Abstract
Cycloserine (CS, 4-amino-3-isoxazolidone) is a cyclic amino acid mimic that is known to inhibit many essential pyridoxal 5′-phosphate (PLP)-dependent enzymes. Two CS enantiomers are known; d-cycloserine (DCS, also known as Seromycin), is a natural product that is used to treat resistant Mycobacterium tuberculosis infections as well as neurological disorders since it is a potent NMDA receptor agonist, and l-cycloserine (LCS), is a synthetic enantiomer whose usefulness as a drug has been hampered by its inherent toxicity arising through inhibition of sphingolipid metabolism. Previous studies on various PLP-dependent enzymes revealed a common mechanism of inhibition by both enantiomers of CS; the PLP cofactor is disabled by forming a stable 3-hydroxyisoxazole/pyridoxamine 5′-phosphate (PMP) adduct at the active site where the cycloserine ring remains intact. Here we describe a novel mechanism of CS inactivation of the PLP-dependent enzyme serine palmitoyltransferase (SPT) from Sphingomonas paucimobilis. SPT catalyses the condensation of l-serine and palmitoyl-CoA, the first step in the de novo sphingolipid biosynthetic pathway. We have used a range of kinetic, spectroscopic and structural techniques to postulate that both LCS and DCS inactivate SPT by transamination to form a free pyridoxamine 5′-phosphate (PMP) and β-aminooxyacetaldehyde that remain bound at the active site. We suggest this occurs by ring opening of the cycloserine ring followed by decarboxylation. Enzyme kinetics show that inhibition is reversed by incubation with excess PLP and that LCS is a more effective SPT inhibitor than DCS. UV-visible spectroscopic data, combined with site-directed mutagenesis, suggest that a mobile Arg378 residue is involved in cycloserine inactivation of SPT.
Introduction
Sphingolipids are a large family of bioactive molecules that are found in all eukaryotic and some prokaryotic membranes. An important example is sphingomyelin, a constituent of the protective myelin sheath that surrounds nerve cells (1). Sphingolipids can associate with cholesterol to form ‘lipid rafts’ or sphingolipid-based microdomains necessary for signal transduction and membrane trafficking (2). Sphingolipid metabolites such as ceramides and sphingosine-1-phosphate play important roles in cell proliferation, differentiation and apoptosis (3-5) and it follows that pharmaceutical intervention that regulates the sphingolipid metabolic pathway could help to combat pathological processes such as carcinogenesis (6), atherosclerosis (7) and Parkinson’s disease (8).
The de novo biosynthetic pathway for sphingolipids varies from one organism to another but the first and rate-limiting step is common to all: condensation of l-serine with palmitoyl-CoA to form 3-ketodihydrosphingosine (9). This step is catalysed by the pyridoxal 5′-phosphate (PLP)-dependent enzyme serine palmitoyltransferase (SPT), a member of the α-oxoamine synthase (AOS) subfamily. Other members of this subfamily that have been well characterised include 8-amino-7-oxononanoate synthase (AONS) (10,11), 5-aminolevulinate synthase (ALAS) (12,13) and 2-amino-3-ketobutyrate-CoA ligase (KBL) (14). These enzymes catalyse reactions in heme biosynthesis, biotin biosynthesis and threonine degradation, respectively. Recent additions to the growing list of identified AOS enzymes are the bacterial quorum-sensing autoinducer synthases CqsA in Vibrio cholerae (15-17) and LqsA in Legionella pneumophila (17).
The AOS enzymes utilise a PLP cofactor at the active site to catalyse the Claisen-like condensation between an amino acid and an acyl-CoA substrate. The first high resolution crystal structure of the holo-form of a bacterial, homodimeric SPT from Sphingomonas paucimobilis clearly shows the cofactor covalently attached to the side-chain of a conserved Lys265 residue via a Schiff’s base (also known as an internal aldimine) at the dimer interface (18). Transaldimination occurs when the l-serine substrate binds at the active site to form an external aldimine that is stabilised by other conserved residues; a His159 that stacks above the PLP ring and His234 that hydrogen bonds with the PLP-bound intermediate (19). The crystal structure of the enzyme-bound PLP:l-serine external aldimine complex has recently been resolved for SPTs from the sphingolipid-producing bacteria S. paucimobilis (20) and Sphingobacterium multivorum (21). The proposed steps subsequent to formation of the external aldimine are: deprotonation at Cα of the external aldimine complex to form a quinonoid (carbanion equivalent) intermediate; a Claisen condensation with the acyl-CoA substrate and loss of free CoASH to form a β-ketoacid intermediate; decarboxylation to form a product quinonoid; protonation of this quinonoid to form the product external aldimine; release of the α-oxoamine product and regeneration of the enzyme PLP internal aldimine (Fig. 1).
Fig. 1.
Catalytic mechanism of SPT.
Both enantiomers of cycloserine (Fig. 2A) can be thought of as cyclic analogues of serine and/or alanine and have been shown to be irreversible inhibitors of many PLP-dependent transaminases (22), racemases (23) and decarboxylases (24). l-cycloserine (LCS) is prepared synthetically whereas d-cycloserine (DCS) is a natural product isolated from Streptomyces strains (25) and is a broad spectrum antibiotic. Due to its severe side effects DCS is most commonly used as a second-line drug in combination therapy to treat tuberculosis (26). DCS is also used in neurological studies since it is a potent agonist of N-methyl-d-aspartic acid (NMDA) receptors that are involved in neurotransmission (27). One of its main antibacterial targets is the PLP-dependent alanine racemase (28,29), an essential enzyme that generates d-alanine for the formation of the d-alanyl-d-alanine dipeptide incorporated into the bacterial peptidoglycan layer. Unlike many irreversible inhibitors that inactivate their protein targets by covalent modification, CS renders its targets inactive by forming a stable adduct with the essential PLP cofactor. It is not surprising that CS has also been shown to be a potent inhibitor of bacterial SPT (30) as well as mammalian SPT in mouse brain microsomes (31). Although LCS is commonly used as a regulator of lipid metabolism in biological research, its mechanism of inhibition is still unknown. In this paper we used a combination of x-ray crystallography, mass spectrometry, UV-Vis spectroscopy and enzyme kinetics to elucidate the mechanism of SPT inactivation by both enantiomers of CS. We highlight differences in the enantiospecific inhibition observed for both LCS and DCS and provide further insight into PLP-dependent chemistry.
Fig. 2.
(A) Structures of SPT substrates and inhibitors. (B) Mechanism of formation of the aromatic adduct observed in each of the crystal structures shown in Table 1. (C) Proposed mechanism of LCS inactivation of SPT by Ikushiro et al. (30).
Results
Both LCS and DCS are irreversible inhibitors of serine palmitoyltransferase
When either LCS or DCS was incubated with SPT, enzyme activity decreased over time. Log plots of % activity remaining versus time, according to the method of Kitz and Wilson (32), were linear for each enantiomer indicating classical time-dependent inactivation of SPT activity (Fig. 3A and 3B). We noted that the Kitz and Wilson secondary plots (Fig. 3C) appeared to pass very close to the origin - there are many examples of this in the literature such as the inhibition of the thiamine-dependent enzyme benzaldehyde lyase (BAL) by methyl benzoylphosphonate (33) and the inhibition of the PLP-dependent γ-aminobutyric acid aminotransferase (GABA-AT) by (1S,3S)-3-amino-4-difluoromethylene-cyclopentanecarboxylic acid (34). Kitz and Wilson replots of this nature suggest that both DCS and LCS form very weak complexes with SPT but inactivation is fast compared with the formation of the CS:SPT complexes. Moreover, these prevent the estimation of the kinact and KI values directly. In comparison with LCS, we found that approximately 15-fold higher concentrations of DCS were required to inactivate SPT activity to the same extent and over a similar period of time. From the secondary plot of 1/kapp versus 1/[cycloserine] the second-order rate constant of inactivation kinact/KI was calculated as 1/slope for each inhibitor (Fig. 3C). LCS (kinact/KI = 0.83 ± 0.5 M−1 s−1) was found to be ~14-fold more effective at inactivating SPT than DCS (kinact/KI = 0.06 ± 0.002 M−1 s−1).
Fig. 3.
(A) Inactivation of SPT by LCS at inhibitor concentrations of 0, 0.625, 1.25, 2.5 and 5 mM. (B) Inactivation of SPT by DCS at inhibitor concentrations of 0, 10, 20, 40 and 80 mM. (C) Secondary plot of 1/kapp versus 1/[inhibitor] for LCS and DCS.
Activity of SPT, measured by monitoring formation of the free thiol of CoASH using a continuous DTNB assay (20) was reduced to 1 % and 22 % after 2 hours inhibition with 5 mM LCS and DCS, respectively (Fig. 4). When these samples were dialysed against buffer in the absence of free PLP we noted that enzyme activity was not recovered. In contrast, activity returned to 83 % (LCS) and 79 % (DCS) of the original activity after dialysis against buffer containing 25 μM PLP. The fact that activity can be recovered upon dialysis against PLP shows that inactivation by both enantiomers does not occur through covalent modification of the protein but rather by disabling the PLP cofactor.
Fig. 4.
Inhibition of SPT activity by LCS and DCS and regeneration of activity by dialysis against PLP buffer. SPT activity of the following samples was measured by DTNB assay: (A). 20 μM SPT (B). 20μM SPT and 5 mM LCS at 25°C after 2 hour incubation (C) Sample B after dialysis against 20 mM potassium phosphate buffer pH 7.5 (D) Sample B after dialysis against 20 mM potassium phosphate pH 7.5 and containing 25 μM PLP (E) 20 μM SPT and 5 mM DCS at 25°C after 2 hour incubation (F) Sample E after dialysis against 20 mM potassium phosphate buffer pH 7.5 (G) Sample E after dialysis against 20 mM potassium phosphate pH 7.5 and containing 25 μM PLP.
UV-visible spectroscopy analysis of LCS and DCS binding to holo- SPT
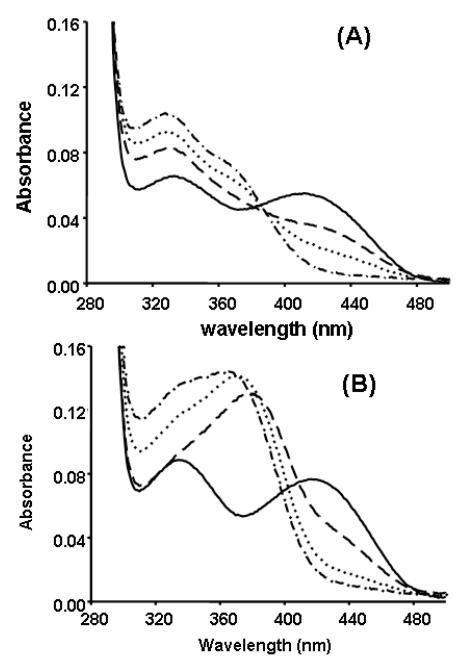
The UV-visible spectrum of holo-SPT displays absorbance maxima at 336 nm and 425 nm corresponding to the enolimine and ketoenamine forms of the PLP-bound enzyme, respectively (Fig. 5A and 5B, solid line). When LCS (5 mM) was added to the enzyme at pH 7.5 and 25°C, notable changes in these peaks occurred suggesting that LCS interacts with the PLP cofactor (Fig. 5A, dashed lines). Over a period of ~30 minutes the ketoenamine peak (425 nm) was reduced to ~10 % of its original value with concomitant growth of new peaks at 330 nm and 380 nm. No further changes in this UV-vis spectrum were observed when the sample was incubated for ~8hrs. This suggests that the PLP cofactor bound to residue Lys265 as an internal aldimine is displaced and one or more new species are formed.
Fig. 5.
(A) SPT and 5 mM LCS at time 0 (solid line), 30 seconds (long dash), 1 minute (dotted line) and 30 minutes (dash dot). (B) SPT and 5 mM DCS at time 0 (solid line), 30 seconds (long dash), 2 hours (dotted line) and 10 hours (dash dot).
A number of mechanisms have been proposed for cycloserine inhibition of a range of PLP-dependent enzymes and have been collated by Olson et al. (35). In one mechanism originally proposed for the inhibition of alanine racemase, the authors speculate that the cycloserine ring of the inhibitor-PLP adduct is opened after attack by either an active site nucleophile on the enzyme or hydroxide ion (36,37). In the case of S. paucimobilis SPT Ikushiro et al. (30) proposed that hydrolysis of the ring-open adduct leads to formation of free pyridoxamine 5′-phosphate (PMP) and beta-aminooxypyruvate (Fig. 2C). They speculated that the 380 nm peak that they observed upon incubation of SPT with LCS could be due to formation of a transient oxime intermediate while the 330 nm peak is due to the ring-opened adduct prior to hydrolysis (Fig. 2C). Of interest, a similar UV-vis spectrum was obtained in a study of the interaction of LCS with ArnB, the PLP-dependent aminotransferase involved in amino arabinose biosynthesis (38). X-ray structure analysis of that inactivated enzyme revealed a bound hydroxyisoxazole-PLP adduct at the ArnB active site. Therefore it appears that LCS can interact with its target enzymes in different ways with the cycloserine ring remaining intact or undergoing ring cleavage.
Dramatic changes in the PLP absorbance spectrum of holo-SPT were also observed within 30 seconds after addition of DCS (Fig. 5B, dashed line) but changes thereafter were much slower compared to LCS, occurring over a period of ~10 hours (Fig. 5B, dotted and dot-dash lines). These spectroscopic changes correlate with the observation that DCS inactivates SPT activity at a slower rate than LCS (see Fig. 3). Loss of the 425 nm peak was accompanied by appearance of a new peak at 380 nm. Over time (~2-10 hrs) this peak shifted to 365 nm along with the slow appearance of a pronounced, broad shoulder with λmax at 330 nm. In contrast to the changes that occurred in the presence of LCS the 380 nm peak was the most dominant and not the 330 nm peak. Since DCS is a much slower inhibitor than LCS a build-up of intermediates along with slower formation of products might be expected and tends to agree with Ikushiro’s proposition (30) that the LCS-derived 380 nm peak could be that of an oxime intermediate, while the slow build-up of products is represented by the slowly-formed 330 nm shoulder.
The UV-vis spectra of holo-SPT in the presence of LCS and DCS (Fig. 5A and 5B) are very different to each other. In contrast, the spectra of free PLP in the presence of both CS enantiomers are identical and display maxima at 360 nm (Supplementary Fig. 1A and 1B). Therefore it is clear that the enzyme does not simply accommodate a PLP-cycloserine aldimine that is formed between the cofactor and inhibitor in solution. The new 330 nm peak observed in the spectrum of the enzyme in the presence of both CS enantiomers is similar to the λmax (320 nm) for free PMP (Supplementary Fig. 1C) and could be due to bound PMP.
Identification of SPT-cycloserine reaction products by LC-MS
The reaction products after inactivation of SPT by both CS enantiomers were analysed by LC ESI-MS (Supplementary Fig. 2). After 30 mins inactivation, low molecular weight products were separated from the protein by centrifugation through a filter with a 3 kDa cut-off. Analysis of this sample by LC ESI-MS showed a peak in the single ion monitoring (SIM) at m/z = 248 with a total ion count (TIC) above background at 2.8 min that corresponds to PMP (Supplementary Fig. 2B). We were unable to detect an ion above background corresponding to the aminooxyaldehyde so the filtrate was derivatised with 2,4-dinitrophenylhydrazine (2,4-DNP) to form the corresponding hydrazone (chemical formula C8H9N5O5 = 255.06, Supplementary Fig. 2A). When the 2,4-DNP derivatised sample was analysed by LC-MS a clear peak eluted from the C18 column at 3.1 minutes that ionized very well above background (Supplementary Fig. 2C). Electrospray MS analysis of this peak revealed it to have m/z = 256 (Supplementary Fig. 2C) which corresponds to the [M+H]+ ion of the 2,4-DNP derivative of β-aminooxyacetaldehyde derived from LCS. No peaks were observed for free PLP and a 2,4-DNP derivative of the PLP aldehyde. These peaks were not observed for control SPT samples treated in the same way but in the absence of cycloserine (data not shown). We carried out a similar analysis of SPT after incubation with DCS. We observed the same peaks as seen in the LCS analysis but the total ion counts were much lower overall and the UV trace was more complicated, unsurprising given the slower rate of reaction with this enantiomer (data not shown). We conclude that LCS inactivation of SPT results in the formation of PMP and β-aminooxyacetaldehyde from the cycloserine ring.
Structure of S. paucimobilis SPT after inactivation by l-cycloserine
Crystals of SPT:LCS-derived complex were prepared by co-crystallisation in the presence of 1 mM LCS. Colourless crystals were obtained that diffracted to a resolution of 1.4 Å. We used our previous 1.3 Å resolution structure of the holo-form of S. paucimobilis SPT with the PLP and water molecules to solve the structure of this new complex (18). It was clear that in the SPT:LCS complex the PLP cofactor is no longer covalently attached to the sidechain of Lys265 (contoured at 3σ and 0.2 Å−3) and that the lysine side-chain adopts a conformation similar to that observed in the SPT:l-ser external aldimine complex (20) (Fig. 6A). The experimental electron density cannot definitively assign the co-factor as either PLP (an aldehyde) or PMP (amine), as the density is very weak for the terminal atom. However, in light of the mass spectrometry results we have modeled the co-factor as PMP. There is no additional density that could be modeled by either free cycloserine or an isoxazole ring, which have been observed in other cycloserine-inactivated PLP dependent enzymes (Table 1). Nor could such additional groups fit into the structure given the position of the side chains and a clearly identified water molecule (Fig. 6B). We do note that unusually the side chain of Tyr73 is disordered in the structure. The aromatic ring of the pyridoxal co-factor essentially rotates ~23 ° about an axis which links the C2 and C5 atoms. The phosphate group has not moved and the interactions with the protein are largely conserved, although there are some changes in water structure. In this new structure the side chains of Arg378 and Gln357 which were noted to make a salt interaction in the holo form (18) are disordered confirming this is a flexible region of the protein. In contrast, in the PLP:l-ser complex the loop containing Arg378 moves into the active site to allow the Arg378 side-chain to make a salt contact with l-ser carboxylate (Fig. 6B). Interestingly there is an additional ‘blob’ of difference electron density lying adjacent to the PMP cofactor and parallel to Lys265. This blob sits at the interface of the two subunits of the functional SPT dimer. If the density is contoured at 0.16 e Å−3 (2.7σ ) it has a continuous nature (Fig. 6B). We have not fitted this density in the final model but in light of the mass spectrometry data have rendered it as the β-aminooxyacetaldehyde. We have been unable to grow x-ray diffraction quality crystals of DCS-inactivated SPT and this is the focus of current research.
Fig. 6.
(A) Fo-Fc electron density for the PMP molecule. This was calculated from molecular replacement model which was refined and had omitted the co-factor. The map is contoured in green at 3σ (0.2 Å−3). Also shown are the side chains of Lys265, His159, Asp231 and His234. Monomer A is shown in ribbon in cyan and monomer B in magenta. The additional Fo-Fc electron density “blob” is shown in blue, contoured at 2.7 σ (0.2 Å−3). A molecule of the β-aminooxyacetalaldehyde identified by mass spectrometry is placed in the density. Carbons are colored yellow, nitrogen blue, oxygen red and phosphorous orange. (B) Overlay of SPT:l-ser (2bwj) and the LCS inactivated form. The loop containing R378 adopts the “swung out” conformation in the LCS form, in contrast to the “swung in” conformation in SPT:l-ser. The color scheme for the LCS inhibited form is as Fig. 6A. For the SPT:l-ser structure, monomer A is colored light blue. Carbons are colored green, other atoms are colored the same as in Fig. 6A. The main chain of R378 adopts a very different conformation from the SPT:L-ser structure because the salt contact with l-ser is missing. A well ordered molecule (red sphere) in the LCS structure is found in the same location as the l-ser carboxylate. The side chain of l-ser points towards the unfitted blob at the active site. This figure was prepared using PyMol (http://www.pymol.org/).
Table 1. PDB entries for PLP-dependent enzymes inactivated by CS.
| Enzyme | Organism | Enantiomer | PDB | Reference |
|---|---|---|---|---|
| Alanine racemase | Enterococcus faecalis | DCS | 3E6E | (60) |
| Alanine racemase | Escherichia coli | DCS | 2RJH | (61) |
| Alanine racemase | Streptomyces lavendulae | LCS | 1VFT | (62) |
| Alanine racemase | Streptomyces lavendulae | DCS | 1VFS | (62) |
| Alanine racemase | Bacillus stearothermophilus | LCS | 1NIU | (23) |
| Alanine racemase | Bacillus stearothermophilus | DCS | 1EPV | (23) |
| Aminodeoxychorismate lyase | Escherichia coli | DCS | 1I2L | (63) |
| ArnB aminotransferase | Salmonella typhimurium | LCS | 1MDZ | (38) |
| Dialkylglycine decarboxylase | Pseudomonas cepacia | DCS & LCS | 1D7S | (24) |
| D-amino acid transferase | Bacillus species | DCS | 2DAA | (46) |
Inactivation of a SPT R378N mutant by LCS and DCS
We recently determined the crystal structure of the SPT:l-serine external aldimine complex that allowed us to identify any changes that occur during formation of this key intermediate from the internal aldimine holo-SPT form. The major difference between the two forms was the movement of Arg378 from a “swung-out” position (bound to the side-chain of Gln357) to a “swung in” position allowing it to form a salt bridge with the −CO2− group of the l-ser substrate (20) (Fig. 6B). This Arg378 residue appears to be highly mobile in S. paucimobilis SPT and we had probed its role in catalysis and found that a SPT R378N mutant was still active albeit at ~40x fold reduced rate when compared with the wild-type enzyme. We inhibited the SPT R378N mutant with LCS and DCS to investigate the mechanism of cycloserine inhibition. The UV-visible spectrum of the SPT R378N holo-enzyme is similar to that of the wild-type SPT showing absorbance maxima at 336 nm and 415 nm corresponding to the enolimine and ketoenamine tautomers of the PLP-bound enzyme, respectively (Fig. 7A and 7B, solid line). When LCS (5 mM) was added to the mutant enzyme, a decrease in the 415 nm peak was observed over 30 mins along with the appearance of a new peak at 330 nm and a small shoulder at 380 nm (Fig. 7A). Upon completion of the LCS inactivation of the SPT R378N mutant we noted that the 380 nm peak (Fig. 7A) is notably smaller than that observed in the wild-type enzyme (Fig. 5A, dashed line) suggesting that in the absence of Arg378 the mutant enzyme is unable to stabilize the putative oxime intermediate.
Fig. 7.
(A) R378N and 5mM LCS at time 0 (solid line), 30 seconds (long dash), 4 minutes (dotted line) and 30 minutes (dot dash). (B) R378N and 5 mM DCS at time 0 (solid line), mixing time (long dash), 30 min (dot dash).
In contrast to the LCS enantiomer, when DCS was added to the SPT R378N mutant, dramatic changes in the UV-visible spectrum occurred within the mixing time (Fig. 7B, dashed line). Most noticeable was the appearance of new a peak at 495 nm which was transient and disappeared after 30 seconds. This could be due the formation of a quinonoid species – these have been observed with SPT and other members of the AOS family only when incubated with substrates and substrate analogues and products, and have characteristic absorbance maxima >480 nm (11,15,16,39,40). In the case of SPT, Ikushiro et al. (39) used a palmitoyl-CoA thioether analogue to initiate deprotonation of the PLP:l-ser external aldimine and the resulting quinonoid was observed as a new peak in the UV-visible spectrum with λmax = 493 nm. In addition to the 495 nm peak, DCS binding to SPT R378N induced formation of a broad peak with a plateau from 340 nm to 380 nm along with a small shoulder at 425 nm (Fig. 7B). We suggest that the broad peak is likely to be a combination of overlapping bands due to a number of PLP-derived species. The peak shifted only slightly over the following 30 minutes and was an indication that maybe a DCS-derived intermediate becomes ‘stalled’ in the SPT R378N mutant. LC ESI-MS analyses of the low molecular weight fraction from both the CS enantiomer-inactivated R378N incubations were inconclusive e.g. no ions corresponding to the PMP and β-aminooxyacetaldehyde products could be detected (data not shown). This is unsurprising since the R378N mutant has such a slow turnover compared with the wild-type enzyme. Taken together, the analyses of the SPT R378N mutant with both LCS and DCS provide further evidence that the SPT enzyme is controlling the reaction pathway of each inhibitor in a stereospecific manner that requires residues not simply involved in binding the PLP cofactor directly. Current efforts are focused on capturing CS-derived intermediates in the mutant enzyme by x-ray crystallography.
Discussion
The antibiotic DCS was first isolated from the soil bacterium Streptomyces and its structure and activity determined over 50 years ago (41-45). Cycloserine inactivates many PLP-dependent enzymes by disabling the essential PLP cofactor at the active site and a list of the x-ray structures from the Protein DataBank with cycloserine adducts bound in the active site is presented in Table 1. In each case inhibitor binding results in transaldimination of the PLP bound, holo-enzyme and leads to formation of a PLP:cycloserine external aldimine. Deprotonation of this species gives rise to a stable 3-hydroxyisoxazole-PMP adduct whereby the cycloserine ring remains intact and covalently linked to the PLP cofactor – the so-called “aromatisation mechanism” (35) (Fig. 2B). Olson et al. studied LCS inactivation of γ-aminobutyric acid aminotransferase and Peisach et al., investigated D-amino acid aminotransferase inhibition by DCS (46). The aromatized CS adduct was observed in both cases where the active site lysine residue was inferred as the base which removes the CS α-proton. It is interesting that inhibition by both enantiomers of CS leads to formation of the same isoxazole-PMP adduct in the alanine racemase that was studied by Fenn et al. (23). This could be due to the alternating acid/base nature of the residues (Lys39, that is also involved in formation of the initial internal aldimine with PLP, and the side-chain of Tyr265) from an enzyme that catalyses racemisation of both enantiomers of alanine. However, this study revealed that the kinetics of inactivation of the two CS enantiomers was different; DCS inactivated alanine racemase much faster than the LCS. This explains why DCS (seromycin) is the better antibiotic. We also found different kinetics for each CS enantiomer with SPT; the apparent half-life of inactivation was approximately 15 minutes for 1.25mM LCS and 20mM DCS. Kitz and Wilson analysis also revealed both enantiomers inhibit via a bimolecular mechanism i.e. they are weak, but fast inhibitors, and suggests that carrying out incubations at lower temperatures would provide further insight into the inhibition mechanism (32).
Since SPT only catalyses condensation between palmitoyl-CoA and l-serine (although it does bind d-serine and forms an external aldimine) we expected to observe differences in SPT inactivation by each CS enantiomer. These were manifested in differences in both the rates of inactivation, with LCS being 14 times faster than DCS, and time-dependent changes in the UV-vis spectra. We could not detect an aromatised CS derivative after inactivation, but we did identify PMP and the derivitised form of a novel aldehyde by mass spectrometry. This CS-derived product was formed from both enantiomers but to a greater extent (based on LC ESI-MS profiles) from LCS inactivation. Moreover, we also identified a PMP form of the cofactor in crystals of the LCS-inactivated SPT and a low molecular weight species non-covalently bound at the active site which clearly no longer contains a cycloserine ring and whose size and shape is consistent with the proposed β-aminoacetaldehyde product.
This combined data leads us to propose a novel decarboxylative, ring-opening mechanism for inactivation of SPT by cycloserine (Fig. 8). The first step in the normal catalytic cycle of SPT is a transaldimination step when the substrate l-Ser replaces the PLP-Lys265 internal aldimine to form a PLP:l-ser external aldimine (Fig. 1). Deprotonation of this species does not occur until the second substrate palmitoyl-CoA binds. It is interesting to note that SPT acts upon both CS enantiomers in the absence of palmitoyl-CoA suggesting that the CS binds in the SPT active site and forms a PLP:CS external aldimine complex which then reacts readily. Previously, we captured the PLP:l-ser external aldmine and the structure revealed the Lys265 poised below this intermediate at a position where it would act as the base that removes the Cα proton (to form a quinonoid) once binding of the thioester causes rotation around the Cα-N bond (20). Interestingly, d-ser can form an external aldimine with SPT but is a poor substrate presumably because the α-H is not in the correct position when palmitoyl-CoA binds (47). To rationalise why we did not observe aromatization of the LCS inhibitor (which has the same S-configuration as l-ser) we suggest that the Cα proton in the PLP:LCS external aldimine cannot be in the optimal orientation to be removed by the lysine (Fig. 1 and Fig. 8A). Instead we propose ring-opening of the LCS ring by cleavage of the amide bond. Amide bond hydrolysis of peptides is catalysed by various proteases which use a powerful nucleophile (e.g. serine or cysteine in the serine or cysteine proteases) to attack this bond and generate an acylated intermediate which is subsequently hydrolysed. We cannot identify such a residue in the active site of SPT at present but conformational changes upon ligand binding may bring such a residue into play. An acylated ring-opened adduct was recently captured by Macheboeuf et al who studied the inhibition of penicillin-binding protein (PBP) by the natural product lactivicin (LTV) (48). This interesting molecule contains separate cycloserine and γ-lactone rings and is the only known natural PBP inhibitor that does not contain a β-lactam. Crystallographic analysis of the PBP revealed that LTV inhibition involves opening of both the cycloserine and γ-lactone rings upon attack by the active site serine nucleophile. We have accommodated this possible route (Fig. 8, path (a)) into the mechanism and the acylated intermediate formed could be hydrolysed to give the carboxylated PLP intermediate.
Fig. 8.
(A) Novel ring-opening, decarboxylative mechanism for inactivation of SPT by LCS. Path (a) denotes an enzymatic, nucleophile(Nu)-mediated mechanism with an acylated intermediate. Path (b) is the direct hydrolytic mechanism. (B) Inactivation of SPT by DCS highlighting a proposed quinonoid intermediate observed in the SPT R378N mutant. The paths are the same as in (A).
We also suggest an alternative mechanism involving direct attack of water on the CS ring (Fig. 8, path (b)). This is justified when one considers the properties of cycloserine. During the initial characterization of the DCS natural product Kuehl et al and Hidy et al found the CS ring opened upon treatment with methanol and HCl to afford the β-amino-D-alanine methyl ester (43,44). Treatment of this ester with alkali converted it back to DCS. Further studies revealed unusual chemical properties of DCS; it was found to dimerise to give a six-membered cis-3-6-bis(aminooxymethyl)-2,5-piperazinedione adduct in acid (44,49). The proposed mechanism of adduct formation involves nucleophilic attack of one CS amino group at the carbonyl position of another CS molecule, which then ring opens at the amide bond to generate the aminooxy form. This reaction occurs again in an intramolecular fashion to generate the piperazine ring. So, in the context of a SPT catalysed mechanism ring-opening could also proceed through acid catalysis and a number of potential proton donors are found in the active site (e.g. His159).
We predict two possible routes for the ring-opened adduct. Firstly, it can form an oxime intermediate, observed as a 380 nm peak in the UV-visible spectrum (Fig. 2C and Fig. 5A). In the second, novel route, the ring-open adduct decarboxylates, which is then followed by imine hydrolysis to form the products, PMP and β-aminooxyacetaldehyde (Fig. 8). This fits well with the SPT mechanism since the enzyme catalyses decarboxylation of the putative β-keto acid Claisen condensation intermediate to give a product external aldimine which is then hydrolysed to release KDS (Fig. 1). Since LCS effectively acts as an amino donor to generate PMP, the enzyme has to be regenerated by addition of excess PLP (Fig. 4). In contrast to LCS, DCS is a much slower inactivator of SPT and this suggests it forms a PLP:DCS external aldimine which can react via a similar decarboxylative mechanism, possibly requiring some conformational change of the ring opened intermediate to promote this step (Fig. 8B). To probe the formation of intermediates during SPT inactivation by both enantiomers we utilised a SPT R378N mutant since this residue forms a salt bridge with the carboxylate of the external aldimine with l-ser. Again, we saw clear enantiospecific differences in the behaviour of this mutant with LCS and DCS by UV-vis spectroscopy (Fig. 7). Incubation of SPT R378N with LCS gave rise to a similar spectrum to the wild-type enzyme but lacked a shoulder at 380nm (Fig. 7A). In contrast, we were excited to observe transient formation of a quinonoid species at 510 nm during DCS inactivation of the R378N mutant (Fig. 7B) and we suggest that this is due to the quinonoid formed by decarboxylation (Fig. 8B). Of interest in a previous study Fenn et al., used a Y265F mutant of alanine racemase to probe the different LCS and DCS inactivation mechanisms of this enzyme (37). The Y265F mutant produced only the aromatised isoxazole PLP adduct with DCS, whereas LCS inactivation led to formation of a range of products including PMP and acetate. They concluded that this active site residue “steers” the pathway of inactivation. In our work, mutation of Arg378 has revealed the presence of transient species and may be useful to capture intermediates in future.
LCS is commonly used in biological research to inhibit de novo biosynthesis of sphingolipids in mammalian cells but it remains debatable whether it specifically targets SPT. LCS was shown to be a potent inhibitor of SPT activity derived from mouse brain microsomes (50) and inhibited mouse brain SPT activity in vivo after intraperitoneal injection (51). On the other hand cycloserine toxicity in Chinese hamster ovary cells was not attributed to SPT inhibition since exogenous sphingosine could not rescue cell death (52). Further research is required to examine whether LCS targets SPT alone or can inhibit other PLP-dependent enzymes on the sphingolipid biosynthetic pathway such as sphingosine-1-phosphate lyase. With knowledge gained from our study of the bacterial, homodimeric SPT, it will be interesting to characterise the products of LCS and DCS inhibition of the heterodimeric human enzyme. Whether the β-aminooxyacetaldehyde derivative is produced by the more complex mammalian isoforms remains to be seen. The identification of two small subunits (ssSPTa and ssSPTb) that stimulate human SPT activity >100 fold should facilitate the analysis of cycloserine inactivation products (53).
Interestingly DCS is a much more potent inhibitor of alanine racemase compared to the synthetic LCS (23). When the antibacterial effect of DCS and LCS was tested on a large number of bacterial strains, most had lower tolerance towards DCS (54). In contrast, the sphingolipid-producing bacterium Bacteroides levii was much more susceptible to LCS than DCS which correlated to inhibition of SPT (50). Future investigations are necessary to determine whether LCS could target sphingolipid-producing bacterial pathogens.
Materials
Plasmids and Escherichia coli competent cells were purchased from Novagen, and all chromatography columns were from GE Healthcare. All buffers and reagents including LCS and DCS were from Sigma. Palmitoyl CoA was from Avanti Lipids.
Methods
Cloning and expression of S paucimobilis SPT wild-type and R378N mutant
The SPT wild-type gene and R378N mutant were cloned in pET28a expression vector (Novagen) as previously described (20). The plasmids were transformed into E coli BL21 (DE3) competent cells and selection was carried out on LB agar containing 30 μg/ml kanamycin. A single colony was used to inoculate an overnight culture grown at 37°C in 500 ml 2YT broth (16 g/litre Bacto-tryptone, 10 g/litre Bacto-yeast extract, 5 g/litre sodium chloride (pH 7.5)). This culture was added to 4 litres of 2YT supplemented with kanamycin and grown to OD600 of 0.6 before addition of 0.1 mM isopropyl 1-thio-β-d-galactopyranoside to induce protein expression. Growth was continued for 5 hours at 30°C. Cells were harvested (Sorvall RC5B centrifuge) by centrifugation at 3500 rpm for 20 minutes at 4°C. The enzyme was purified using IMAC on nickel resin (Invitrogen) as well as size exclusion chromatography (Sephadex S200HR, GE Healthcare). Before UV-vis spectroscopy and inactivation assays, the enzyme was freshly converted to the PLP-bound form by dialysis against buffers containing 25 μM PLP.
UV-visible spectroscopy of SPT inhibition by cycloserine
All UV-visible spectra were recorded on a Cary 50 UV-visible spectrophotometer (Varian) and analysed using Cary WinUV software (Varian). Enzyme was dialysed against 20 mM potassium phosphate (pH 7.5) containing 150 mM NaCl and 25 μM PLP for 4 hours at 4°C. Excess PLP was removed on a PD-10 (Sephadex G-25M) desalting column (GE Healthcare). For UV-visible assays, the concentration of recombinant protein was 20 μM. The spectrophotometer was blanked with 20 mM potassium phosphate (pH 7.5) containing 150 mM NaCl and spectra were collected from 800 nm to 200 nm. Quartz cuvettes from NSG Precision Cells, Type 18-BM (Material quartz, Lightpath 10mm) had a sample volume of 500 μl. By setting the instrument to cycle mode and collecting spectra at time intervals, changes in the UV-visible spectrum were monitored after addition of cycloserine.
Rates of SPT inactivation by cycloserine using DTNB assay
SPT activity was measured using the DTNB assay as previously described (20). Assays contained enzyme, substrates and DTNB with final concentrations as follows: 1 μM enzyme, 25 mM l-serine, 250 μM palmitoyl-CoA, 0.2 mM DTNB in 20 mM HEPES buffer, pH 8.0. SPT was incubated with LCS (0 mM, 0.63 mM, 1.25 mM, 2.5 mM and 5 mM) and DCS (0 mM, 10 mM, 20 mM, 40 mM and 80 mM). At time intervals an aliquot of the enzyme plus inhibitor solution was diluted into the assay buffer to record the activity. The remaining activity was calculated as a percentage of the total activity in the absence of inhibitor i.e. at time zero. Kitz and Wilson plots of ln % remaining activity against time were linear (32). Graphs were plotted using WinCurveFit software, Kevin Raner, Australia.
Isolation and derivitisation of aldehyde followed by detection by LC-MS
SPT (100μM) was inactivated with 5 mM LCS for 2 hours followed by centrifugation at 13000 rpm for 10 minutes in a Vivaspin 500 concentrators (VWR) to separate the low and high molecular weight material. The filtrate (0.1 ml) was added to a 2, 4-DNP solution (0.5 ml) prepared in acetonitrile and containing 0.5 % HCl. The solution was incubated at 37°C for 30 minutes to allow derivitisation of the aldehyde. The hydrazone product was detected by LC-MS on a 6130 quadrupole mass spectrometer with a 1200 Series quaternary LC system (Agilent Technologies). Sample volumes were typically 10 μl under automated injection into a LC system equipped with a C18 column followed by separation in a methanol:water:formic acid (80:19:1) mobile phase. The mass spectrometer was set up in selected ion monitoring (SIM) mode to search for ions with masses corresponding to the β-aminoacetaldehyde and its 2, 4-DNP hydrazone derivative (Supplementary Fig. 2).
Structural biology
Crystals of LCS-inactivated SPT were prepared by co-crystallisation. A 10mM stock solution of LCS was prepared and added to freshly prepared holo-SPT (20 μM, in 10 mM Tris 7.5, 150 mM NaCl and 25 μM PLP) to give a final LCS concentration of 1mM . After overnight incubation at 4 °C, the protein was concentrated to 20 mg/ml and this solution was used to set up crystal trials. Colourless SPT:LCS crystals grew from a 2 μl drop consisting of 1 μl of the SPT/LCS solution and 1 μl of well solution (100 mM HEPES pH 6.5, 110 mM MgCl2, 21.5 % PEG 3350). A single crystal was mounted in a cryo-loop and cryo-protected in 22% PEG 3350, 120 mM MgCl2, 100 mM HEPES pH 6.5, 20% PEG400. The crystal was frozen and data obtained that diffracted to a resolution of 1.40 Å on beamline ID14-1 (wavelength = 0.934 Å) at 100 K. Data were collected on and processed to 1.40 Å using XDS (55). The structure was solved by molecular replacement using the 1.3 Å holo form as the model (with co-factor and water molecules removed). Due to an oversight a new Free-R set was devised for the structure, more rigorous would have to be use the same as the isomorphous search model. To overcome the bias in the Free-R set, all the ligands and waters were removed, the B-factors reset to a common value and then the structure was refined. TLS groups were defined using the TLS server (56) and once again the B-factors reset to a single value. The asymmetric unit contains a monomer of protein, but as with the native contains a functional dimer. he structure was refined using REFMAC5 (57), TLS groups were used and in the final steps of refinement anisotropic thermal factors were refined for all atoms. Inclusion of anisotropic B-factors decreases R-free by 0.6% and was therefore judged appropriate. PMP was modelled into the difference electron density immediately after molecular replacement, the density was unambiguous. The choice of PMP not PLP was made based on mass spectrometric data not the X-ray data themselves. Water molecules were added manually and checked using COOT (58). A dictionary of the aldehyde was made using the PRODRG server (59). The aldehyde was not included in the final model. Its inclusion did not perturb either the R-factor or the stereochemical quality of the model. The data collection and refinement statistics are given in Supplementary Table.
Supplementary Material
Acknowledgements
This work was supported by Biotechnology and Biological Sciences Research Council (BBSRC) grants BB/F009739/1 (supporting JL) and BBS/B/14434 to DJC and JHN. VTB and MCCR were supported by Univeristy of Edinburgh, EastChem studentships. BAY was supported by a University of Edinburgh/Syngenta PhD studentship. We thank Dr. Mike Greaney for access to mass spectrometry instrumentation.
Abbreviations
- 2,4-DNP
2,4-dinitrophenylhydrazine
- DCS
d-cycloserine
- DTNB
dithionitrobenzoic acid
- LC ESI-MS
liquid chromatography electrospray ionisation mass spectrometry
- LCS
l-cycloserine
- PLP
pyridoxal 5′-phosphate
- PMP
pyridoxamine 5′-phosphate
- SPT
serine palmitoyltransferase
Footnotes
The atomic coordinates and structure factors of the structure reported in this paper have been submitted to the Protein Data Bank and are awaiting acceptance.
References
- 1.O’Brien JS, Sampson EL. J Lipid Res. 1965;6:537–544. [PubMed] [Google Scholar]
- 2.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 3.Futerman AH, Hannun YA. EMBO Rep. 2004;5:777–782. doi: 10.1038/sj.embor.7400208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hannun YA, Obeid LM. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 5.Pruett ST, Bushnev A, Hagedorn K, Adiga M, Haynes CA, Sullards MC, Liotta DC, Merrill AHJ. J. Lipid Res. 2008;49:1621–1639. doi: 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fyrst H, Oskouian B, Bandhuvula P, Gong Y, Byun HS, Bittman R, Lee AR, Saba JD. Cancer Res. 2009;69:9457–9464. doi: 10.1158/0008-5472.CAN-09-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park TS, Rosebury W, Kindt EK, Kowala MC, Panek RL. Pharmacol Res. 2008;58:45–51. doi: 10.1016/j.phrs.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 8.Bras J, Singleton A, Cookson MR, Hardy J. Febs J. 2008;275:5767–5773. doi: 10.1111/j.1742-4658.2008.06709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanada K. Biochim Biophys Acta. 2003;1632:16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 10.Alexeev D, Alexeeva M, Baxter RL, Campopiano DJ, Webster SP, Sawyer L. J Mol Biol. 1998;284:401–419. doi: 10.1006/jmbi.1998.2086. [DOI] [PubMed] [Google Scholar]
- 11.Webster SP, Alexeev D, Campopiano DJ, Watt RM, Alexeeva M, Sawyer L, Baxter RL. Biochemistry. 2000;39:516–528. doi: 10.1021/bi991620j. [DOI] [PubMed] [Google Scholar]
- 12.Jordan PM. Biosynthesis of Tetrapyrroles. Elsevier; Amsterdam: 1991. [Google Scholar]
- 13.Ferreira GC, Gong J. J Bioenerg Biomembr. 1995;27:151–159. doi: 10.1007/BF02110030. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt A, Sivaraman J, Li Y, Larocque R, Barbosa JA, Smith C, Matte A, Schrag JD, Cygler M. Biochemistry. 2001;40:5151–5160. doi: 10.1021/bi002204y. [DOI] [PubMed] [Google Scholar]
- 15.Jahan N, Potter JA, Sheikh MA, Botting CH, Shirran SL, Westwood NJ, Taylor GL. J Mol Biol. 2009;392:763–773. doi: 10.1016/j.jmb.2009.07.042. [DOI] [PubMed] [Google Scholar]
- 16.Kelly RC, Bolitho ME, Higgins DA, Lu W, Ng WL, Jeffrey PD, Rabinowitz JD, Semmelhack MF, Hughson FM, Bassler BL. Nat Chem Biol. 2009;5:891–895. doi: 10.1038/nchembio.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spirig T, Tiaden A, Kiefer P, Buchrieser C, Vorholt JA, Hilbi H. J Biol Chem. 2008;283:18113–18123. doi: 10.1074/jbc.M801929200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yard BA, Carter LG, Johnson KA, Overton IM, Dorward M, Liu H, McMahon SA, Oke M, Puech D, Barton GJ, Naismith JH, Campopiano DJ. J Mol Biol. 2007;370:870–886. doi: 10.1016/j.jmb.2007.04.086. [DOI] [PubMed] [Google Scholar]
- 19.Eliot AC, Kirsch JF. Annu Rev Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 20.Raman MC, Johnson KA, Yard BA, Lowther J, Carter LG, Naismith JH, Campopiano DJ. J Biol Chem. 2009;284:17328–17339. doi: 10.1074/jbc.M109.008680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikushiro H, Islam MM, Okamoto A, Hoseki J, Murakawa T, Fujii S, Miyahara I, Hayashi H. J Biochem. 2009;146:549–562. doi: 10.1093/jb/mvp100. [DOI] [PubMed] [Google Scholar]
- 22.Soper TS, Manning JM. J Biol Chem. 1981;256:4263–4268. [PubMed] [Google Scholar]
- 23.Fenn TD, Stamper GF, Morollo AA, Ringe D. Biochemistry. 2003;42:5775–5783. doi: 10.1021/bi027022d. [DOI] [PubMed] [Google Scholar]
- 24.Malashkevich VN, Strop P, Keller JW, Jansonius JN, Toney MD. J Mol Biol. 1999;294:193–200. doi: 10.1006/jmbi.1999.3254. [DOI] [PubMed] [Google Scholar]
- 25.Svensson ML, Gatenbeck S. Arch. Microbiol. 1982;131:129–131. doi: 10.1007/BF00405880. [DOI] [PubMed] [Google Scholar]
- 26.Di Perri G, Bonora S. J Antimicrob Chemother. 2004;54:593–602. doi: 10.1093/jac/dkh377. [DOI] [PubMed] [Google Scholar]
- 27.Sheinin A, Shavit S, Benveniste M. Neuropharmacology. 2001;41:151–158. doi: 10.1016/s0028-3908(01)00073-9. [DOI] [PubMed] [Google Scholar]
- 28.Strominger JL IE, Threnn RH. J Am Chem Soc. 1960;82:998–999. [Google Scholar]
- 29.Neuhaus FC, Lynch JL. Biochemistry. 1964;3:471–480. doi: 10.1021/bi00892a001. [DOI] [PubMed] [Google Scholar]
- 30.Ikushiro H, Hayashi H, Kagamiyama H. Biochemistry. 2004;43:1082–1092. doi: 10.1021/bi035706v. [DOI] [PubMed] [Google Scholar]
- 31.Sundaram KS, Lev M. J Neurochem. 1984;42:577–581. doi: 10.1111/j.1471-4159.1984.tb02716.x. [DOI] [PubMed] [Google Scholar]
- 32.Kitz R, Wilson IB. J Biol Chem. 1962;237:3245–3249. [PubMed] [Google Scholar]
- 33.Brandt GS, Nemeria N, Chakraborty S, McLeish MJ, Yep A, Kenyon GL, Petsko GA, Jordan F, Ringe D. Biochemistry. 2008;47:7734–7743. doi: 10.1021/bi8004413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan H, Silverman RB. Bioorg Med Chem Lett. 2007;17:1651–1654. doi: 10.1016/j.bmcl.2006.12.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson GT, Fu M, Lau S, Rinehart KL, Silverman RB. J Am Chem Soc. 1998;120:2256–2267. [Google Scholar]
- 36.Rando RR. Biochem Pharmacol. 1975;24:1153–1160. doi: 10.1016/0006-2952(75)90055-6. [DOI] [PubMed] [Google Scholar]
- 37.Fenn TD, Holyoak T, Stamper GF, Ringe D. Biochemistry. 2005;44:5317–5327. doi: 10.1021/bi047842l. [DOI] [PubMed] [Google Scholar]
- 38.Noland BW, Newman JM, Hendle J, Badger J, Christopher JA, Tresser J, Buchanan MD, Wright TA, Rutter ME, Sanderson WE, Muller-Dieckmann HJ, Gajiwala KS, Buchanan SG. Structure. 2002;10:1569–1580. doi: 10.1016/s0969-2126(02)00879-1. [DOI] [PubMed] [Google Scholar]
- 39.Ikushiro H, Fujii S, Shiraiwa Y, Hayashi H. J Biol Chem. 2008;283:7542–7553. doi: 10.1074/jbc.M706874200. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Ferreira GC. J Biol Chem. 2002;277:44660–44669. doi: 10.1074/jbc.M203584200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harned RL, Hidy PH, La Baw EK. Antibiotics & Chemotherapy. 1955;5:204–205. [PubMed] [Google Scholar]
- 42.Harris DA, Ruger M, Reagan MA, Wolf FJ, Peck RL, Walick H, Woodroff HB. Antibiotics & Chemotherapy. 1955;5:183–190. [PubMed] [Google Scholar]
- 43.Kuehl FA, Jr., Wolf FJ, Trenner NR, Peck RL, Buhs RP, Howe E, Putter I, Hunnewell BD, Ormond R, Downing G, Lyons JE, Newstead E, Chaiet L, Folkers K. J. Am. Chem. Soc. 1955;77:2344–2345. [Google Scholar]
- 44.Hidy PH, H. HE, Young VV, Harned RL, Brewer GA, Phillips WF, Runge WF, Stavely HE, Pohland A, Boaz H, Sullivan HR. J. Am. Chem. Soc. 1955;77:2345–2346. [Google Scholar]
- 45.Stammer CH, Wilson AN, Holly FW, Folkers K. J. Am. Chem. Soc. 1955;77 [Google Scholar]
- 46.Peisach D, Chipman DM, Van Ophem P, Manning JM, Ringe D. J Am Chem Soc. 1998;120:2268–2274. [Google Scholar]
- 47.Hanada K, Hara T, Nishijima M. FEBS Lett. 2000;474:63–65. doi: 10.1016/s0014-5793(00)01579-9. [DOI] [PubMed] [Google Scholar]
- 48.Macheboeuf P, Fischer DS, Brown T, Jr., Zervosen A, Luxen A, Joris B, Dessen A, Schofield CJ. Nat Chem Biol. 2007;3:565–569. doi: 10.1038/nchembio.2007.21. [DOI] [PubMed] [Google Scholar]
- 49.Lassen FO, Stammer CH. J. Org. Chem. 1971;36:2631–2634. doi: 10.1021/jo00817a015. [DOI] [PubMed] [Google Scholar]
- 50.Sundaram KS, Lev M. Antimicrob Agents Chemother. 1984;26:211–213. doi: 10.1128/aac.26.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sundaram KS, Lev M. J Lipid Res. 1985;26:473–477. [PubMed] [Google Scholar]
- 52.Hanada K, Nishijima M, Fujita T, Kobayashi S. Biochem Pharmacol. 2000;59:1211–1216. doi: 10.1016/s0006-2952(00)00251-3. [DOI] [PubMed] [Google Scholar]
- 53.Han G, Gupta SD, Gable K, Niranjanakumari S, Moitra P, Eichler F, Brown RH, Jr., Harmon JM, Dunn TM. Proc Natl Acad Sci U S A. 2009;106:8186–8191. doi: 10.1073/pnas.0811269106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neuhaus FC. D-cycloserine and O-carbamyl-D-serine. Springer-Verlag; New York: 1967. [Google Scholar]
- 55.Kabsch W. J. Appl. Cryst. 1993;26:795–800. [Google Scholar]
- 56.Painter J, Merritt EA. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 57.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 58.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 59.Schuettelkopf AW, van Aalten DMF. Acta Crystallographica. 2004;D60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 60.Priyadarshi A, Lee EH, Sung MW, Nam KH, Lee WH, Kim EE, Hwang KY. Biochim Biophys Acta. 2009;1794:1030–1040. doi: 10.1016/j.bbapap.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 61.Wu D, Hu T, Zhang L, Chen J, Du J, Ding J, Jiang H, Shen X. Protein Sci. 2008;17:1066–1076. doi: 10.1110/ps.083495908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Noda M, Matoba Y, Kumagai T, Sugiyama M. J Biol Chem. 2004;279:46153–46161. doi: 10.1074/jbc.M404605200. [DOI] [PubMed] [Google Scholar]
- 63.Jensen PY, Parsons JF, Fisher KE, Pachikara AS, Tordova M, Howard AJ, Eisenstein E, Ladner JE. 2003. 10.2210/pdb1i2l/pdb.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.