation on the structure and dynamics of nucleosomes
A major field of study in chromatin biology is to understand the role(s) that histone and DNA modifications serve in gene regulation. In the past 15 years, many labs have suggested that post-translational modifications (PTMs) of histones (e.g. acetylation) and DNA modifications (e.g. methylation), which can be evolutionarily conserved, act as codes that recruit factors to modulate local chromatin structure [50]. Another major action of these modifications might be to alter nucleosome dynamics, rendering the local chromatin more or less accessible by DNA binding factors [51, 52]. This section discusses the single molecule studies exploring this possibility.
DNA methylation in mammals typically takes place at cytosine in a CpG dinucleotide context. CpG methylation is associated with gene silencing and repression. Hypermethylation of CpGs in tumor suppressor promoters is found in many cancer cells [53]. It is unclear whether DNA methylation is a means to recruit transcription factors that cause repression, directly alter the conformation and/or dynamics of nucleosomes, or both. Toward understanding what consequence DNA methylation might have on nucleosome structure and dynamics, smFRET measurements have been performed by Choy et al. They simultaneously measured FRET efficiency and fluorescence polarization of surface immobilized nucleosomes [54], which revealed that DNA methylation induces a more compact and rigid nucleosome structure. More recently, the Lee group showed that DNA methylation is associated with tighter wrapping of the internal regions of nucleosomal DNA, and an accompanying topology change [55]. These results might appear to contradict the accepted view that methylation increases DNA stiffness. However, DNA flexibility (bendability/twistability) and the extent of DNA bendedness/twistedness are two independent properties of DNA allowing for more rigid DNA (lower bendability/twistability) to form more stable bent/twisted conformations [56]. These results show that DNA methylation is not merely used as a `scaffold' for methylated DNA binding proteins, but also imparts an effect on the inherent dynamics of the nucleosome. According to these studies, DNA methylation can directly change the DNA wrapping such that the nucleosome adopts a more rigid `closed' conformation. This would add more energy barriers for proteins to overcome during their transactions with DNA, such as when RNA polymerase transcribes genes assembled in nucleosomes, which might at least in part explain how DNA hypermethylation renders genes transcriptionally silent.
Histone acetylation is typically associated with gene activation [57]. Many groups have conducted biochemical studies to explore the effects of histone acetylation on nucleosome stability [58–61]. In general, it appears that histone acetylation leads to some destabilization of nucleosomes and/or chromatin folding depending on the acetylation site. Histone acetylation on histone H4 lysine 16 (H4K16) has been reported to inhibit inter- and intra-nucleosomal interactions [60, 62, 63]. Efforts have been made by the single molecule community to characterize the changes in the structure and dynamics of nucleosomes upon histone acetylation. Although an early study by the Langowski group showed that histone acetylation alters nucleosome stability, the low signal-to-noise level of these results did not allow them to draw any conclusions about the nucleosome conformational changes induced by histone acetylation [64]. A recent study by Lee et al. showed that histones H2A and H4 acetylation by Piccolo NuA4 leads to unwrapping of nucleosomal DNA, accompanied by a topology change, consistent with weakened DNA-histone interactions [65]. The unwrapping of DNA upon histone acetylation is subtle, yet significant, and would not have been possible to detect with ensemble-averaging measurements. In line with these results, force spectroscopic studies showed a decrease in nucleosome stability when H2A-H2B tails were acetylated [66]. Molecular and genetic studies have shown that acetylation of histone H3 on lysine 56 (H3K56) is correlated with DNA replication. However, the physical consequences of this modification that might help facilitate replication are unknown. Two recent studies using FRET and magnetic tweezers elegantly showed that acetylation on H3K56 increases the spontaneous un- and rewrapping dynamics of the terminal regions of nucleosomal DNA [67, 68]. These studies show that acetylation can directly alter the structure and dynamics of nucleosomes, rendering the DNA more accessible, and support the idea that the inherent physical properties are changed in a way that facilitates targeting of DNA by DNA binding factors. Previously, the `charge neutralization' hypothesis postulated that acetylation of e-amino groups on lysines neutralizes their charge, and causes lysines to no longer strongly interact with the backbone of DNA, thereby promoting a more open chromatin conformation. Other hypotheses include disruption of H4 tail interactions with neighboring histones and disruption of H4 tail helix formation upon H4K16 acetylation [69, 70]. These hypotheses are supported by the above single molecule studies revealing that changes in nucleosome structure and dynamics are the result of altering the energy barriers of histone-DNA or histone-histone interactions upon histone acetylation.
Considering that genome transactions occur typically on DNA that is assembled into many nucleosomes, how histone acetylation affects inter-nucleosome interactions is another key question that needs to be addressed. The Hayes laboratory found, using a crosslinking strategy, that the N-terminal tail of histone H4 can interact with the C-terminal region of histone H2A in neighboring nucleosomes [62]. The nucleosomal packing shown from the crystal structure reported by Luger and Richmond in 1997 also suggested the presence of this interaction. Neumann et al. addressed part of this question by showing that H3K56 acetylation does not have any significant impact on the folding of nucleosome arrays [67]. However, Lee et al. reported that spontaneous formation of dinucleosomes by freely diffusing nucleosomes in solution is inhibited by acetylation of histones H4 and H2A [65]. This result suggests that histone acetylation attenuates internucleosomal interactions, which would contribute to the formation of open chromatin structures. Recent technological advances in native chemical ligation allows for the construction of nucleosomes and nucleosome arrays with specific acetylated residues [71]. Combined with single molecule studies, these tools will help elucidate role of acetylation of each lysine for all histones in regulating histone-DNA and histone-histone interactions in inter- and intra-nucleosomal contexts.
The structure and dynamics of nucleosomes clearly participate in the regulation of DNA transactions such as transcription and replication. Histone and/or DNA modifications can modulate the structure and dynamics of nucleosomes and consequently alter the transcriptional outcome. These events can be thought of as intrinsic dynamics, but directed structural changes of chromatin at a nucleosome level also take place and are crucial in regulating many genome transactions. We discuss studies of directed structural changes in the following section.
ATP-dependent chromatin remodeling
Another way to regulate DNA accessibility is to slide away or evict nucleosomes. Multi-subunit chromatin remodeling complexes can directly alter nucleosome positions and/or structures. To do so requires overcoming energy barriers that are posed by the DNA-histone interactions within one nucleosome and/or between nucleosomes in an array. Chromatin remodelers are widely hypothesized to provide the energy by hydrolyzing ATP. These ATP-dependent chromatin remodelers are found throughout eukaryotes and many labs have developed biochemical and single molecule assays to study their mechanisms of action [72]. There are at least four families of chromatin remodelers identified: Switch/Sucrose non-fermentable (SWI/SNF), Imitation Switch (ISWI), Chromodomain Helicase DNA-binding (CHD), and Inositol Requiring 80 (INO80) remodelers [73]. It is well established that remodelers can either disorganize nucleosomes by random positioning (e.g. SWI/SNF family) or organize nucleosomes by equally spacing them within nucleosome arrays (e.g. ISWI family). In a single-molecule study by Blosser et al. [74], surface-immobilized mononucleosomes with both DNA and histones fluorescently labeled were used to monitor the process of ATP-dependent chromatin remodeling by a ISWI remodeler complex, ACF (ATP-utilizing chromatin assembly and remodeling factor). By monitoring the actions of individual ACF complexes, they revealed that ACF is a highly processive and bidirectional nucleosome translocase, which might help explain how ACF leads to equally spaced, highly organized nucleosome arrays. The yeast SWI/SNF and RSC (remodel the structure of chromatin) complexes have also been studied using optical and magnetic tweezers [75, 76], sometimes in combination with AFM [77]. These studies revealed nucleosome- and ATP-dependent real-time DNA looping by the ySWI/SNF and RSC complexes. These measurements validated a long-standing hypothesis that chromatin remodelers are ATP-driven motors that translocate DNA by looping. The real-time observation of DNA looping gave valuable insights into how the remodelers unravel DNA from the histone core as it repositions nucleosomes. These studies also directly measured the mechanical force involved in breaking DNA-histone interactions during chromatin remodeling, which is another important factor in the remodeling mechanism. The forces measured were ~12pN, which is well below the 20pN total rupture force required for complete disruption of DNA from the histone octamer. This helps explain how remodelers can reposition nucleosomes without complete disassembly. Two potentially important future directions for these studies are to elucidate how these different remodelers differ in the level of the mechanical force, and to define how their activity correlates with the nucleosome positioning strength of the DNA sequence. As different remodelers are reported to act on different regions of chromatin and their abundance in cells varies widely, discriminating remodelers according to their general and sequence-specific force generating properties will be crucial to elucidate their detailed roles and mechanisms. Another important direction is to discern the relationship between chromatin modifications and remodeling. Thanks to the technical advance in chromatin biochemistry and biology, and single molecule methods, it is now feasible to construct a system comprising nucleosome arrays with endogenous DNA sequences that contain chromatin modifications and chromatin remodelers. This setup would help identify the relevant factors that recruit chromatin remodelers in a DNA sequence- or epigenetic mark-dependent manner, and understand how remodeling complexes act in concert with chromatin modifications to modulate nucleosome structure and dynamics to facilitate various genome processes.
Concluding remarks
Despite the success in identifying and characterizing factors affecting chromatin structures, biophysical insights into how these structures work to facilitate genome transactions are only beginning to emerge. In particular, measuring the intrinsic nucleosome dynamics of mononucleosomes shows how spontaneous nucleosomal DNA un- and rewrapping excursions can regulate DNA-binding protein access to target sequences. Experiments that explored how DNA methylation and histone acetylation affect the structure of nucleosomes and internucleosomal dynamics gave us a first look at how simple chemical changes can alter these dynamics to modulate access to the DNA. These studies provide a foundation to study more complex chromatin problems such as chromatin remodeling. One focus in the near future might be to elucidate how the structural stability of nucleosomes and their dynamics are regulated within a framework that integrates chromatin modifications and ATP-dependent remodeling. Furthermore, model nucleosome arrays can be used in combination with chromatin modifications and remodelers to measure the nucleosome dynamics that are encountered during polymerase binding and processing of DNA. This could open up an opportunity to develop a precise mechanistic understanding of how the inherent thermodynamics and kinetics of nucleosome conformations contribute to gene regulation activities in vivo. Achieving this goal can be greatly facilitated by improving the spatio-temporal resolution of single molecule measurements, aided by innovative electronics technology and/or development of bright and stable fluorophores in conjunction with advances in chromatin biology. Overall, the future holds exciting new promise for single molecule experiments to build on our current understanding in chromatin biology and to expand our view of how chromatin dynamics can have a profound effect on the control of virtually all genome processes.
Box 1. Fluorescence and force methods to study nucleosome structures and dynamics.
FRET [78]
Fluorescence resonance energy transfer. A fluorescently excited fluorophore (donor) in close proximity to another fluorophore (acceptor) whose absorption spectrum overlaps with the emission spectrum of the donor, transfers some of its excited state energy to the acceptor non-radiatively via resonance. Therefore, the distance between the two fluorophores can be estimated based on their fluorescence intensities.
smFRET or spFRET [79]
Single molecule FRET or single pair FRET. FRET between a pair of single fluorophore molecules. By recording the fluorescence intensities of a single FRET pair labeled at a single molecule or complex, one can monitor the FRET distance in a single molecule. If the molecule of interest is immobilized on a surface, one can construct a FRET time trajectory of a single molecule, which represents the time-resolved dynamics of the molecule.
FCS [80]
Fluorescence correlation spectroscopy is a method used to measure the time scales of dynamic processes that modulate the fluorescence intensity of individual fluorophore molecules. This time scale contains information on the dynamics of the molecule attached to the fluorophore. Fluorescence correlation value at time t is a measure of fluorescence recurrence at a time interval t and indicates the extent of the dynamics that yield the fluorescence fluctuation on the time scale t. In general, decreases in the correlation value at a particular time point indicate dynamics on that time scale. For example, if nucleosomes are labeled with a FRET pair and the fluorescence intensities of the two fluorophores change upon the dynamic motions of nucleosomal DNA, in a time scale shorter than the residence time of nucleosomes in the focal volume of a laser beam, the correlation value of the fluorophore intensities would decay at this time scale.
Force measurements with an optical trap [81]
Light exerts force as it refracts off an object, due to conservation of momentum. This principle enables a focused light beam to trap an object that refracts the light to an extent large enough to inhibit its Brownian motion. By measuring the displacement of the trapped object from the center of the trap, one can estimate the force on the object that displaces it. This technique can be applied to measure forces that bind two objects: one component is typically anchored at a fixed position while the other is attached to an object that can be optically trapped. Either the anchored or trapped component can be moved to separate them. At the moment of rupture (when they separate), displacement of the trapped object is measured to estimate the `rupture force'. The rupture force is pulling-rate and trap stiffness (i.e. the spring constant of the trap) dependent, but it is still a good relative measure of the interaction strength between the ruptured components. An optical trap can also be used to monitor the conformational dynamics of a system at a constant trapping force. Here, the displacement of the trapped object is held constant by employing a feedback mechanism to maintain a constant position of the trap relative to the anchored part.
Force measurements with a magnetic trap [82]
The experimental schemes described in the optical trap section can also be implemented with a magnetic trap, except that the trap is a magnetic force applied to an object with a magnetic dipole. In addition to measuring/controlling force based on the linear displacement of the trapped object, one can also measure/control the rotational degree of a trapped object in a magnetic trap, which is the basis of the topology measurements/control in double stranded DNA systems.
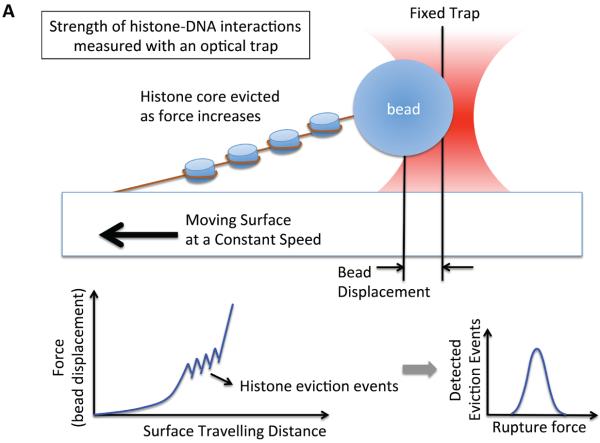
Figure 2. Single molecule studies of nucleosomes with optical tweezers.
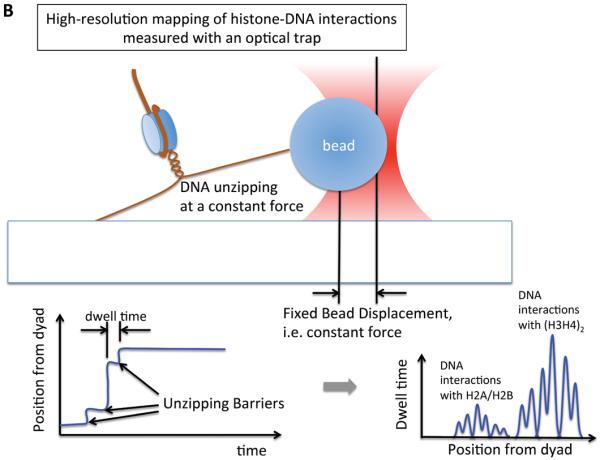
(a) A setup based on using optical trapping to study the interaction strengths between DNA and histones in a nucleosome array. One end of the array is attached to a glass surface and the other end is attached to a plastic bead that is trapped within a focus of a laser beam. By moving the slide in one direction, the force to elongate the array is gradually increased. As the force increases, histones are evicted and the force (i.e. the distance between the two points of attachment) decreases sharply at the moment of eviction because histone eviction yields increases in the length of the array. Multiple histone eviction events are observed within an array as depicted in the chart. (b) A setup based on optical trapping that gives a high-resolution map of interactions between nucleosomal DNA and core histones [31]. Each end of the nucleosomal DNA is anchored and the measurements were made for a single nucleosome. The force to unzip the DNA is held constant by maintaining the distance between the two anchored points constant. When the applied force is slightly above the unzipping force, DNA is unzipped stepwise through multiple unzipping barriers. When the resolution of force control/measurement is high, the unzipping barriers would be detected as schematized in the chart and bear information on the interactions between the DNA and histones within a nucleosome.
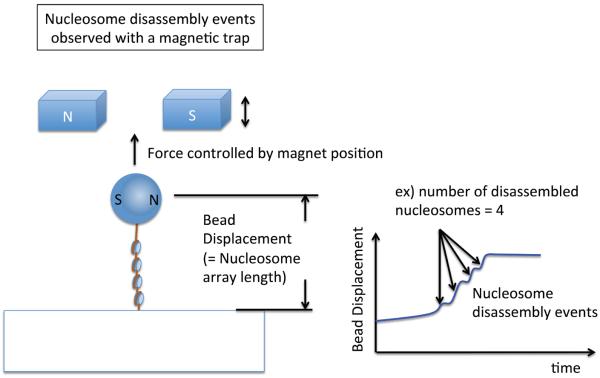
Figure 3. Single molecule studies of nucleosome structure with magnetic tweezers.
A magnetic tweezers setup to observe nucleosome eviction events in a nucleosome array. One end of the array is attached to a glass surface and the other end is attached to a magnetic bead trapped in a magnetic field. The vertical position of the magnet is held constant to apply a constant force along the array. When the force is slightly above the nucleosome rupture force, multiple nucleosome dissociation events are detected as depicted in the chart.
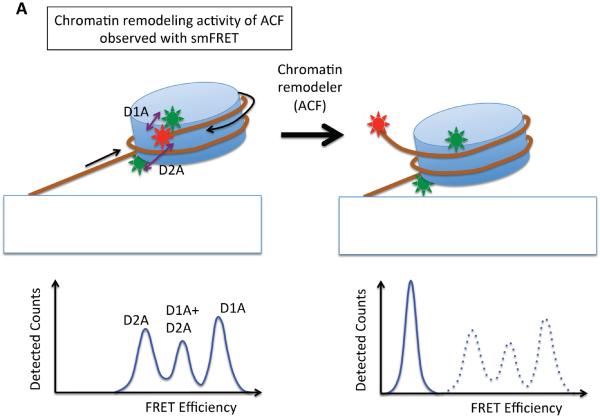
Figure 4. Single molecule studies of ATP-dependent chromatin remodeling.
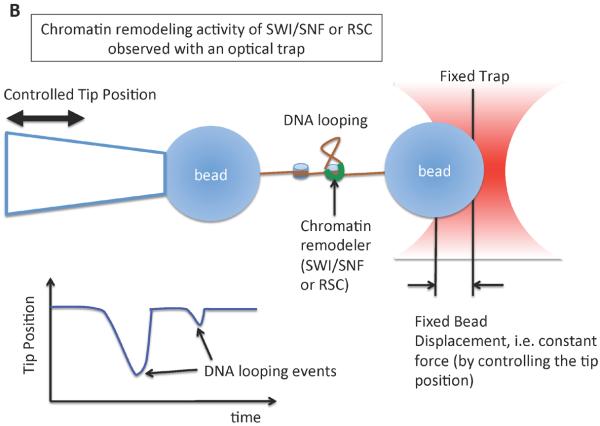
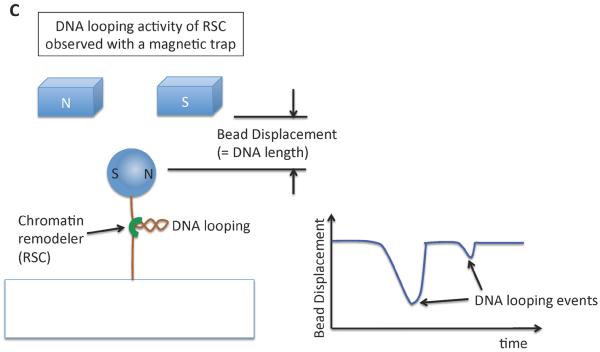
(a) A single molecule fluorescence resonance energy transfer (FRET) setup to study nucleosome sliding by the ACF remodeling complex [74]. Three FRET populations were observed from nucleosomes: one from D1A FRET pair (the peak marked as D1A in the FRET histogram), another from D2A pair (peak D2A in the histogram) and the third from the two-donor and one-acceptor FRET (some nucleosomes have two donor fluorophores according to the stoichiometry of the labeled histone in the histone core, marked D1A+D2A in the histogram). Consistent with nucleosome sliding mediated by ACF, a dramatic FRET change was observed upon the addition of ACF and ATP. (b) An optical tweezers setup to monitor the structure of a short nucleosome array altered by SWI/SNF or RSC remodeling complexes in the presence of ATP [75]. Several reversible changes in the length of the array were observed upon the addition of SWI/SNF (or RSC) and ATP, suggesting DNA looping events induced by SWI/SNF remodeling. (c) A magnetic tweezers setup used to study the topology change of DNA caused by the remodeling activity of the RSC complex [77]. The study revealed that RSC induces superhelical loops of DNA in an ATP dependent manner.
Glossary Box
- Ensemble-averaging
The term `ensemble' means a set of an often infinitely large number of imaginary copies of a system in all states accessible by the system. The averaged properties of an ensemble represent the averaged properties of many identical systems, or the properties of one system averaged over a long period of time.
- Fluorophore blinking
Fluorescence typically involves transitions between different energy states of a molecule. When excited molecules immediately decays by emitting a photon, this process is called fluorescence. However, an excited molecule can also be trapped for an extended period of time in an intermediate state without any radiative decay path to the ground state leaving the molecule non-fluorescent for that time. The molecule can escape the trapped state and become fluorescent again. Reversible trapping and escaping causes blinking of the fluorescenct molecule. Blinking can be observed only at a single fluorophore level and is not observable in an ensemble.
- Reaction coordinate
A reaction coordinate is typically a one-dimensional coordinate along which the change in the property of a system relevant to the reaction can be described. Measuring the rupture force of a molecular complex with optical tweezers, the reaction coordinate could be the overall stretch of all the chemical and physical bonds along the pulling direction between the two anchored points if all the bond strengths are equal to that direction. Alternatively, it could be the combined length of a few specific bonds that are considerably weaker than the others and consequently most directly relevant to the rupture process.
- Fluorescence polarization (anisotropy)
An electromagnetic wave (light) can oscillate along two perpendicular primitive axes.. The orientation of the oscillation determines the polarization state of light. The polarization state of light is altered when the source of light changes its rotational position. If a polarized excitation wave is used, the fluorescence from the excited molecules will also be polarized if the molecules do not rotate during the emission cycle. If the molecule rotates during the excitation-emission cycle, the fluorescence polarization level will be reduced. The level of reduction can be used to study the rotational motion of a fluorophore or a fluorophore-labeled molecule in the time scale of the fluorescence excitation-emission cycle.
- Native chemical ligation
Native chemical ligation is a technique to construct a large polypeptide from smaller peptide fragments [71]. This technique requires thioester modification at the C terminus of a peptide and a cysteine residue at the N terminus of the other peptide to be ligated. Upon ligation, a desulfurization reaction can be employed to convert the ligation point to an alanine residue [83].
References
- 1.Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD, Thomas JO. Chromatin structure; oligomers of the histones. Science. 1974;184:865–868. doi: 10.1126/science.184.4139.865. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 4.Richmond TJ, et al. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 5.Klug A, et al. A low resolution structure for the histone core of the nucleosome. Nature. 1980;287:509–516. doi: 10.1038/287509a0. [DOI] [PubMed] [Google Scholar]
- 6.Luger K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 7.Vettese-Dadey M, et al. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 8.Simpson RT. Structure of chromatin containing extensively acetylated H3 and H4. Cell. 1978;13:691–699. doi: 10.1016/0092-8674(78)90219-2. [DOI] [PubMed] [Google Scholar]
- 9.Simpson RT, et al. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985;42:799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- 10.Chahal SS, et al. Acetylation of histone H4 and its role in chromatin structure and function. Nature. 1980;287:76–79. doi: 10.1038/287076a0. [DOI] [PubMed] [Google Scholar]
- 11.Antequera F, et al. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989;58:509–517. doi: 10.1016/0092-8674(89)90431-5. [DOI] [PubMed] [Google Scholar]
- 12.Ausio J, van Holde KE. Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry. 1986;25:1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- 13.Bauer WR, et al. Nucleosome structural changes due to acetylation. J Mol Biol. 1994;236:685–690. doi: 10.1006/jmbi.1994.1180. [DOI] [PubMed] [Google Scholar]
- 14.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 15.Pikaart MJ, et al. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thastrom A, et al. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J Mol Biol. 1999;288:213–229. doi: 10.1006/jmbi.1999.2686. [DOI] [PubMed] [Google Scholar]
- 17.Gottesfeld JM, Luger K. Energetics and affinity of the histone octamer for defined DNA sequences. Biochemistry. 2001;40:10927–10933. doi: 10.1021/bi0109966. [DOI] [PubMed] [Google Scholar]
- 18.Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 19.Glatt S, et al. Recognizing and remodeling the nucleosome. Curr Opin Struct Biol. 21:335–341. doi: 10.1016/j.sbi.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Bhaumik SR, et al. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- 21.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 22.Li G, et al. Rapid spontaneous accessibility of nucleosomal DNA. Nature Struct. Mol. Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 23.Tims HS, et al. Dynamics of nucleosome invasion by DNA binding proteins. J Mol Biol. 411:430–448. doi: 10.1016/j.jmb.2011.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gansen A, et al. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc Natl Acad Sci U S A. 2009;106:15308–15313. doi: 10.1073/pnas.0903005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomschik M, et al. Nucleosome Dynamics as Studied by Single-pair Fluorescence Resonance Energy Transfer: A Reevaluation. Journal of Fluorescence. 2009;19:53–62. doi: 10.1007/s10895-008-0379-1. [DOI] [PubMed] [Google Scholar]
- 26.Tomschik M, et al. Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 2005;102:3278–3283. doi: 10.1073/pnas.0500189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bohm V, et al. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2010;39:3093–3102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koopmans WJ, et al. Single-pair FRET microscopy reveals mononucleosome dynamics. J Fluoresc. 2007;17:785–795. doi: 10.1007/s10895-007-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luger K, et al. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 1999;119:1. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 30.Koopmans WJA, et al. spFRET Using Alternating Excitation and FCS Reveals Progressive DNA Unwrapping in Nucleosomes. Biophys J. 2009;97:195–204. doi: 10.1016/j.bpj.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall MA, et al. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Forties RA, et al. A quantitative model of nucleosome dynamics. Nucleic Acids Res. 2011;39:8306–8313. doi: 10.1093/nar/gkr422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brower-Toland BD, et al. Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc Natl Acad Sci U S A. 2002;99:1960–1965. doi: 10.1073/pnas.022638399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennink ML, et al. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat Struct Biol. 2001;8:606–610. doi: 10.1038/89646. [DOI] [PubMed] [Google Scholar]
- 35.Bancaud A, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 36.Shlyakhtenko LS, et al. Dynamics of nucleosomes revealed by time-lapse atomic force microscopy. Biochemistry. 2009;48:7842–7848. doi: 10.1021/bi900977t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mihardja S, et al. Effect of force on mononucleosomal dynamics. Proc Natl Acad Sci U S A. 2006;103:15871–15876. doi: 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gemmen GJ, et al. Forced unraveling of nucleosomes assembled on heterogeneous DNA using core histones, NAP-1, and ACF. J Mol Biol. 2005;351:89–99. doi: 10.1016/j.jmb.2005.05.058. [DOI] [PubMed] [Google Scholar]
- 39.Claudet C, et al. Histone octamer instability under single molecule experiment conditions. J Biol Chem. 2005;280:19958–19965. doi: 10.1074/jbc.M500121200. [DOI] [PubMed] [Google Scholar]
- 40.Andrews AJ, et al. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37:834–842. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park YJ, Luger K. The structure of nucleosome assembly protein 1. Proc Natl Acad Sci U S A. 2006;103:1248–1253. doi: 10.1073/pnas.0508002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowman A, et al. The histone chaperones Nap1 and Vps75 bind histones H3 and H4 in a tetrameric conformation. Mol Cell. 2011;41:398–408. doi: 10.1016/j.molcel.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poirier MG, et al. Spontaneous access to DNA target sites in folded chromatin fibers. J Mol Biol. 2008;379:772–786. doi: 10.1016/j.jmb.2008.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poirier MG, et al. Dynamics and function of compact nucleosome arrays. Nat Struct Mol Biol. 2009;16:938–944. doi: 10.1038/nsmb.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Studitsky VM, et al. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- 46.Studitsky VM, et al. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- 47.Clark DJ, Felsenfeld G. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell. 1992;71:11–22. doi: 10.1016/0092-8674(92)90262-b. [DOI] [PubMed] [Google Scholar]
- 48.O'Neill TE, et al. Histone octamer dissociation is not required for transcript elongation through arrays of nucleosome cores by phage T7 RNA polymerase in vitro. Proc Natl Acad Sci U S A. 1993;90:6203–6207. doi: 10.1073/pnas.90.13.6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodges C, et al. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 51.Muthurajan UM, et al. Structure and dynamics of nucleosomal DNA. Biopolymers. 2003;68:547–556. doi: 10.1002/bip.10317. [DOI] [PubMed] [Google Scholar]
- 52.Blossey R, Schiessel H. The dynamics of the nucleosome: thermal effects, external forces and ATP. FEBS J. 278:3619–3632. doi: 10.1111/j.1742-4658.2011.08283.x. [DOI] [PubMed] [Google Scholar]
- 53.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16(Spec No 1):R50–59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 54.Choy JS, et al. DNA Methylation Increases Nucleosome Compaction and Rigidity. Journal of the American Chemical Society. 2010;132:1782. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee J, Lee T-H. Effects of DNA methylation on the structure of nucleosomes. J Am Chem Soc. 2011 doi: 10.1021/ja210273w. in pres. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Widom J. Role of DNA sequence in nucleosome stability and dynamics. Quarterly reviews of biophysics. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 57.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Hayes JJ. Site-specific binding affinities within the H2B tail domain indicate specific effects of lysine acetylation. J Biol Chem. 2007;282:32867–32876. doi: 10.1074/jbc.M706035200. [DOI] [PubMed] [Google Scholar]
- 59.Wang X, Hayes JJ. Acetylation mimics within individual core histone tail domains indicate distinct roles in regulating the stability of higher-order chromatin structure. Mol Cell Biol. 2008;28:227–236. doi: 10.1128/MCB.01245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 61.Loyola A, et al. Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription. Genes Dev. 2001;15:2837–2851. doi: 10.1101/gad.937401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kan PY, et al. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol. 2009;29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sinha D, Shogren-Knaak MA. Role of Direct Interactions between the Histone H4 Tail and the H2A Core in Long Range Nucleosome Contacts. Journal of Biological Chemistry. 2010;285:16572. doi: 10.1074/jbc.M109.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gansen A, et al. Structural variability of nucleosomes detected by single-pair Forster resonance energy transfer: histone acetylation, sequence variation, and salt effects. J Phys Chem B. 2009;113:2604–2613. doi: 10.1021/jp7114737. [DOI] [PubMed] [Google Scholar]
- 65.Lee JY, et al. Effects of histone acetylation by Piccolo NuA4 on the structure of a nucleosome and the interactions between two nucleosomes. J Biol Chem. 2011;286:11099–11109. doi: 10.1074/jbc.M110.192047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brower-Toland B, et al. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J Mol Biol. 2005;346:135–146. doi: 10.1016/j.jmb.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 67.Neumann H, et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol Cell. 2009;36:153–163. doi: 10.1016/j.molcel.2009.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon M, et al. Histone fold modifications control nucleosome unwrapping and disassembly. Proc Natl Acad Sci U S A. 2011;108:12711–12716. doi: 10.1073/pnas.1106264108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Allahverdi A, et al. The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 2011;39:1680–1691. doi: 10.1093/nar/gkq900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang D, Arya G. Structure and binding of the H4 histone tail and the effects of lysine 16 acetylation. Phys Chem Chem Phys. 2011;13:2911–2921. doi: 10.1039/c0cp01487g. [DOI] [PubMed] [Google Scholar]
- 71.Camarero JA, Muir TW. Native chemical ligation of polypeptides. Curr Protoc Protein Sci. 2001;Chapter 18(Unit18):14. doi: 10.1002/0471140864.ps1804s15. [DOI] [PubMed] [Google Scholar]
- 72.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 73.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 74.Blosser TR, et al. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, et al. DNA Translocation and Loop Formation Mechanism of Chromatin Remodeling by SWI/SNF and RSC. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sirinakis G, et al. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. EMBO J. 2011;30:2364–2372. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lia G, et al. Direct Observation of DNA Distortion by the RSC Complex. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Förster T. Zwischenmolekulare energiewanderung und fluoreszenz. Annalen der Physik. 1948;437:55–75. [Google Scholar]
- 79.Ha T, et al. Probing the interaction between two single molecules: fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci U S A. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ehrenberg M, Rigler R. Fluorescence correlation spectroscopy applied to rotational diffusion of macromolecules. Q Rev Biophys. 1976;9:69–81. doi: 10.1017/s003358350000216x. [DOI] [PubMed] [Google Scholar]
- 81.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lionnet T, et al. Single-molecule studies using magnetic traps. Cold Spring Harb Protoc. 2012;2012:34–49. doi: 10.1101/pdb.top067488. [DOI] [PubMed] [Google Scholar]
- 83.Wan Q, Danishefsky SJ. Free-radical-based, specific desulfurization of cysteine: a powerful advance in the synthesis of polypeptides and glycopolypeptides. Angew Chem Int Ed Engl. 2007;46:9248–9252. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]