Abstract
Background
Degradation of extracellular matrix support in the large abdominal arteries contribute to abnormal dilation of aorta, leading to abdominal aortic aneurysms, and matrix metalloproteinase-9 (MMP-9) is the predominant enzyme targeting elastin and collagen present in the walls of the abdominal aorta. Previous studies have suggested a potential association between MMP-9 genotype and abdominal aortic aneurysm, but these studies have been limited only to the p-1562 and (CA) dinucleotide repeat microsatellite polymorphisms in the promoter region of the MMP-9 gene. We determined the functional alterations caused by 15 MMP-9 single-nucleotide polymorphisms (SNPs) reported to be relatively abundant in the human genome through Western blots, gelatinase, and promoter–reporter assays and incorporated this information to perform a logistic-regression analysis of MMP-9 SNPs in 336 human abdominal aortic aneurysm cases and controls.
Methods and Results
Significant functional alterations were observed for 6 exon SNPs and 4 promoter SNPs. Genotype analysis of frequency-matched (age, sex, history of hypertension, hypercholesterolemia, and smoking) cases and controls revealed significant genetic heterogeneity exceeding 20% observed for 6 SNPs in our population of mostly white subjects from Northern Wisconsin. A step-wise logistic-regression analysis with 6 functional SNPs, where weakly contributing confounds were eliminated using Akaike information criteria, gave a final 2 SNP (D165N and p-2502) model with an overall odds ratio of 2.45 (95% confidence interval, 1.06–5.70).
Conclusions
The combined approach of direct experimental confirmation of the functional alterations of MMP-9 SNPs and logistic-regression analysis revealed significant association between MMP-9 genotype and abdominal aortic aneurysm.
Keywords: aneurysm, case-control study, genetic association, MMP, single nucleotide polymorphism genetics
The incidence of abdominal aortic aneurysm (AAA) has been increasing in recent years and is now thought to affect 8% of men over 60 years of age.1 Diseased patients are frequently asymptomatic at the time of presentation and the aneurysms only found during the course of an unrelated workup. Male sex, hypertension, hypercholesterolemia, history of smoking, and history of coronary artery disease are clinical risk factors associated with most cardiovascular diseases, including AAAs.2 A genetic basis for AAA was first reported by the observation that a positive history of AAA in a first-degree relative increased the risk of AAA 10-fold.3 A recent meta-analysis supported the genetic influence of AAA, although a distinct class of sporadic AAA also appears to exist.1 Thompson et al4 reviewed for potential genes affecting the pathogenesis of AAA on the basis of a logical 4-step mechanistic progression of initiation, formation, growth, and rupture.4 A meta-analysis of 6 gene polymorphisms identified the angiotensin-converting enzyme (287 bp insertion–deletion polymorphism), methylenetetrahydrofolate reductase (+677C>T polymorphism), and matrix metalloproteinase-9 (MMP-9; −1562 C>T polymorphism) as associated with an increased risk for AAA,4 although McColgan et al,5 in another meta-analysis, failed to replicate the association between MMP-9 p-1562 single-nucleotide polymorphism (SNP) and AAA.5 Of these, the MMP-9 polymorphism lends to a simplistic mechanistic interpretation as the p-1562 C>T SNP in the promoter increases the gene transcription, resulting in an enhanced production of the proteolytic MMP-9 able to be secreted in the extracellular space, where, upon further regulation, is able to exacerbate the degradation of the extracellular matrix support of the abdominal aorta.6
MMPs, a family of Zn2+-dependent endopeptidases, play a key role in extracellular matrix remodeling, especially elastin and collagen turnover.7 Increased levels of the gelatinase subset (MMP-2 and -9) of this protease family are found in developing AAA.8,9 Despite the early enthusiasm on the utility of a simple plasma MMP-9 level as a promising biomarker for a variety of cardiovascular diseases, including AAA, cumulative data suggest lack of utility for this simple approach. This failure is not surprising given the complex regulation of MMP-9 activity at several levels, including gene expression, activation of precursor proenzymes by proteolytic cleavage, and inhibition by endogenous tissue inhibitors of MMP (TIMPs).7 The biological significance of MMP-9 and its role in AAA pathogenesis is most likely the enzymatic activity of available MMP-9 and not the simple abundance of the protein. The MMP-9 gene (≈9 kb) has numerous documented nucleotide variants in the promoter and the coding exons. Genetic polymorphism and the consequence of such genetic variation on the MMP-9 expression and enzymatic function must be accounted for to effectively use the MMP-9 genotype as a biomarker for AAA. We selected 5 promoter SNPs and 9 exon SNPs that result in an amino acid change in the encoded protein, and investigated the functional consequences of the SNPs with an in vitro enzymatic assay of the expressed MMP-9 cDNA with the exon SNPs and a promoter–reporter assay for the promoter SNPs. A genotype analysis of the SNPs with functional consequences was performed on cases, and clinical risk factor frequency-matched controls selected from the data accumulated in the Marshfield Clinic Personalized Medicine Research Project (PMRP),10 and a MMP-9 genotype–AAA association model identified by a logistic-regression analysis. Our results identified a 2 SNP MMP-9 genotype–AAA association model with an odds ratio (OR) of 2.45 (95% confidence interval, 1.06–5.70).
Materials and Methods
Selection of MMP-9 SNPs to be Studied
The NCBI database (http://www.ncbi.nlm.nih.gov/SNP/snp_ref.cgi?locusId=4318) documents hundreds of MMP-9 SNPs with constantly changing information as new information is added to the database. We based our selection of the SNPs to be studied on information provided at the initiation of this study (September 2010). We restricted the exon SNPs to missense mutations resulting in an amino acid change. Five SNPs in the genomic promoter region of the gene and 9 SNPs in the coding regions were further selected on the basis of the reported abundance (heterogeneity) of the SNP (Table 1). The microsatellite CA repeats in the promoter region (rs2234681), previously reported to increase the promoter activity with increasing number of repeats,11,12 was not included because prior studies suggested that this genetic polymorphism did not show association with AAA.13-15
Table 1. Selected MMP-9 Single Nucleotide Polymorphisms Investigated.
| Heterozygosity | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Name | rs# ID | SNP | Mutation | October 2010 | January 2012 | Domain | Protein Secretion | Total Enz. Activity |
Promoter Activity |
|
|
|||||||||
| A20V | rs1805088 | C>T | Ala>Val | 0.057 | 0.051 | Sig.pept. | Down | ||
| N38S | rs41427445 | A>G | Asn>Ser | 0.133 | 0.023 | Propep.N-glyco. | Down | Down | |
| E82K | rs1805089 | G>A | Glu>Lys | N.D. | 0.000 | Propep. | Down | Down | |
| D165N* | rs8125581 | G>A | Asp>Asn | N.D. | 0.002 | Catalytic | Down | Down | |
| L187F | rs55789927 | C>T | Leu>Phe | N.D. | 0.001 | Catalytic | Down | ||
| T258I | rs41529445 | C>T | Thr>IIe | 0.039 | 0.003 | Catalytic | |||
| R279Q | rs17576 | G>A | Arg>GIn | ND | 0.440 | Catalytic | Up | ||
| P574R | rs2250889 | C>G | Pro>Arg | 0.293 | 0.213 | Hemopexin | Down | ||
| R668Q | rs17577 | G>A | Arg>Gln | 0.287 | 0.280 | Hemopexin | Up | ||
| p-1562 | rs3918242 | C>T | 0.288 | 0.290 | Promoter | Up | |||
| p-1702 | rs3918241 | T>A | 0.258 | 0.249 | Promoter | Down | |||
| p-2024 | rs6104421 | C>T | 0.146 | 0.151 | Promoter | ||||
| p-2118 | rs3918240 | C>T | 0.470 | 0.463 | Promoter | Up | |||
| p-2502* | rs8113877 | G>T | 0.423 | 0.426 | Promoter | Down | |||
MMP-9 indicates matrix metalloproteinase-9; SNP, single nucleotide polymorphism; Total enz. activity, total enzymatic activity; Sig. pept, signal peptide; Propep, propeptide; N-glyco, N-linked glycosylation; and ND, no data.
The common nomenclature used in our study denoting the amino acids changed for the exon SNPs and the location of the SNPs with respect to the start ATG for the promoter SNPs.
Indicates the 2 SNPs that were used in the final logistic-regression model. The SNP heterozygosity column lists the numbers available through NCBI at the time of initiation of this study (2010) and the numbers listed at the most recent access (2012). Domain lists the functional domain of the gene where the SNPs are located. Protein secretion, total nz. activity, and promoter activity, lists the functional consequences of the SNPs.
Creation of MMP-9 Clones With the Desired SNPs and Enzymatic and Promoter Activity Assays
Starting with the wild-type human MMP-9 cDNA (Accession BC006093, Image Clone MGC: 12688) we created the less prevalent exon genotypes by introducing the desired nucleotide switch by overlap PCR using the primers listed in online-only Data Supplement Table SI. All final constructs were sequenced to confirm the presence of the desired mutagenesis and the lack of unintended base changes as a result of PCR.
The cDNAs containing the exon SNPs were subcloned into an expression vector (pCI/neo, Promega, Madison, WI) and the protein expressed through transient transfection of the human embryonic kidney (HEK) 293 cells (ATCC, Manassas, VA) using Lipofectamine 2000 (Life Technologies, Grand Island, NY). HEK293 cells were grown in Dulbecco’s Modification of Eagle’s Medium (Mediatech, Inc, Manassas, VA) with 4.5 g/L glucose, l-glutamine, sodium pyruvate, and supplemented with 10% heat inactivated fetal bovine serum (Mediatech Inc), 100 U/mL penicillin, and 0.1 mg/mL streptomycin. The culture supernatant was harvested 24 hours after transfection and MMP-9 activated by in vitro treatment with 4-aminophenylmercuric acetate which activates the proenzyme by unfolding the protein. The MMP-9 enzymatic activity was assayed by gelatin zymography and quantified with gel densitometry. Briefly, 0.75 mm SDS-PAGE gels were prepared with the incorporation of gelatin (1 mg/mL) before casting. Denatured but nonreduced samples and standards were run at constant voltage. Gels were allowed to renature by 4 washes in an enzyme renaturing buffer containing 2.5% v/v Triton X-100 for 1 hour. Gels were then incubated at 37°C for an extended period of time, followed by staining with Coomassie Brilliant Blue 250-R. After destaining, zones of enzyme activity showed up as clear bands and quantified by densitometric analysis of the inverted image. Preliminary studies confirmed that the amount of transfected plasmids (0.5 μg/L well of a 6-well plate) and gel loading all fell within a linear range of densitometry (online-only Data Supplement Figure SI) and in general gave a more reproducible result compared with activity assessment with a kinetic measurement of the cleavage of a fluorogenic MMP-9 substrate (Enzo-Biomol, Farmingdale, NY; data not shown). Gelatin zymography had the advantage of concurrently measuring the activity of monomeric and dimeric MMP-9. A Western blot of culture supernatant with the anti-MMP-9 (1:1000 in 1% milk TBST, Clone L51/82, NeuroMab, Davis, CA) antibody followed by densitometric quantification documented the amount of secreted MMP-9 protein. Total secreted MMP-9 was also quantified by the hMMP-9 ELISA kit (#KHC3061, Invitrogen, Camarillo CA). ELISA was carried out according to vendor’s recommendation and the linearity of the assay confirmed for MMP-9 concentrations within 0 to 1500 pg/mL. HEK293 supernatant was diluted (1:1000) to attain an appropriate concentration within the linear range and loaded in triplicate in a 96-well format. MMP-9 protein amount was quantified via colorimetric detection using a microplate reader.
The MMP-9 promoter and 5′untranslated region region consisting of ≈2.8 kb upstream of the start methionine was PCR amplified from genomic DNA isolated from HEK293 cells and inserted into the pGL4.10 luciferase reporter (Promega, Madison, WI). SNPs were introduced by a PCR-based mutagenesis strategy as above. The MMP-9 promoter–reporters were transfected, along with a Renilla luciferase expressing plasmid (pGL4.75, Promega), into HEK293 cells and lysates analyzed for the promoter activity (firefly luciferase) were normalized for potential transfection variability by Renilla luciferase using a Dual-Glo Luciferase Reporter kit (Promega) using the Synergy 2 microplate reader (BioTek Inc, Winooski, VT).
Statistical Analysis
Individual measures of the enzymatic activity by zymography were repeated 5 times and presented as mean±SEM compared with the wild-type activity, and a statistically significant difference tested by the nonparametric Mann–Whitney test at P<0.05. Triplicate measures of the promoter activity were repeated 6 times and presented as mean±SEM compared with the wild-type activity, and a statistically significant difference tested by the nonparametric Mann–Whitney test at P<0.05. Two-way ANOVA (replicates and repeated experiments) indicated no significant source of variation within the triplicate promoter–reporter assays but with significant variance between experiments. Triplicate measures were treated as 1 mean data point and the final overall mean and SD calculated from repeated 6 experiments.
Selection of Subjects With AAA and Frequency-Matched Controls
The PMRP clinical database search and genetic material transfer from the Marshfield Clinics to UW Madison was approved by the University of Wisconsin IRB protocol M2010-1315. The sole inclusion criterion was patients with a diagnosis of aortic aneurysm (International Classification of Diseases, Ninth Revision [ICD-9]) code between 441.1 and 441.7) at 2 dates and at least 1 confirmatory scan (MRI, contrast computed tomography, or angiography) of the AAA with a maximal dimension measurement of 3 cm or greater or at least 1 AAA rupture diagnosis code. Exclusion criteria included history of syndromic connective tissue diseases (eg, Marfan and Ehlers-Danlos syndromes), presence of any nonaortic aneurysms, abdominal trauma, and syphilis. The same exclusion criteria applied to the control population derived from subjects enrolled in the PMRP. Because the subjects were selected from a well-defined patient population thoroughly evaluated for the presence of aortic aneurysm, the incidence was 100% for this case–control genetic analysis circumventing loss of statistical power as a result of misdiagnosed subjects. Control subjects had no diagnoses of aortic aneurysm and must have had at least 1 scan with no aortic measurements >2.5 cm. The well-known confounds for aortic aneurysm, such as male sex, patient age, hypertension, hypercholesterolemia, history of coronary artery disease, and history of smoking,2 were frequency matched in the selection of the controls to maximize detection of unaccounted genetic contribution to the disease.
Genotyping and Calculation of the Hardy-Weinberg Equilibrium, Linkage Disequilibrium, and Logistic Regression
Genomic DNA of the case and control subjects (n=168 each) were genotyped for the SNPs of interest by the University of Wisconsin Biotechnology Center using the Kasper method (www.kbioscience.co.uk/reagents/KASP.html) with the primers listed in online-only Data Supplement Table SII. Ambiguous calls were regenotyped resulting in detection of all genotypes for all subjects with the exception of 1 failed call for rs41427445 for 1 control subject.
Of the 6 exon and 4 promoter SNPs with functional consequences (see Results) 4 (p-1562, N38S, E82K, L187F) with observed minor allele frequency of <20% were removed from further analysis as rare alleles are unlikely to contribute to a model discriminating cases and controls. Hardy–Weinberg equilibrium was calculated for the remaining 6 SNPs. We observed 1 SNP (D165N) at a significant deviation from Hardy–Weinberg equilibrium at P=0.05, but none at P=0.01. However, this SNP was retained in the analysis because the statistical tests performed in this study are valid in the presence of a departure from Hardy–Weinberg equilibrium. Linkage disequilibrium was calculated between each pair of SNPs on the basis of observed and expected genotypes using the linkage disequilibrium function in R and several SNPs demonstrated moderate linkage disequilibrium indicating potential for colinearity between these confounds. Five SNPs (p-2502, p-2118, p-1702, R279Q, and R668Q) displayed all 3 genotypes (AA, Aa, and aa), whereas 1 (D165N) displayed only the heterozygote and major homozygote genotypes (AA and Aa). “A” denotes the major allele and “a” the minor allele. Coding nomenclature was given from the perspective of the minor allele. Dominant (dominant coding being 0 for AA and 1 for Aa or aa) and recessive (recessive coding being 0 for AA or Aa and 1 for aa) forms of each SNP were examined where the minor homozygote genotype was observed. Additionally, an additive coding model was analyzed (AA=2, Aa=1, aa=0), but it did not yield any improvements or changes to the final model. For the final model, dominant forms of all SNPs except the recessive form of p-2502 were used. The 6 selected SNPs were tested for AAA association using step-wise logistic regression using the generalized linear model (glm) function in R based on the binomial family and coefficients of regression considered significant for P<0.10. Determination of whether to keep or eliminate a given confound in step-wise logistic regression was based on the Akaike information criteria threshold of 0.8.16 This goodness-of-fit measure penalizes for increase in the number of confounds and likely to lead to a compact model. Statistical inference for the individual coefficients and OR of the final logistic-regression model were based on a large sample approximation to the sample distribution, where standard error of the estimate SE=sqrt (∑1/ni), where ni are the numbers of observed data entries in a 2 × 2 contingency table.17 A receiver operating characteristic was constructed for the final logistic-regression model using the data from the 336 human cases and controls with a logit cutoff ≥0.799.
Results
Bioinformatics scanning of the MMP-9 gene revealed many annotated SNPs. Of these we chose 5 SNPs in the MMP-9 promoter and 9 SNPs in the coding region based on the relative abundance of the occurrence and the SNPs being missense mutations resulting in a change in the encoded amino acids. Table 1 lists the SNPs investigated with the common nomenclature used in this work and the corresponding SNP identification number. The heterozygosity information indicating the prevalence of the SNPs lists the historical data available around the time of initiation of this study and the current updated information (accessed January 20, 2012).
Our first assumption was that a given MMP-9 genotype could potentially serve as a biomarker for AAA only if the SNP resulted in a functional alteration either at the transcriptional level or at the enzymatic function level. We created MMP-9 cDNAs or promoter–reporter constructs encoding the nucleotide changes observed in the SNPs using the 2-step PCR technology. The primers used for the mutagenesis and creation of the starting wild-type 2.8 kb promoter–reporter construct are listed in online-only Data Supplement Table SI.
The functional consequences of exon SNPs were examined by expression of the cDNAs by transient transfection of HEK293 cells. The MMP-9 protein is secreted as a proenzyme, therefore, the culture supernatant was harvested and the protein abundance examined by Western blot and the enzymatic function quantified by gelatin zymography.
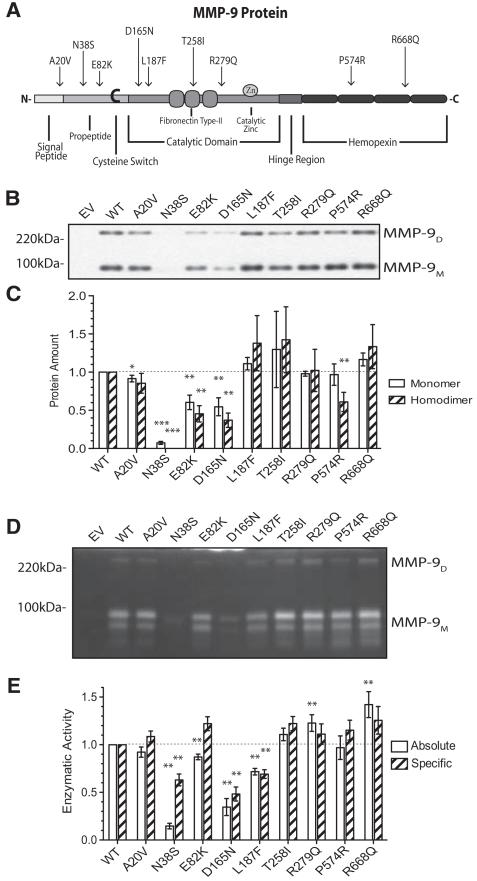
Of the 9 exon SNPs examined (Figure 1A), several showed a reduced amount of secreted protein. MMP-9 is secreted as monomers and dimers with potentially different properties influencing the overall activity of this enzyme.18 A separate densitometric analysis of dimeric and monomeric forms of the secreted protein amounts by nonreducing Western blot did not reveal a significant effect of any of the SNPs indicating no effect of the SNPs on MMP-9 dimerization (Figure 1B and 1C). Therefore, we used the ELISA assay for total MMP-9 protein in subsequent studies. Gelatin zymography of the culture supernatant demonstrated enzymatic activity of both the dimeric and monomeric MMP-9 for all SNPs with no difference in the dimer/monomer ratio between the constructs (data not shown). Densitometric quantitation demonstrated statistically significant variations in the total enzymatic activity (Figure 1D and 1E, open bars) with N38S, D165N, and L187F significantly lower, and R668Q with higher absolute activity. Two other SNPs demonstrated small (decrease for E82K and increase for R279Q) but statistically significant difference from the wild-type total enzymatic activity. Of note, the R279Q SNP previously suggested as possibly having reduced specific enzymatic activity due to the location of the amino acid change in the catalytic domain of MMP-919 showed a small increase in the total specific enzymatic activity compared with the wild-type. The specific enzymatic activity (total activity/secreted protein amount) was lower for N38S, D165N, and L187F suggesting a significant effect of these amino acid changes on the enzymatic activity in addition to the alteration of the protein secretion (Figure 1E, hatched bars).
Figure 1.
Functional consequences of MMP-9 Exon SNPs. A, A cartoon of the MMP-9 protein denoting the various functional domains and the relative locations of the SNPs investigated resulting in amino acid changes. C indicates the cysteine switch, Zn the zinc binding site, and the various functional domains are indicated by the different shades. B, Nonreducing PAGE of the culture supernatants collected 24 hours after transfection of HEK293 cells with the denoted constructs. The membrane after transfer was blotted with anti–MMP-9 antibody (1:1000 in TBST containing 1% dry milk) and shows the upper and lower-MMP-9 immunoreactive bands corresponding to the dimeric and monomeric MMP-9 protein. C, A bar plot summary of densitometric analysis of monomeric (open bars) and dimeric (hatched bars) protein amount normalized by the respective values for the wild-type. D, Gelatin zymography of cell culture supernatant of transfected cells, as in B. The enzymatic activity was quantified by densitometric analysis of the gelatin zymography (see online-only Data Supplement Figure SI). E, Absolute (open bars) and specific (hatched bars) enzymatic activity normalized to that of the wt-MMP-9 transfected experiment (first columns). The enzymatic activity quantified is the total activity including both the monomeric and dimeric MMP-9 activity. Bar plots are mean±SEM from 5 independent experiments. *P<0.05, **P<0.01, and ***P<0.001 by Mann–Whitney nonparametric test. MMP-9 indicates matrix metalloproteinase-9; SNP, single nucleotide polymorphism; and HEK, human embryonic kidney.
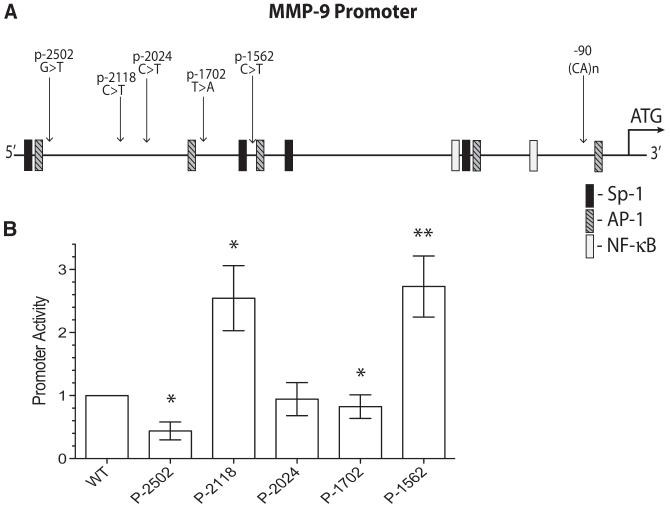
The 2.8 kb sequence of the MMP-9 gene upstream of the start methionine has potential consensus binding sites for many transcription factors (Figure 2A). The potential effects of SNPs in the promoter region of the MMP-9 gene were evaluated by the promoter–reporter constructs. The constructs were transfected into HEK293 cells and the firefly luciferase activity reflecting the promoter activity was normalized by the Renilla luciferase expressed by the cotransfected Renilla luciferase plasmid to control for potential variations in transfection efficiency and difference in the cell number in the wells. Increased promoter activity for p-2118 and p-1562, and reduced promoter activity for p-2502 and p-1702 were observed (Figure 2B). Except for the previously reported increase in the promoter activity for the p-1562 SNP, the functional consequences of the other SNPs revealed above have not been reported in the literature.
Figure 2.
Basal promoter activity of MMP-9 promoter SNPs. A, A cartoon of the MMP-9 genomic sequence upstream of the start methionine depicting the relative location of the 5 promoter SNPs and the (CA)n microsatellite polymorphism. Locations of the DNA consensus binding motifs for some of the common transcription factors are also noted. B, Relative promoter activity normalized to the wt promoter–reporter construct. HEK293 cells transfected with the noted promoter–reporter constructs in a 24-well dish (0.4 μg promoter–reporter plasmid + 0.013 μg Renilla plasmid/well) were harvested 24 hours later and replated into 3-replicate wells of a 96-well plate at an approximate cell density of 104 cells/well in DMEM supplemented with 1% FBS. The firefly luciferase promoter–reporter activity normalized by the Renilla luciferase activity to compensate for potential transfection efficiency and cell density differences were assessed 24 hours later. Bars represent mean±SEM from 6 independent experiments each with triplicate readings. *P<0.05 and **P<0.01 by Mann–Whitney nonparametric test. MMP-9 indicates matrix metalloproteinase-9; SNP, single nucleotide polymorphism; HEK, human embryonic kidney; FBS, fetal bovine serum.
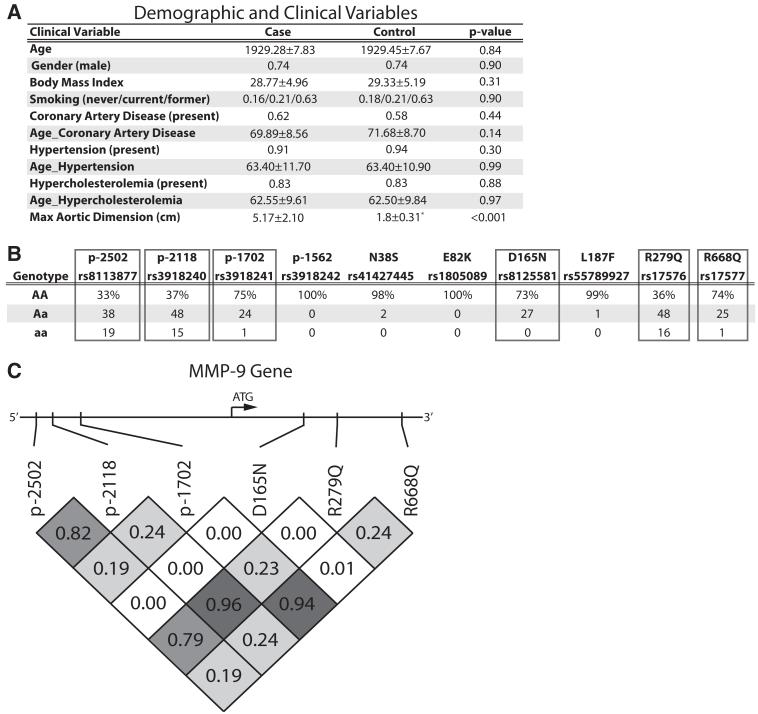
SNPs with functional consequences may alter the MMP-9 activity and may be of utility as biomarkers of diseases where the pathogenesis might correlate with the gene activity. We selected subjects with confirmed AAA (cases) and controls from the Marshfield Clinic’s PMRP database. The cases were selected on the basis of the documentation of AAA by 2 consecutive diagnoses based on ICD-9 code at different time points and with at least 1 imaging study confirming dilated abdominal aorta. The controls were selected from all those who have had at least 1 imaging study of the abdomen revealing no reported abnormality of the aorta. Manual abstraction of ≈10% of the medical records revealed no classification errors. To maximize the chances of revealing the role of MMP-9 genotype in AAA, we further matched our controls for the well-documented confounds of male sex and history of smoking, hypertension, hypercholesterolemia, and coronary artery disease positively correlating with AAA.2 Figure 3A lists the demographic and clinical data for the cases and controls confirming matching of the clinical confounds.
Figure 3.
Demographic, clinical, and genotype data for the case: control study. A, A table comparing the demographic and clinical variables of the cases and controls. Date of birth rather than the current age was used because the former is a better measure of age at the entry of the subjects into the PMRP database and a better measure allowing direct comparison between the cases and controls in this retrospective database study. Date of birth is given as year and a decimal value corresponding to the month/12. Age deceased for the case: control was 81.4 ± 7.96 and 80.68 ± 6.86, respectively (P=0.635 by t test). The maximum abdominal aortic dimension for the control group (*) is based on 66 subjects with reported abdominal aortic dimensions stated on the imaging reports and all subjects in this group had no mention of abdominal aortic abnormality on at least 1 imaging study of the abdominal aorta. Nominal variables are displayed as a percentage of total. B, Genotype distribution of the relevant SNPs in the combined population. The 6 enclosed in the box demonstrated MAF >20%. C, Linkage disequilibrium of the selected SNPs. The bar denotes the relative locations of the SNPs in the MMP-9 gene and the boxes denote the linkage disequilibrium measure D between the relevant SNPs. The D index of 0 indicates no linkage and 1.0 indicates perfect linkage. D165N has a D of 0.00 between other SNPs. MMP-9 indicates matrix metalloproteinase-9; SNP, single nucleotide polymorphism; PMRP, Personalized Medicine Research Project; and MAF, minor allele frequency.
The genomic DNA samples from cases and controls were genotyped for the newly identified functionally significant SNPs using the Kasper genotyping primers listed in online-only Data Supplement Table SII. Significant polymorphism exceeding 20% was observed for 6 of 10 SNPs confirmed to exhibit functional changes from the wild-type (enclosed in a box in Figure 3B). The 4 SNPs with insufficient frequency of occurrence in this population provided no information with regards to potential association with AAA and deleted from further analysis. The genotypes at the remaining 6 functionally significant SNPs were in Hardy–Weinberg equilibrium. A number of the SNPs selected did show moderate linkage disequilibrium indicating potential colinearity providing redundant information as biomarkers for AAA; however, D165N displayed no linkage disequilibrium with the other sites (Figure 3C). A step-wise logistic-regression analysis of the 6 SNPs revealed a significant association with AAA with the minimal model including only D165N (rs8125581) and p-2502 (rs8113877) SNPs (Equation 1) yielding an OR of 2.45 (95% confidence interval, 1.06–5.70) predictive of AAA. D165N reduces the enzymatic activity and p-2502 reduces the promoter activity both reducing the association with AAA in the logistic-regression model. A false positive rate of 5.4% and a positive predictive value of 69% were determined from the receiver–operator plot analysis.
| (Equation 1) |
The estimates of the model coefficients and the statistical properties are summarized in Table 2. The final model retains the nonspecified nonlinear interaction between rs8125581 (D165N) and rs8113877 (p-2502), most likely indicating that the decrease in the gene expression afforded by these 2 SNPs do not add linearly. Such nonlinear interaction is expected as the association with AAA and MMP-9 genotype is most likely through the final enzymatic activity of this elastolytic enzyme regardless of the genetic basis of the alteration in the gene expression.
Table 2. Summary of Logistic-Regression Model Included in Equation 1.
| Coefficients | Estimate | Std Err | P Value | Coding | Genotype Model |
|---|---|---|---|---|---|
| A0 (intercept) | 0.799 | 0.401 | 0.047 | — | — |
| A1 (rs8125581) | −0.799 | 0.458 | 0.081 | Dominant | GC vs AG |
| A2 (rs8113877) | −0.983 | 0.492 | 0.046 | Recessive | GG+GT vs TT |
| A3 (rs8125581: rs8113877) |
0.799 | 0.556 | 0.151 | Dominant: Recessive |
— |
Std Err indicate standard error.
Discussion
MMP-9 levels in abdominal aortic walls correlate with rupture and MMP-2 with the expansion of the aneurysm20 and the levels of circulating elastin and collagen degradation products has been suggested as a useful predictor of AAA expansion with potential of clinical utility in predicting those that will need intervention within 5 years.21 Consistent with a role of MMP in AAA, inhibition of the enzymatic activity with doxy-cycline has been shown to reduce the aneurysmal dilation or MMP-9 activity in several experimental models of AAA and in small human clinical trials.22,23 Despite the abundance of data implicating MMP-9 activity in AAA, association studies between MMP-9 genotype and AAA have not revealed a significant association. The problem, we believe, is in the selection of the MMP-9 SNPs studied.
Starting with the assumption that SNPs must have a functional consequence on either the promoter or the enzymatic activity of MMP-9 to be a viable candidate for an association study with AAA, we studied the effects of selected MMP-9 SNPs using Western blot, gelatin zymography, and a promoter–reporter assay. Because of the tight posttranscriptional and posttranslational regulation of MMP-9 (eg, miRNA targeting, latent pro-MMP-9 activation and TIMP inhibition) alterations in promoter activity and enzymatic activity provides only an initial approximation of the fundamental effect SNPs have on MMP-9 biology, nevertheless, providing critical a priori information to perform a guided genotype association study. We documented altered specific enzymatic activity for 3 exon SNPs and altered promoter activity in 4 promoter SNPs. The exon N38S SNP results in the loss of an N-glycosylation site and the decreased secretion of this mutant may be because of altered subcellular targeting. The D165N and L187F SNP is located within the enzyme catalytic domain close to the residues responsible for Ca2+ chelation, whereas the R668Q resides within the hemopexin domain mediating homo-oligomerization and interaction with TIMPs. On the promoter side, nothing mechanistic is known about the reasons for the altered promoter activity for any of the SNPs. The p-1562 SNP results in an altered mobility shift on the electrophoretic mobility shift assay,6 but the identity of this SNP nucleotide specific DNA-binding protein has not been reported. Furthermore, an increase in the transcript in itself does not guarantee increased enzymatic activity and further studies are necessary to better understand how these newly identified exon and promoter SNPs alter MMP-9 expression and function.
Genotyping of AAA cases and controls at these 10 SNPs showed sufficient genotype heterogeneity for 6 variants, and a logistic-regression analysis based on these 6 SNPs revealed a regression model with a modest association between MMP-9 SNPs and AAA. Further step-wise logistic-regression analysis reduced the significant confounds to p-2502 and D165N with an overall OR=2.45. Our study differs from prior association studies in 2 important respects: (1) we focused our association with SNPs identified as being functionally significant, and (2) we selected a frequency-matched control population matching demographic and clinical confounds known to be associated with AAA. These 2 methodological differences, we believe, might have allowed us to reveal the significant MMP-9 genotype association with AAA.
Our analysis revealed a significant correlation between MMP-9 genotype and AAA with a large OR compared with the previous reports. A meta-analysis seeking association between various genotypes and AAA reported a relative risk of 1.094 and a more recent meta-analysis failed to reveal any significant influence of the MMP-9 gene.5 Both analyses, however, were based on the meta-analysis of only 3 original reports investigating for potential association between MMP-9 genotype and AAA and all reports only looked at the MMP-9 p-1562 promoter polymorphism. The largest of these reports by Jones et al24 compared 414 subjects with AAA to 172 subjects with peripheral vascular diseases and 203 healthy control subjects.24 Their final analysis revealed a model with an OR of 2.41 (AAA versus healthy controls) and OR of 2.94 (AAA versus peripheral vascular diseases). This report of a positive association is balanced by the conclusion from 2 reports of no association between the same MMP-9 SNP and AAA.15,25 Our functional analysis did indicate an increased promoter activity with the less frequent T compared with the C allele for the p-1562 SNP in agreement with a prior report6 although a recent report did not observe an alteration of MMP-9 expression in vascular smooth muscle cells by this promoter polymorphism.26 In our sample population of an entirely white cohort from northern Wisconsin, there was insufficient heterogeneity at this SNP to provide any information.
In general, the strongest final logistic-regression model only consists of terms with P<0.05, and this approach should be taken in the absence of any additional information. However, biochemical experiments support the importance of the SNPs rs8125581 (D165N) and rs8113877 (p-2502) in MMP-9 function lending additional confidence to inclusion of these 2 terms which display P values suggestive of significance (0.05≈P<0.10) in the final model (Table 2). In addition, these 2 SNPs only achieve this level of significance when the interaction term (rs8125581: rs8113877) is included in the model. Therefore, the interaction term must be included in the final model although it is outside of the conventional significance threshold. In a hypothetical, analogous situation wherein an interaction is significant, but ≥1 of the main terms are not, it is standard practice to include all of the main terms regardless because the main terms and interaction form a unit. Importantly, these 3 terms (rs8125581, rs8113877, and rs8125581:rs8113877) consist of the 3 strongest P values of the terms considered in the model. A mechanistic interpretation of this phenomenon can be explained by the functional alterations these SNPs impart. The SNP, rs8125581 (D165N), results in a variant that decreases enzymatic function whereas rs8113877 (p-2502) results in a variant that leads to a reduction in promoter activity of MMP-9. By impairing both the expression and activity of MMP-9, individuals with both of these SNPs exhibit a decrease in AAA risk that exceeds an additive model. Unfortunately, only 29 of the 336 subjects in our study possess both SNP alleles, resulting in a weaker P value. A simulation assuming the same proportion of the genotypes observed indicates that an additional 100 cases and controls for a total sample size of 436 should yield a P<0.05 for the 2 SNPs in equation 1. With twice the sample size (total of 672 subjects), we can expect a P<0.01 for the 2 main SNPs and a P<0.05 for the interaction term. We believe the marginal P values obtained in our analysis is because of the limited sample size but based on our biochemical evidence, we believe these SNPs as well as the interaction term to be real.
Despite the significant OR in our final 2 SNP model with p-2502 and D165N, the large A0 coefficient points to the fact that a large number of the observed subjects with AAA is not explained by the MMP-9 genotype even in the frequency-matched cases and controls. AAA is clearly a complex disease with many likely genes involved in its pathogenesis. Expression of MMP-9 itself is regulated by mechanisms such as promoter methylation and microRNA inhibition27-29 not captured by a simple genotype analysis and inhibition of the enzymatic activity by circulating factors such as TIMP is also well described.30 Nevertheless, we believe our study brings back MMP-9 as a relevant gene with a strong association with AAA, potentially useful in the clinical diagnosis of the disease. Future studies should address the limitations of our present study, such as the mostly homogeneous racial distribution to whites and the inclusion of non–MMP-9 genotypes such as TIMP, methylenetetrahydrofolate reductase, and angiotensin-converting enzyme to obtain an association model with a greater ability to discriminate those with AAA. Validity of the 2 SNP model demonstrating correlation between MMP-9 genotype and AAA must be confirmed in an independent larger cohort of subjects and controls.
The main utility of any biomarker study for AAA (or any disease for that matter) is in guiding rational utilization of medical resources. A periodic follow-up ultrasound of AAA that is the current standard of care is noninvasive and cost-effective in reducing the chances of an unexpected rupture and a surgical emergency. The practical question is who should be screened with an ultrasound study and how frequently should the follow up imaging study be performed? We believe a robust biomarker for AAA could serve as an additional criterion for selecting those asymptomatic individuals who might be at an increased risk. We do not think biomarkers alone will provide sufficient sensitivity to replace imaging that can provide definitive diagnosis and prognosis of AAA, however, a systematic search for a better comprehensive biomarker model based on functionally confirmed SNPs of contributory genes should move us toward rational utilization of imaging studies for selecting those asymptomatic individuals that might be at an increased risk and in need of therapeutic intervention.
In conclusion, we provide evidence for the association of MMP-9 genotype with AAA using a guided approach with functional knowledge of the direct SNP effects. With this a priori information we were able to generate a more concise, relevant genotype association with AAA.
Supplementary Material
CLINICAL PERSPECTIVE.
Abdominal aortic aneurysm (AAA) is the 10th-leading cause of death in men over the age of 50. Aortic rupture causes as many as 30 000 deaths annually, rivaling prostate or breast cancer. Matrix metalloproteinase 9 (MMP9) that targets extracellular matrix proteins has been implicated in the breakdown of the aortic wall leading to AAA formation. Previous genotype association studies between AAA and MMP9 focused on a few select single nucleotide polymorphisms with variable results. This work examines multiple MMP9 single nucleotide polymorphisms in a well-defined human population comprising AAA case and controls and provides a multiple allele-association model with AAA with possible utility as a genetic biomarker. Development of such a biomarker could serve as an additional criterion, in conjunction with other known clinical risk factors, for identifying asymptomatic individuals that might be at an increased risk for AAA.
Acknowledgments
We thank the Wisconsin Genomic Initiative Demonstration Project Grant from the State of Wisconsin and the Betty Bamforth Endowment Fund from the Department of Anesthesiology for providing partial support for this study. The members of the Biomedical Informatics Research Center at Marshfield Clinics Research Foundation performed screening and data extraction using the Marshfield Clinics and PMRP databases. University of Wisconsin Office of Clinical Trials is partly supported by grant 1UL1RR025011 from the Clinical and Translational Science Award program of the National Center for Research Resources, NIH.
Sources of Funding
This work was supported by the Genomics Institute Demonstration Project Grant, State of Wisconsin; Betty Bamforth Endowment Fund, Department of Anesthesiology, University of Wisconsin–Madison.
Footnotes
Disclosures
None.
The online-only Data Supplement is available at http://circgenetics.ahajournals.org/lookup/suppl/CIRCGENETICS.112.963082/-/DC1.
Contributor Information
Tyler Duellman, Molecular and Cellular Pharmacology Graduate Program, University of Wisconsin School of Medicine and Public Health, Madison.
Christopher L. Warren, Illumavista Biosciences LLC, Madison, WI.
Peggy Peissig, Biomedical Informatics Research Center, Marshfield Clinics Research Foundation, Marshfield, WI.
Martha Wynn, Department of Anesthesiology, University of Wisconsin School of Medicine and Public Health, Madison.
Jay Yang, Molecular and Cellular Pharmacology Graduate Program, University of Wisconsin School of Medicine and Public Health, Madison; Department of Anesthesiology, University of Wisconsin School of Medicine and Public Health, Madison.
References
- 1.Sandford RM, Bown MJ, London NJ, Sayers RD. The genetic basis of abdominal aortic aneurysms: a review. Eur J Vasc Endovasc Surg. 2007;33:381–390. doi: 10.1016/j.ejvs.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 2.Kent KC, Zwolak RM, Egorova NN, Riles TS, Manganaro A, Moskowitz AJ, et al. Analysis of risk factors for abdominal aortic aneurysm in a cohort of more than 3 million individuals. J Vasc Surg. 2010;52:539–548. doi: 10.1016/j.jvs.2010.05.090. [DOI] [PubMed] [Google Scholar]
- 3.Johansen K, Koepsell T. Familial tendency for abdominal aortic aneurysms. JAMA. 1986;256:1934–1936. [PubMed] [Google Scholar]
- 4.Thompson AR, Drenos F, Hafez H, Humphries SE. Candidate gene association studies in abdominal aortic aneurysm disease: a review and meta-analysis. Eur J Vasc Endovasc Surg. 2008;35:19–30. doi: 10.1016/j.ejvs.2007.07.022. [DOI] [PubMed] [Google Scholar]
- 5.McColgan P, Peck GE, Greenhalgh RM, Sharma P. The genetics of abdominal aortic aneurysms: a comprehensive meta-analysis involving eight candidate genes in over 16,700 patients. Int Surg. 2009;94:350–358. [PubMed] [Google Scholar]
- 6.Zhang B, Ye S, Herrmann SM, Eriksson P, de Maat M, Evans A, et al. Functional polymorphism in the regulatory region of gelatinase B gene in relation to severity of coronary atherosclerosis. Circulation. 1999;99:1788–1794. doi: 10.1161/01.cir.99.14.1788. [DOI] [PubMed] [Google Scholar]
- 7.Janssens S, Lijnen HR. What has been learned about the cardiovascular effects of matrix metalloproteinases from mouse models? Cardiovasc Res. 2006;69:585–594. doi: 10.1016/j.cardiores.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Koullias GJ, Ravichandran P, Korkolis DP, Rimm DL, Elefteriades JA. Increased tissue microarray matrix metalloproteinase expression favors proteolysis in thoracic aortic aneurysms and dissections. Ann Thorac Surg. 2004;78:2106–10. doi: 10.1016/j.athoracsur.2004.05.088. discussion 2110. [DOI] [PubMed] [Google Scholar]
- 9.McMillan WD, Pearce WH. Increased plasma levels of metalloproteinase-9 are associated with abdominal aortic aneurysms. J Vasc Surg. 1999;29:122–7. doi: 10.1016/s0741-5214(99)70363-0. discussion 127. [DOI] [PubMed] [Google Scholar]
- 10.McCarty CAWR, Giampietrao PF, Wesbrook SD, Caldwell MD. Marshfield clinics personalized medicine research project (pmrp): Design methods and recruitment for a large population-based biobank. Personalized Medicine. 2005;2:49–79. doi: 10.1517/17410541.2.1.49. [DOI] [PubMed] [Google Scholar]
- 11.Peters DG, Kassam A, St Jean PL, Yonas H, Ferrell RE. Functional polymorphism in the matrix metalloproteinase-9 promoter as a potential risk factor for intracranial aneurysm. Stroke. 1999;30:2612–2616. doi: 10.1161/01.str.30.12.2612. [DOI] [PubMed] [Google Scholar]
- 12.Shimajiri S, Arima N, Tanimoto A, Murata Y, Hamada T, Wang KY, et al. Shortened microsatellite d(CA)21 sequence down-regulates promoter activity of matrix metalloproteinase 9 gene. FEBS Lett. 1999;455:70–74. doi: 10.1016/s0014-5793(99)00863-7. [DOI] [PubMed] [Google Scholar]
- 13.St Jean PL, Zhang XC, Hart BK, Lamlum H, Webster MW, Steed DL, et al. Characterization of a dinucleotide repeat in the 92 kDa type IV collagenase gene (CLG4B), localization of CLG4B to chromosome 20 and the role of CLG4B in aortic aneurysmal disease. Ann Hum Genet. 1995;59(Pt 1):17–24. doi: 10.1111/j.1469-1809.1995.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 14.Yoon S, Tromp G, Vongpunsawad S, Ronkainen A, Juvonen T, Kuivaniemi H. Genetic analysis of MMP3, MMP9, and PAI-1 in Finnish patients with abdominal aortic or intracranial aneurysms. Biochem Biophys Res Commun. 1999;265:563–568. doi: 10.1006/bbrc.1999.1721. [DOI] [PubMed] [Google Scholar]
- 15.Armani C, Curcio M, Barsotti MC, Santoni T, Di Stefano R, Dell’omodarme M, et al. Polymorphic analysis of the matrix metalloproteinase-9 gene and susceptibility to sporadic abdominal aortic aneurysm. Biomed Pharmacother. 2007;61:268–271. doi: 10.1016/j.biopha.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Akaike H. A new look at the statistical model identification. IEEE Trans Aut Cont. 1974:716–723. [Google Scholar]
- 17.Mosteller F. Association and estimation in contingency tables. J Am Stat Assoc. 1968:1–28. [Google Scholar]
- 18.Olson MW, Bernardo MM, Pietila M, Gervasi DC, Toth M, Kotra LP, et al. Characterization of the monomeric and dimeric forms of latent and active matrix metalloproteinase-9. Differential rates for activation by stromelysin 1. J Biol Chem. 2000;275:2661–2668. doi: 10.1074/jbc.275.4.2661. [DOI] [PubMed] [Google Scholar]
- 19.Zhang B, Henney A, Eriksson P, Hamsten A, Watkins H, Ye S. Genetic variation at the matrix metalloproteinase-9 locus on chromosome 20q12.2-13.1. Hum Genet. 1999;105:418–423. doi: 10.1007/s004390051124. [DOI] [PubMed] [Google Scholar]
- 20.Petersen E, Wågberg F, Angquist KA. Proteolysis of the abdominal aortic aneurysm wall and the association with rupture. Eur J Vasc Endovasc Surg. 2002;23:153–157. doi: 10.1053/ejvs.2001.1572. [DOI] [PubMed] [Google Scholar]
- 21.Lindholt JS, Heickendorff L, Vammen S, Fasting H, Henneberg EW. Five-year results of elastin and collagen markers as predictive tools in the management of small abdominal aortic aneurysms. Eur J Vasc Endovasc Surg. 2001;21:235–240. doi: 10.1053/ejvs.2001.1329. [DOI] [PubMed] [Google Scholar]
- 22.Baxter BT, Terrin MC, Dalman RL. Medical management of small abdominal aortic aneurysms. Circulation. 2008;117:1883–1889. doi: 10.1161/CIRCULATIONAHA.107.735274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abdul-Hussien H, Hanemaaijer R, Verheijen JH, van Bockel JH, Geelkerken RH, Lindeman JH. Doxycycline therapy for abdominal aneurysm: Improved proteolytic balance through reduced neutrophil content. J Vasc Surg. 2009;49:741–749. doi: 10.1016/j.jvs.2008.09.055. [DOI] [PubMed] [Google Scholar]
- 24.Jones GT, Phillips VL, Harris EL, Rossaak JI, van Rij AM. Functional matrix metalloproteinase-9 polymorphism (C-1562T) associated with abdominal aortic aneurysm. J Vasc Surg. 2003;38:1363–1367. doi: 10.1016/s0741-5214(03)01027-9. [DOI] [PubMed] [Google Scholar]
- 25.Ogata T, Shibamura H, Tromp G, Sinha M, Goddard KA, Sakalihasan N, et al. Genetic analysis of polymorphisms in biologically relevant candidate genes in patients with abdominal aortic aneurysms. J Vasc Surg. 2005;41:1036–1042. doi: 10.1016/j.jvs.2005.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maqbool A, Turner NA, Galloway S, Riches K, O’Regan DJ, Porter KE. The −1562C/T MMP-9 promoter polymorphism does not predict MMP-9 expression levels or invasive capacity in saphenous vein smooth muscle cells cultured from different patients. Atherosclerosis. 2009;207:458–465. doi: 10.1016/j.atherosclerosis.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z, Lu S, Liu C, Zhao B, Pei K, Tian L, et al. Expressional and epigenetic alterations of placental matrix metalloproteinase 9 in preeclampsia. Gynecol Endocrinol. 2010;26:96–102. doi: 10.3109/09513590903184100. [DOI] [PubMed] [Google Scholar]
- 28.Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, et al. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis Rheum. 2005;52:3110–3124. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 29.Jones AESR, O’Quinn EC, Black LE, Barth JL, Elefteriades JA, Bavaria JE, et al. Selective microrna suprresion in human thoracic aneurysms: Relationship of mir-29a to aortic size and proteoltic induction. Circulation Cardiovascular Genetics. 2012:605–613. doi: 10.1161/CIRCGENETICS.111.960419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta. 2010;1803:55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.