Abstract
Cytoplasmic dynein plays important roles in mitosis and the intracellular transport of organelles, proteins, and mRNAs. Dynein function is particularly critical for survival of neurons, as mutations in dynein are linked to neurodegenerative diseases. Dynein function is also implicated in neuronal regeneration, driving the active transport of signaling molecules following injury of peripheral neurons. To enhance our understanding of dynein function and regulation in neurons, we established a novel knock-in mouse line in which the neuron-specific cytoplasmic dynein 1 intermediate chain 1 (IC-1) is tagged with both GFP and a 3xFLAG tag at its C-terminus. The fusion gene is under the control of IC-1’s endogenous promoter and is integrated at the endogenous locus of the IC-1-encoding gene Dync1i1. The IC-1-GFP-3xFLAG fusion protein is incorporated into the endogenous dynein complex, and movements of GFP-labeled dynein expressed at endogenous levels can be observed in cultured neurons for the first time. The knock-in mouse line also allows isolation and analysis of dynein-bound proteins specifically from neurons. Using this mouse line we have found proteins, including 14-3-3 zeta, which physically interact with dynein upon injury of the brain cortex. Thus, we have created a useful tool for studying dynein function in the central nervous system under normal and pathologic conditions.
Introduction
Cytoplasmic dynein plays a variety of roles in the cell ranging from mitosis to the transport of many cellular cargoes such as organelles/vesicles, proteins and mRNAs [Karki and Holzbaur 1999; Kardon and Vale 2009; Vaughan 2011; Moore and Cooper 2010; Vallee et al., 2012]. The function of cytoplasmic dynein is especially important for the survival of neuronal cells, and defects in dynein and its accessory complex, dynactin, are linked to neuron degeneration [LaMonte et al., 2002; Heerssen et al., 2004; Munch et al., 2004; Puls et al., 2003; 2005; Chen et al., 2007; Lai et al., 2007; Lambrechts et al., 2007; Chevalier-Larsen et al., 2008; Cosker et al., 2008; Ilieva et al., 2008; Perlson et al., 2010; Banks et al., 2011; Weedon et al., 2011; Harms et al., 2012]. Dynein transports a variety of cargoes in neurons including short microtubules, mitochondria, endosomes, autophagosomes, and prion protein vesicles [Bass and Buster 2004; Ha et al., 2008; Zhang et al., 2009; Pandey and Smith 2011; Yi et al., 2011; Encalada et al., 2011; Maday et al., 2012; Sheng and Cai 2012; Zhou et al., 2012; Mitchell et al., 2012], but how dynein interacts with all these cargoes requires further study [Akhmanova and Hammer 2010; Vallee et al., 2012]. Moreover, dynein has also been implicated in cellular responses to neuronal injury. Upon axonal injury of peripheral neurons, dynein is involved in transporting injury signaling molecules toward the nucleus, a process implicated in neuronal regeneration [Cavalli et al., 2005; Perlson et al., 2005; Abe and Cavalli 2008; Perlson et al., 2010]. Whether this happens in the central nervous system is unclear and deserves to be further studied.
Cytoplasmic dynein is a complex of multiple proteins including heavy chains (HCs), intermediate chains (ICs), light intermediate chains (LICs) and light chains (LCs) [Pfister et al., 2005]. The two HCs form a homodimer whose motor heads are responsible for ATP-hydrolysis-coupled motility along microtubules [Karki and Holzbaur 1999; Reck-Peterson et al., 2011; Höök and Vallee 2012]. The HCs of cytoplasmic dynein 1, the major form of cytoplasmic dynein in mammalian organisms, are encoded by a single gene in mammalian genomes [Pfister et al., 2005]. Because it is required for many cellular tasks, it was not surprising that homozygous knockout of dynein HC results in an embryonic lethal phenotype [Harada et al., 1998]. The ICs bind to the tail of the HCs [Habura et al., 1999; Tynan et al., 2000], and the ICs are known to bind the dynactin complex important for dynein function in cells [Karki and Holzbaur 1995; Vaughan and Vallee 1995; Plamann et al., 1994; Schroer 2004; Kim et al., 2007; Moore et al., 2009; Zhang et al., 2011; Lloyd et al., 2012; Moughamian and Holzbaur 2012; Yeh et al., 2012]. There are two cytoplasmic dynein 1 IC genes in the mouse, rat and human genomes [Vaughan and Vallee 1995; Pfister et al., 2005]. In mice, one gene encodes for IC-1 whose expression is only detected in brain, testis and ovary [Kuta et al., 2010], and another gene encodes for IC-2 whose expression is detected in all tissues [Kuta et al., 2010]. In the brain, IC-1 appears to be expressed specifically in neurons, while IC-2 is expressed in both neurons and glia [Pfister et al., 1996; Ha et al., 2008].
Despite the importance of cytoplasmic dynein in neurons, tools for studying neuronal dynein have been limited. For example, there has never been a mouse line in which fluorescence-labeled dynein expressed at its endogenous level can be visualized in neurons. In addition, it has been difficult to isolate neuronal proteins that bind to dynein in response to brain injury or under other pathogenic conditions. Here we have created the first dynein-GFP knock-in mouse line in which the neuron-specific dynein IC-1 is tagged with GFP and 3xFLAG at its C-terminus, and the expression of the IC-1-GFP-3XFLAG fusion is under the control of IC-1’s endogenous promoter. This line should become a very useful tool for the neuronal cell biology community, as it will allow investigators to visualize GFP-labeled dynein in neurons and identify neuronal dynein-bound proteins under various physiological, pathological and pathogenic conditions.
Results
Establishing a homozygous knock-in line of mice in which the dynein IC-1 is tagged with GFP and a 3xFLAG affinity tag
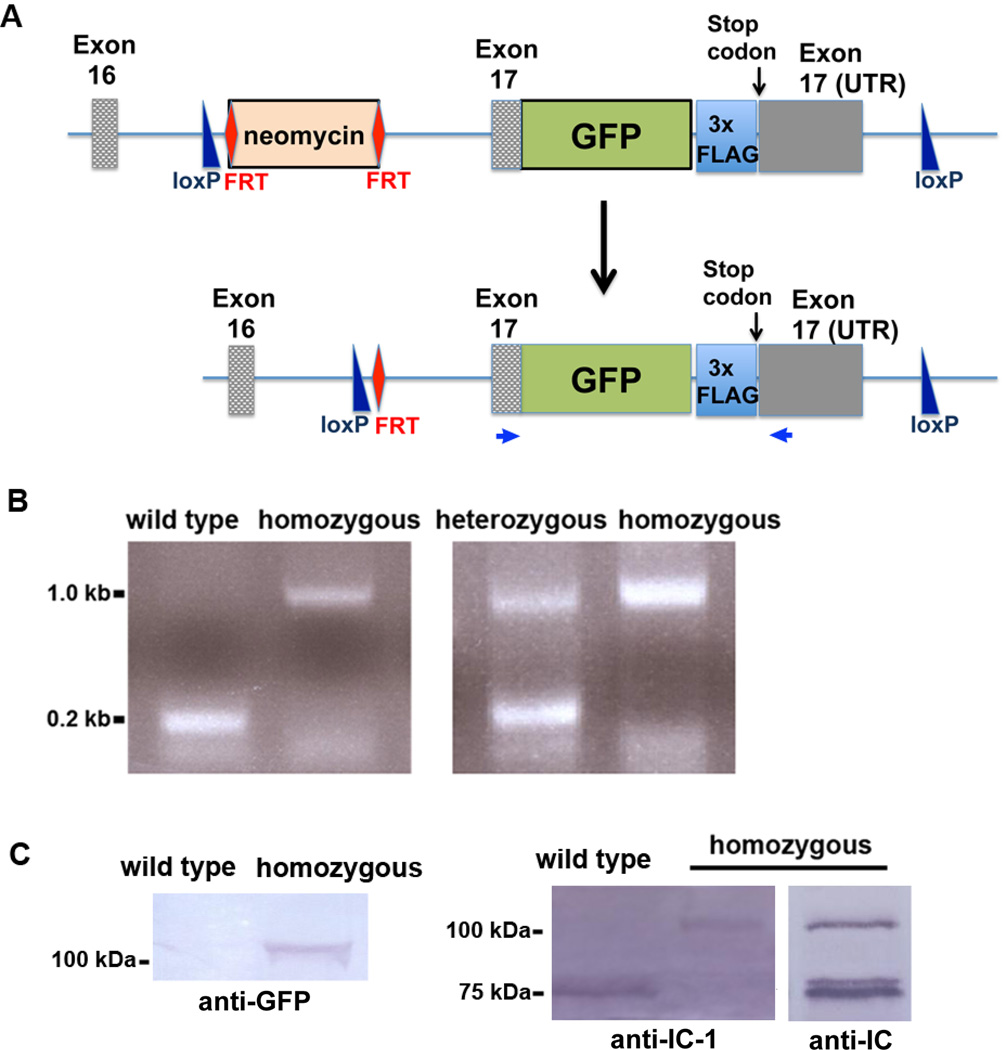
The gene that encodes the IC-1 of cytoplasmic dynein 1 is called Dync1i1 [Pfister et al., 2005]. It is located on chromosome 6 and contains 17 exons. A knock-in strategy was used to insert the GFP tag and the 3xFLAG tag right before the stop codon in the middle of exon 17 of the endogenous Dync1i1 locus (Figure 1A). The knock-in mouse line was created as described in Materials and Methods. Information on the genomic DNA sequence of the knock-in allele of Dync1i1 is presented in Figure S1, and that of the endogenous Dync1i1 allele is presented in Figure S2. Heterozygous mice were crossed to obtain homozygous knock-in mice. Heterozygous mice were verified by both PCR and Southern blot analyses; a PCR-based strategy was used to screen for homozygous progeny (Figure 1B). Primers used for PCRs as well as the stop codon are highlighted in Figure S1 and Figure S2. This pair of primers should generate a 0.2-kb product from the endogenous allele and a 1-kb product from the knock-in allele. In the homozygous knock-in mice, only the 1-kb product was generated (Figure 1B), demonstrating that the endogenous allele had been replaced by the knock-in allele.
Figure 1.
Construction and characterization of the dynein IC-1 knock-in mice. (A) A diagram showing the in-frame insertion of the GFP tag and the 3xFLAG tag before the stop codon of the endogenous Dync1i1 gene. The selection marker, the FRT-site-flanked neomycin gene, was inserted in the intron between exons 16 and 17. Note that the coding region of exon 17 is labeled as “Exon 17” and the 3’ untranslated region (UTR) of exon 17 is labeled as “Exon 17 (UTR)”. Two loxP sites were also inserted to flank exon 17, which would potentially aid in the deletion of the coding region of exon 17 and its 3’ untranslated region for making a partial IC-1 knockout mouse model. The neomycin marker was removed by breeding with Flp delete mice. The two primers used for the PCR analysis shown in B are indicated as small blue arrows. (B) A PCR-based method to screen for homozygous and heterozygous knock-in mice. Positions of the primers are shown in A. This pair of primers generates a 0.2 kb product for the wild type, a 1 kb product for the homozygous knock-in mice and both of these products for the heterozygous knock-in mice. (C) Western blots showing the IC-1-GFP-3xFLAG fusion protein of ~100 kDa recognized by an anti-GFP antibody, the IC-1-specific antibody (labeled as “anti-IC-1”) and the general anti-IC antibody 74.1 (labeled as “anti-IC”). Note that the anti-IC-1 antibody only recognized the ~75 kDa IC-1 protein in the wild type sample and the ~100 kDa fusion protein in the homozygous sample while the anti-IC antibody recognized both the IC-1-GFP-3xFLAG fusion protein (~100 kDa) and the IC-2 protein (~75 kDa) in the homozygous sample.
Homozygous mice were also analyzed by western blot. Total brain extract isolated from either wild type or homozygous mice was probed with several different antibodies. The anti-GFP antibody recognized the ~100-kDa IC-1-GFP-3xFLAG fusion protein in the homozygous sample but not in the wild type sample (Figure 1C). When probed with the IC-1-specific antibody [Mitchell et al., 2012], the ~100 kDa IC-1-GFP-3xFLAG fusion but not the ~75 kDa endogenous IC-1 protein was identified in the homozygous sample (Figure 1C), confirming that the endogenous IC-1 allele has indeed been replaced by the allele encoding IC-1-GFP-3xFLAG. When probed with the general anti-IC antibody 74.1 [Dillman and Pfister 1994], which recognizes both IC-1 and IC-2, both the ~100 kDa IC-1-GFP-3xFLAG fusion protein and the ~75 kDa IC-2 protein were detected in the homozygous sample (Figure 1C). In most western blots described in this paper, we used the commercially available anti-IC antibody 74.1.
IC-1-GFP-3xFLAG incorporates into endogenous dynein and can be present in the same dynein complex with IC-2
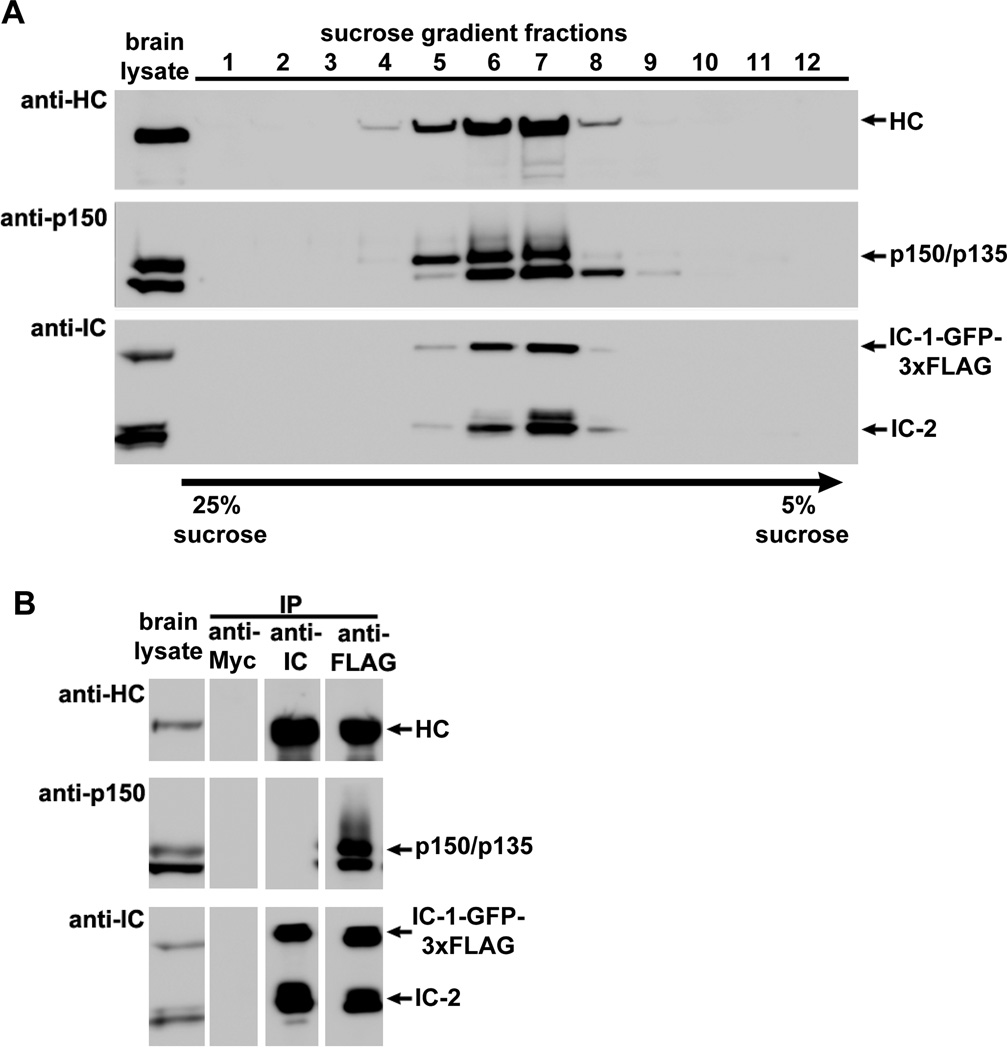
To determine if the IC-1-GFP-3xFLAG fusion protein is incorporated into the endogenous dynein complex, we first performed a sucrose-gradient sedimentation experiment using total brain protein extract from the homozygous knock-in mice. Western analyses of the sucrose-gradient fractions demonstrated that the IC-1-GFP-3xFLAG fusion protein, just like the endogenous IC-2, co-sediments with the dynein HC as well as p150/p135 of the dynactin complex (Figure 2A). We next performed immunoprecipitation experiments using an anti-Myc antibody (as a negative control), the anti-IC antibody 74.1 and an anti-FLAG antibody (Figure 2B). The anti-IC antibody 74.1 was raised against an N-terminal epitope shared by IC-1 and IC-2 [Dillman and Pfister 1994; Vaughan and Vallee 1995], and it blocks the dynactin-IC interaction [McKenney et al., 2011], which involves IC’s N-terminal region [Vaughan and Vallee 1995; King et al., 2003]. Thus, while it co-immunoprecipitated dynein HC with IC-1-GFP-3xFLAG and IC-2, it did not co-immunoprecipitate p150/p135 of the dynactin complex (Figure 2B). In contrast, the antibody against the FLAG tag placed at the C-terminus of IC-1 co-immunoprecipitated p150/p135 proteins (Figure 2B), indicating that the IC-1-GFP-3xFLAG fusion protein is functional in interacting with dynactin. In addition, dynein HC was also co-immunoprecipitated in the same experiment, indicating that the IC-1-GFP-3xFLAG fusion protein is able to bind dynein HC. Together, these results demonstrate that the IC-1-GFP-3xFLAG fusion protein is incorporated into the endogenous dynein complex and is also able to interact with the dynactin complex.
Figure 2.
The dynein IC-1-GFP-3xFLAG fusion protein is incorporated into endogenous dynein and can be present in the same complex with IC-2. (A) Result of a sucrose-gradient sedimentation experiment indicating that both the IC-1-GFP-3xFLAG fusion protein and the native IC-2 co-fractionate with dynein HC and p150/p135 of the dynactin complex. Brain lysate from a homozygous IC-1 knock-in mouse was fractionated over a 5–25% sucrose gradient, and western blots were probed with the anti-HC, anti-IC and anti-p150 antibodies. (B) Results of immunoprecipitation experiments done using brain lysate from a homozygous IC-1 knock-in mouse indicating that the IC-1-GFP-3xFLAG fusion protein is incorporated into endogenous dynein. Immunoprecipitation with an anti-Myc antibody (anti-Myc) is shown as a negative control. Immunoprecipitation with the anti-IC antibody is shown for comparison (this antibody blocks dynactin binding [McKenney et al., 2011], and thus, the p150/p135 bands are not seen in the IP). The IC-1-GFP-3xFLAG fusion protein is incorporated into endogenous dynein as assessed by co-immunoprecipitation of dynein HC and dynactin subunits p150/p135 with an anti-FLAG antibody. Note that in the anti-FLAG IP lane, both the IC-1-GFP-3xFLAG fusion protein and the endogenous IC-2 isoforms were detected, indicating that the IC-1 fusion is able to form heterodimer with IC-2.
IC-1 and IC-2 isoforms are implicated in transporting different cellular cargoes since the neuron-specific IC-1B isoform but not the ubiquitous IC-2C isoform is involved in transporting neurotrophin receptor tyrosine kinase B (TrkB) signaling endosomes in cultured hippocampal neurons [Ha et al., 2008]. However, differentially tagged IC-1 and IC-2, when overexpressed in tissue culture cells, are able to pull down each other [Lo et al., 2006], suggesting that they may be able to associate with each other. In our current immunoprecipitation experiments using the anti-FLAG antibody, IC-2 proteins were clearly co-immunoprecipitated (Figure 2B). These results demonstrate for the first time that IC-1 and IC-2 can be present in the same dynein complex in vivo when they are expressed at endogenous levels. Studies of native and reconstituted mammalian cytoplasmic dynein complexes indicate that ICs are present as a dimer within a mammalian cytoplasmic dynein complex as the IC:HC ratio is about 1:1 [Kini et al., 2001; Trokter et al., 2012] (although IC:HC for budding yeast dynein could possibly be 2:1 [Markus et al., 2011]). Thus, our results suggest that IC-1 and IC-2 proteins can be present in the same complex as a heterodimer, raising the possibility that IC-1 and IC-2 may have independent as well as cooperative functions in the same neuronal cells.
IC-1-GFP-3xFLAG expression levels in hippocampal and cortical neurons increase as cultures mature in vitro
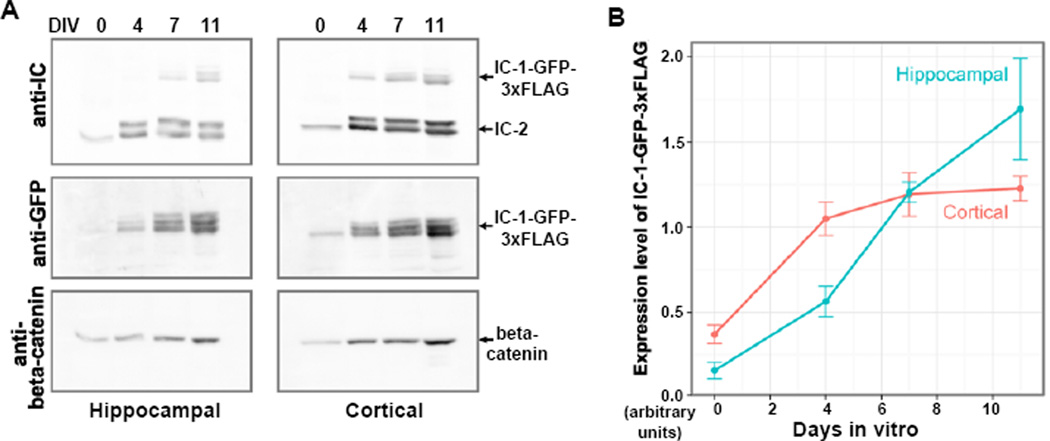
Primary hippocampal and cortical cultures derived from embryonic E15.5 stage homozygous knock-in mice were generated to examine the expression of the IC-1-GFP-3xFLAG fusion protein in primary neurons. These cultures contained mostly neurons with few glia. In both hippocampal and cortical cultures, the IC-1-GFP-3xFLAG fusion protein was detected on western blots using either the anti-IC antibody 74.1 that recognizes both IC-1 and IC-2, or an antibody directed against the GFP tag. Because the endogenous IC-1 was replaced by the GFP-3xFLAG-tagged IC-1 in homozygous mice, western analysis using the 74.1 anti-IC antibody is able to easily distinguish the 100 kDa IC-1-GFP-3xFLAG fusion protein from the ~75 kDa IC-2 protein (Figure 3A). This analysis demonstrated that both IC-1 and IC-2 are expressed in hippocampal and cortical neurons, confirming previous observations from 2D gel analysis of S35-labeled IC proteins in cortical neuron cultures [Pfister et al., 1996]. Expression levels of the IC-1-GFP-3xFLAG fusion protein increased with time in vitro, in relation to that of beta-catenin (Figure 3B). The intensity of the IC-2 signal is higher than that of IC-1-GFP-3xFLAG (Figure 3A). Since the antibody is raised against an epitope shared by the IC-1 and IC-2 proteins [Dillman and Pfister 1994; Vaughan and Vallee 1995], and is unlikely to preferentially bind to IC-2, it is most likely that IC-2 is expressed at a higher level than IC-1-GFP-3xFLAG in these neurons at this stage in development.
Figure 3.
An analysis of the expression levels of the dynein IC-1-GFP-3xFLAG fusion protein in cultured neurons. (A) Western blots of the cytosolic fraction of hippocampal and cortical E15.5 neuronal cultures, collected on the day of preparation (day 0) or after culture for 4, 7 or 11 days in vitro (DIV) with an equal volume of lysis buffer. Samples of equal volume were loaded onto the protein gel for western blot analyses. The blots were probed sequentially by anti-IC, anti-GFP and anti-beta-catenin antibodies. (B) Change in the expression level of IC-1-GFP-3xFLAG relative to that of beta-catenin correlated with culture time. The signal intensity of IC-1-GFP-3xFLAG revealed by the anti-GFP antibody was normalized to that of beta-catenin for each time point and each neuronal population. The means for each time point and neuronal population were plotted (n = 3 cultures, error bars show SEM).
GFP-labeled dynein can be observed in cultured hippocampal neurons derived from the knock-in mice
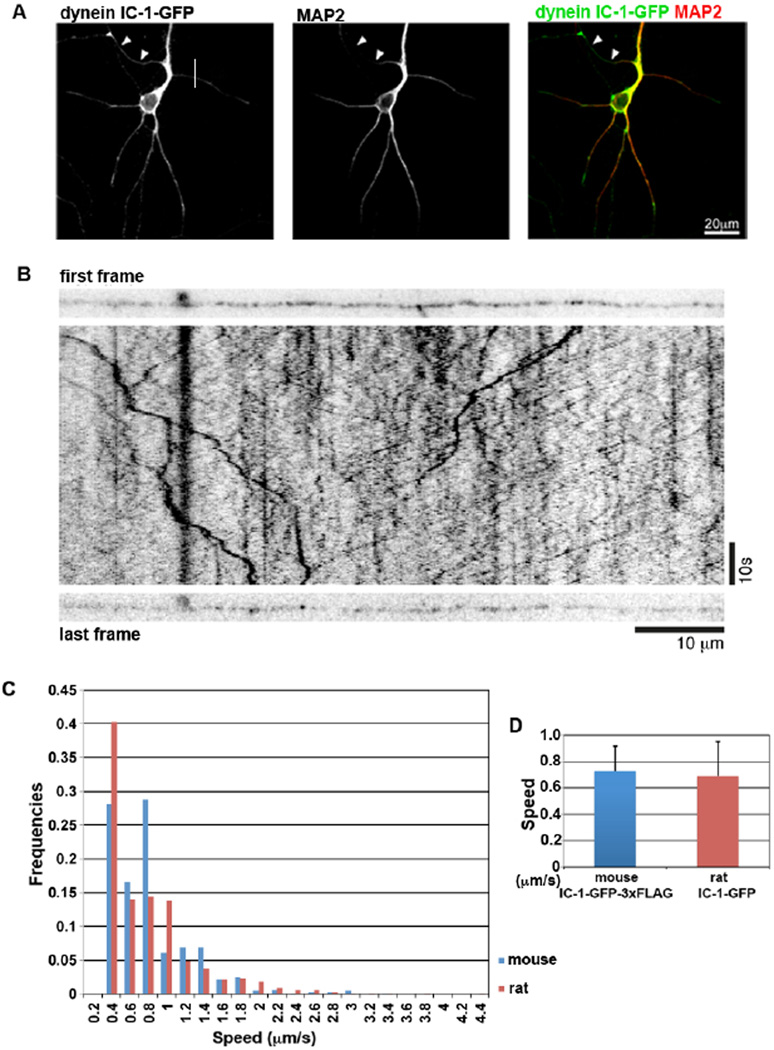
In hippocampal neuronal cultures, IC-1-GFP signals are detectable both by immunofluorescence (Figure 4A) and by live imaging (Figure 4B). IC-1-GFP could be seen throughout the somatodendritic compartment and axonal processes (Figure 4A). In processes that were morphologically identified as axonal, GFP puncta could be seen moving in both directions by live cell imaging (Figure 4B). This is the first time dynein-associated structures have been observed in mammalian neurons where GFP-labeled dynein is expressed at endogenous levels.
Figure 4.
An analysis of the dynein IC-1-GFP-3xFLAG in cultured hippocampal neurons. (A) Immunofluorescent confocal image of a fixed hippocampal neuron from an E15 culture after 10 DIV. Neurons were costained with antibodies to GFP and MAP2. Arrowheads indicate MAP2 negative axon. (B) The first and last frame, and the intervening kymograph generated from a movie of 1C-1GFP-3xFLAG puncta moving within an axonal process of a 14 DIV hippocampal neuron from an E15 culture. IC-1-GFP-3xFLAG-containing puncta move in both directions within the axon. (C) Comparison of velocity distribution of the IC-1-GFP-3xFLAG-containing puncta with that of transiently transfected IC-1-GFP in rat neuronal culture [Ha et al., 2008]. The fractional percent of dots that move at various speeds is shown as the frequency value. (D) Comparison of the average speed of the mouse IC-1-GFP-3xFLAG-containing puncta with that of transiently transfected rat IC-1-GFP that undergo retrograde movement in rat neuronal culture [Ha et al., 2008].
In a separate experiment, the movement of IC-1-GFP puncta was analyzed in the axons of hippocampal cultures from P1/2 mice. Cultures were 7–10 days in vitro at the time of analysis and as axonal processes were very long at this stage, directionality of movement could not be unambiguously identified. Nevertheless, the speed distribution and the average speed of movements of the IC-1-GFP-associated dots in mouse hippocampal neurons derived from the knock-in mice are similar to that of the retrogradely moving puncta labeled with exogenously expressed IC-1B-GFP proteins in rat hippocampal neurons (Figure 4C and D) [Ha et al., 2008]. This result further indicates that the IC-1-GFP-3xFLAG fusion protein is incorporated into a functional dynein complex capable of driving retrograde movements.
The knock-in mice can be used for identifying proteins that bind to dynein upon brain injury
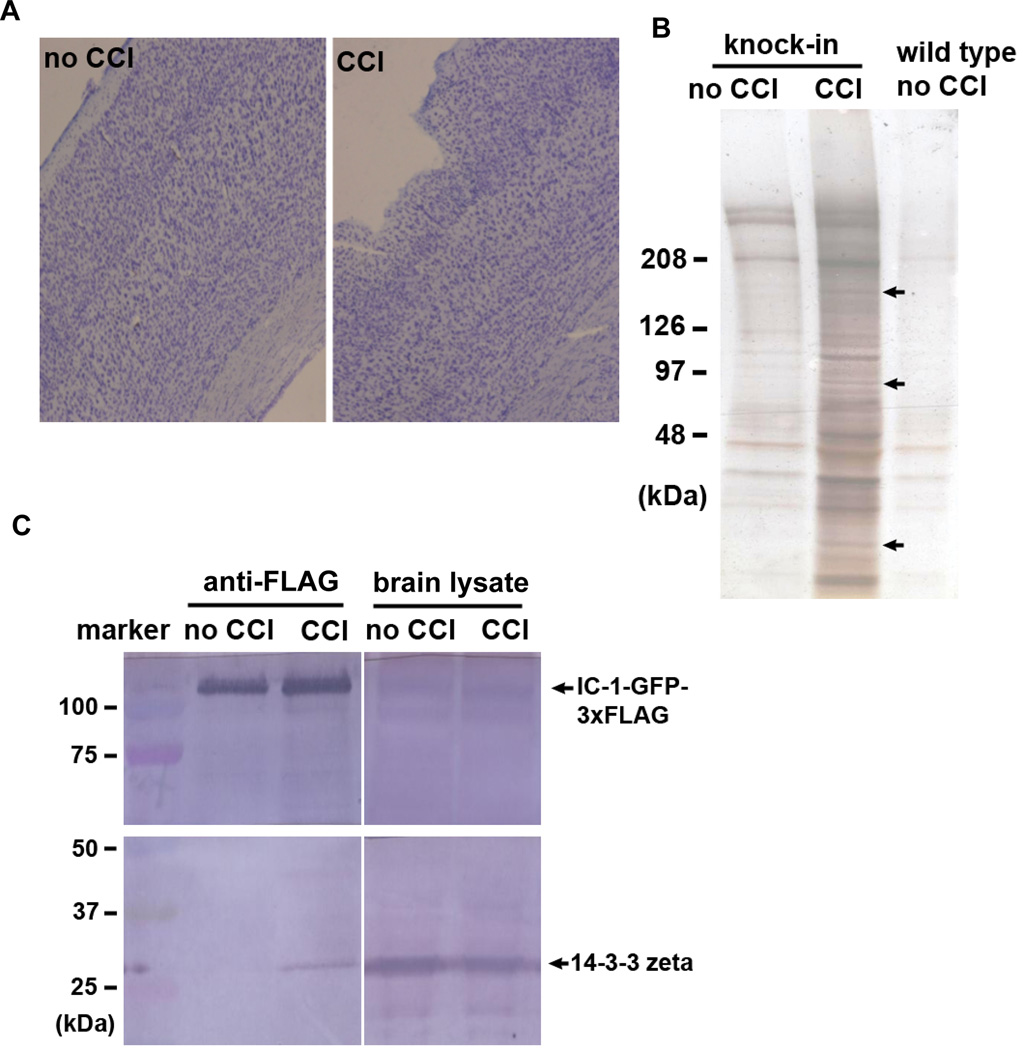
Using either the anti-FLAG antibody or the anti-GFP-antibody, it is possible to isolate neuronal proteins that are associated with the GFP- and FLAG-tagged dynein in either normal or pathogenic conditions. Because dynein is implicated in binding to injury factors upon injury of peripheral neurons [Cavalli et al., 2005; Perlson et al., 2005; Abe and Cavalli 2008; Perlson et al., 2010], we sought to identify proteins that become associated with dynein upon traumatic brain injury. Specifically, we used a “controlled cortical impact” (CCI) procedure to injure the mouse brain cortex, and then analyzed the proteins that were pulled down by the anti-FLAG antibody. Wild type mice without the 3xFLAG tag were used as a negative control during the pull-down experiment. To avoid analyzing known components of the dynein and dynactin complexes, knock-in mice that did not go through the CCI procedure were also used as negative control. The CCI procedure produced a small lesion on the mouse brain (Figure 5A), and mice that went through CCI were alive. Two days after the CCI procedure, the anti-FLAG antibody pulled down several new proteins that were not pulled down from the uninjured negative control (Figure 5B). To this end, we have only analyzed three bands excised from the silver-stained gel using mass-spectrometry (arrows in Figure 5B). The mass-spectrometry data shown in Table 1 indicated that the band at ~170 kDa contained mainly Clathrin heavy chain and a smaller proportion of dynein HC, which presumably was due to degradation of the dynein HC. The band at the ~90 kDa contained mainly the heat shock protein HSP90 and aconitate hydratase, a mitochondrial enzyme. Interestingly, the band at ~27 kDa contained mainly 14-3-3 zeta. Note that the 14-3-3 zeta data were the best in terms of the number of peptides detected (Table 1). To verify its association with dynein after injury, we performed an independent injury experiment in which one knock-in mouse went through the CCI procedure and one knock-in mouse served as negative control (Figure 5C). Instead of using mass-spectrometry analysis, we used a commercially available antibody against 14-3-3 zeta/delta in the experiment. In total brain protein extracts, 14-3-3 zeta is easily detectable by the antibody in brain extracts regardless of whether the mice had been injured with CCI or not (Figure 5C). However, its association with dynein was only detected after injury. The association, however, seemed to only involve a small portion of the 14-3-3 zeta protein. This could be because that the injury is relatively mild and restricted to a small spot on the cortex, while the pull-down experiment was done using the whole brain. As evidenced from the silver-stained gel (Figure 5A), we believe that more dynein-bound proteins could be identified after brain injury, and the availability of this mouse model will be beneficial for a community of researcher who are interested in studying cellular responses upon injury of the central nervous system.
Figure 5.
Isolation and identification of proteins that were pulled down with IC-1-GFP-3xFLAG after brain injury. (A) Images of brain sections stained by cresyl violet showing the injury site of the mouse brain cortex after the Controlled-Cortical-Impact (CCI) procedure. (B) A silver-stained gel showing bands of pulled-down proteins from the injured brain. Three of these bands (indicated by arrows) were analyzed by mass spectrometry. Data of the two top hits from each band are shown in Table 1. (C) A western analysis on brain lysate and proteins pulled down by anti-FLAG antibody. The western blot was probed with the anti-14-3-3 zeta/delta antibody.
Table 1.
Mass spectrometry data from three bands (data for the two top hits from each band are shown).
| Hits | No. of Peptide |
Sequence Header |
|---|---|---|
| 1 | 31 | >gi|51491845|ref|NP_001003908.1| clathrin heavy chain 1 (Mus musculus) |
| 2 | 15 | >gi|134288917|ref|NP_084514.2| cytoplasmic dynein 1 heavy chain 1 (Mus musculus) |
| 1 | 18 | >gi|6754254|ref|NP_034610.1| heat shock protein HSP 90-alpha (Mus musculus) |
| 2 | 16 | >gi|18079339|ref|NP_542364.1| aconitate hydratase, mitochondrial precursor (Mus musculus) |
| 1 | 56 | >gi|148676868|gb|EDL08815.1| tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide, isoform CRA_b (Mus musculus) |
| 2 | 14 | >gi|22094075|ref|NP_031477.1| ADP/ATP translocase 2 (Mus musculus) |
Discussion
In recent years, it has become clear that dynein is critical for the function and survival of neurons, and there is a great need for new tools for studying neuronal dynein. In this report, we describe a newly generated knock-in mouse line in which the neuron-specific dynein IC-1 is tagged with GFP and 3xFLAG at its C-terminus. The fusion gene is under the control of IC-1’s endogenous promoter, and is integrated at the endogenous locus of the IC-1-encoding gene Dync1i1. Importantly, the IC-1-GFP-3xFLAG fusion protein is fully incorporated into the endogenous dynein complex that drives retrograde movements in neurons. This mouse line allows GFP-labeled dynein expressed at the endogenous level to be observed in cultured neurons for the first time. It should potentially also allow GFP-labeled mammalian dynein to be purified for studies of motor behavior in vitro, which should complement and/or improve other existing methods to enhance research in this important area [Ross et al., 2006; Trokter et al., 2012].
The newly generated knock-in mouse line will be an ideal tool for studying dynein function under normal and pathological conditions. Defects in cytoplasmic dynein function are implicated in multiple brain diseases/disorders including the brain development disorder lissencephaly, and neuronal diseases such as ALS, spinal muscular atrophy, Charcot-Marie-Tooth disease and Perry syndrome [Tsai and Gleeson 2005; Wynshaw-Boris 2007; Perlson et al., 2010; Reiner et al., 2006; Farrer et al., 2009; Weedon et al., 2011; Harms et al., 2012]. Previously, it has been shown that dynein-cargo interaction changes in a mouse ALS model [Perlson et al., 2009]. Using GFP and FLAG tags in the neuron-specific dynein should allow isolation of proteins that bind to dynein in neuronal cells specifically. Since the knock-in mice can be easily bred with other mouse models of neuronal diseases, identifying dynein cargoes and regulators in normal mice and in disease models should be possible and should enhance our understanding of disease mechanisms. Moreover, alterations of the microtubule cytoskeleton in general have been implicated in brain development disorders and neurological diseases in humans and mice [Keays 2007; Keays et al., 2007; Poirier et al., 2007; Morris-Rosendahl et al., 2008; Jaglin et al., 2009; Tischfield et al., 2010]. Microtubule dynamics are also implicated in neuronal regeneration after axonal injury in neuronal cultures and in C. elegans [Gomis-Ruth et al., 2008; Sweet and Firestein 2008; Chen et al., 2011; Ghosh-Roy et al., 2012]. However, a knock-in mouse for visualizing fluorescently labeled microtubules in vivo is not available. Since the mouse line we created allows visualization of bi-directional movements of GFP-labeled dynein along microtubules, it will facilitate in vivo studies of neuronal microtubule integrity and function in normal and disease conditions.
Finally, this dynein IC-1 knock-in mouse line will also be ideal for identifying dynein-bound proteins after traumatic brain injury. Our study in this direction was inspired by earlier studies using peripheral neurons, in which dynein is implicated in neuronal regeneration via transporting injury signaling molecules toward the nucleus after axonal injury [Cavalli et al., 2005; Perlson et al., 2005; Abe and Cavalli 2008; Perlson et al., 2010]. At this point, we found that dynein’s physical interaction with 14-3-3 zeta is enhanced 2 days after the controlled-cortical-impact (CCI) procedure applied to the mouse cortex. 14-3-3 zeta has previously been found to be a biomarker for traumatic brain injury (TBI), transient forebrain ischemia and other types of acute brain damage [Siman et al., 2004; 2011]. It is known to be involved in neurodevelopment [Cheah et al., 2012] and is implicated in several neurodegenerative diseases [Steinacker et al., 2011]. It also promotes cell survival and inhibits apoptosis in other cell types [Neal et al., 2012; Yang et al., 2012; Matta et al., 2012]. In addition, 14-3-3 zeta associates with active protein kinase C (PKC) that is important for synaptic plasticity in the brain [Dai and Murakami 2003], and PKC participates in cellular wound healing and axonal injury responses [Kono et al., 2012; Pastuhov et al., 2012]. Future studies will be needed to address whether its association with dynein plays any positive role in the survival of neurons upon injury in the central nerve system.
Materials and Methods
Generating the knock-in mouse line in which the dynein IC-1-coding gene Dync1i1 is replaced by the IC-1-GFP-3xFLAG fusion gene
The project of generating the IC-1 knock-in mouse line was carried out using the transgenic mouse service of genOway in France (www.genoway.com). The targeting vector used to create the knock-in allele contains the two homologous arms, a fragment encoding eGFP-5xGlyAla-3xFLAG inserted in-frame before the stop codon in exon 17. Between the GFP tag and the 3xFLAG tag, 5xGlyAla (GA) repeats were inserted to create a flexible region between the two tags. The same GA repeats were inserted before the GFP-tag or the S-tag at the C-termini of proteins in Aspergillus nidulans [Yang et al., 2004]. The selection marker, the FRT-site-flanked neomycin gene, is inserted in the intron between exons 16 and 17 (Figure 1A). To facilitate the potential selection of homologous recombination events in ES cells, a negative selection marker, Diphtheria Toxin A (DTA) gene, was linked to the 5’ end of the short homologous arm. Two loxP sites were also inserted to flank exon 17, which would potentially aid in the deletion of the coding region of exon 17 and its 3’ untranslated region for making a partial IC-1 knockout mouse model. Standard molecular biology techniques were used to construct this targeting vector. Homolog arms were prepared from 129Sv/Pas genomic DNA by polymerase chain reactions (PCRs) using Accuprime Taq DNA Polymerase High Fidelity (Invitrogen). The amplified DNA fragments were subcloned into the TOPO vector, and at least three independent clones corresponding to each fragment were fully sequenced. Clones that contain no mutations in the coding region were chosen for further construction of the targeting vector. The sequence analysis also showed that within these regions, there is no polymorphism between the genetic backgrounds of C57BL/6 and 129Sv/Pas mice.
After validation of the targeting vector by restriction analysis and partial sequencing, the linearized vector was transfected into ES cells derived from the 129Sv mice, and five ES clones in which the targeting DNA had integrated into the Dync1i1 locus were obtained. For these five ES clones, besides verification of the correct integration by PCR and Southern blot analysis, sequencing of the eGFP-5xGA-3xFLAG cassette was done to confirm its integrity in the genome. All five of these ES clones were injected into C57BL/6J blastocysts, and the injected blastocysts were re-implanted into pseudo-pregnant females. Mice born with both the black (from C57BJ) and agouti (from 129Sv) coat colors were considered as chimeric mice, and four male chimeric mice were used for breeding with C57BL/6 Flp delete females to remove the neomycin selective marker gene in the germline. After breeding for two generations, the desired knock-in mice were obtained and verified by Southern and PCR analysis. For generating homozygous knock-in mice, heterozygous knock-in mice were crossed to each other. Tail DNA was extracted from the progeny and analyzed by PCR using the following pair of primers, KIU (5’- gcttgccgttccacacaatg -3’) and KID (5’-agatgccacggggatactg-3’) that generated a 0.2 kb band for the wild type, a 1 kb band for the homozygous knock-in mice and both of these bands for the heterozygous knock-in mice (Figure 1B).
Protein isolation and analysis
For protein analyses described in Figure 1 and Figure 5, the frozen brains were ground to fine powder in the presence of liquid nitrogen, and mixed with freshly prepared ice-cold lysis buffer (50 mM Tris, pH 8.0, 50 mM NaCl, 1% Triton X-100, 5mM EDTA, 2 mM PMSF and 10 µl/ml protease inhibitor cocktail (Sigma)). The mixtures were centrifuged for 30 minutes (14,000 rpm, 4°C), and supernatant was collected. For western analysis on total lysate, anti-GFP antibody was from Clontech, anti-IC-1 antibody was as described previously [Mitchell et al., 2012], and the anti-IC antibody 74.1 was as described previously [Dillman and Pfister 1994]. A FLAG-tag purification kit (Sigma) was used for pulling down proteins associated with the IC-1-GFP-3xFLAG protein. Elutes were analyzed using a 4%–20% SDS-PAGE gradient gel. For western analysis, the anti-IC antibody 74.1 [Dillman and Pfister 1994] and an anti-14-3-3 zeta/delta polycolonal antibody (Acris Antibodies) were used. The protein identification work was carried out at ProtTech, Inc. by using the NanoLC-ESI-MS/MS peptide sequencing technology (www.ProtTech.com).
To confirm that the IC-1-GFP-3XFLAG fusion protein was incorporated normally into the dynein complex in knock-in mice (described in Figure 2), we used a slightly modified protein-isolation procedure described as follows. Brains were homogenized in ice-cold BRB80 buffer (80 mM PIPES, 1 mM MgCl2, 1 mM EGTA, pH 6.8, supplemented with 1 mM DTT, 1 mM ATP and protease inhibitor cocktail from Roche), clarified by centrifugation, and analyzed by sucrose density centrifugation, performed as described previously [Ross et al., 2006], and by immunoprecipitation. For immunoprecipitation, anti-FLAG (M2, Sigma), anti-IC (MAB1618, clone 74.1, Millipore), and anti-Myc (Invitrogen), were coupled according to manufacturer’s instructions to Protein G dynabeads (Invitrogen). Antibody-bound beads were incubated with equal volumes of clarified lysate at room temperature for 60 minutes with intermittent agitation, washed in homogenization buffer, and eluted by boiling in gel sample buffer. The samples were analyzed by SDS-PAGE and probed with the anti-IC antibody (MAB1618, clone 74.1, Millipore), the anti-HC antibody (R-325, Santa Cruz) and the anti-p150 antibody (UP502) [Tokito et al., 1996].
Analyses of IC-1-GFP-3xFLAG expression and dynamics in neuronal culture
Cortical or hippocampal cells were isolated from E15.5 mice as previously described [Kaech and Banker, 2006; Banker and Goslin, 1998; Twelvetrees et al., 2010]. For preparing cytosolic fractions, approximately 1 million cells were plated per 6 cm dish. After growth for either 0, 4, 7 or 11 days in vitro (DIV), cultures were rinsed briefly in PBS and lysed in 100 µl of lysis buffer (50 mM HEPES, 1 mM EDTA, 1 mM MgCl2, 25 mM NaCl, 0.5% Triton-X-100 with protease inhibitor cocktail) with incubation on ice for 10 minutes. Debris was pelleted at 14000 rpm for 10 minutes. Supernatant was collected and run on a 7% SDS-PAGE gel followed by western blotting. Anti-IC (MAB1618, clone 74.1, Millipore), anti-beta-catenin (610154, BD Transduction Laboratories), and anti-GFP (GFP-1020, Aves Labs) antibodies were used for western analyses. Secondary HRP-conjugated antibodies were from Jackson. Chemiluminescent signal was detected with a Fujifilm LAS3000 imager. Band intensity was quantified using ImageJ. Total band intensity for each set of four samples (0, 4, 7 and 11 DIV) was set to be 100% such that each band was as a proportion of this total, then each GFP intensity was divided by its corresponding beta-catenin intensity.
For immunofluorescence analysis, hippocampal neurons were cultured on coverslips and fixed and stained as previously described [Twelvetrees et al., 2010]. Primary antibodies were chicken anti-GFP and rabbit anti-MAP2 (AB5622, Chemicon). Secondary antibodies were Cy2 conjugated goat anti-chicken (Jackson) and goat anti-rabbit Alexa 594 (Invitrogen). Neurons were imaged using a spinning-disk confocal microscope (UltraVIEW VoX; PerkinElmer) with an Apochromat 100×, 1.49 NA oil immersion objective (Nikon). Digital images were acquired with an EM charge-coupled device camera (C9100; Hamamatsu Photonics) controlled by Velocity software (PerkinElmer). For live cell imaging and analyses of IC-1-GFP-3xFLAG puncta in axons, hippocampal neurons were cultured from P1/2 homozygous knock-in mice and the movement of GFP-dynein was imaged on DIV 7 – 10 as described previously [Mitchell et al., 2012] with a 500 msec frame rate. The movement velocity was analyzed as described previously [Ha et al., 2008; Mitchell et al., 2012].
Controlled cortical impact (CCI) injury
Controlled cortical impact (CCI) injury of mice was done as similarly described recently using the core facility that belongs to the Center for Neuroscience and Regenerative Medicine (CNRM) in USUHS [Budinich et al., 2012; Villapol et al., 2012; Yi et al., 2012; Yu et al., 2012]. The CCI device (Impact One, Leica, Wetzlar, Germany) consists of a computer-controlled, electromagnetically driven impactor with a steel tip mounted on a stereotaxic micromanipulator [Budinich et al., 2012]. For our experiments, we only chose male mice for CCI. After a skull fragment was removed by making an incision, a probe with a tip size of 2 mm in diameter and a movement speed of 5 m/s was used to hit the brain cortex. The dwell time of the tip at the cortex was 100 ms and the hitting depth 2 mm. As described previously [Budinich et al., 2012], the skull fragment was carefully put back after the CCI procedure. Mice were placed in a heated cage to maintain body temperature until fully awake from anesthesia, before they were returned to their home cage. Two days after the procedure, the mice were sacrificed. At approximately the same time, mouse brains were dissected out and frozen quickly on dry ice, and subsequently kept in a -80 °C freezer.
A histochemical analysis of the injured cortex was done using cresyl violet (Nissl) staining similar to what has been described previously [Penney et al., 2002]. Briefly, brain sections mounted on gelatin-coated slides were rehydrated in a graded ethanol series and then stained for 2 min with cresyl violet (1.25 g cresyl violet acetate and 0.75 ml glacial acetic acid were dissolved in deionized water in a total volume of 250 ml). After staining they were washed with deionized water, and dehydrated in a graded ethanol series and preserved on slide with mounting media.
Supplementary Material
Acknowledgements
We thank Sharon Juliano and Zygmunt Galdzicki for advice and support, and Amanda Fu, Hongna Pan and W. Russell Smiley for technical help. The work was funded by the Center for Neuroscience and Regenerative Medicine (CNRM)/Department of Defense at Uniformed Services University of the Health Sciences (to X. X.) and by National Institutes of Health grants RO1 GM086472 (to K.K.P.), RO1 NS060698 (to E.L.F.H.) and RO1 GM097580 (to X.X.). Disclaimer: The views expressed are those of the authors and do not reflect the official policy or position of the Uniformed Services University of the Health Sciences, the Department of the Defense, or the United States government.
References
- Abe N, Cavalli V. Nerve injury signaling. Curr Opin Neurobiol. 2008;18:276–283. doi: 10.1016/j.conb.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A, Hammer JA., 3rd Linking molecular motors to membrane cargo. Curr opin Cell biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker G, Goslin K. Culturing Nerve Cells. The MIT Press; 1998. [Google Scholar]
- Banks GT, Haas MA, Line S, Shepherd HL, Alqatari M, Stewart S, Rishal I, Philpott A, Kalmar B, Kuta A, Groves M, Parkinson N, Acevedo-Arozena A, Brandner S, Bannerman D, Greensmith L, Hafezparast M, Koltzenburg M, Deacon R, Fainzilber M, Fisher EM. Behavioral and other phenotypes in a cytoplasmic dynein light intermediate chain 1 mutant mouse. J Neurosci. 2011;31:5483–5494. doi: 10.1523/JNEUROSCI.5244-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass PW, Buster DW. Slow axonal transport and genesis and neuronal morphology. J Neurobiol. 2004;58:3–17. doi: 10.1002/neu.10281. [DOI] [PubMed] [Google Scholar]
- Budinich CS, Chen H, Lowe D, Rosenberger JG, Bernstock JD, McCabe JT. Mouse brain psa-ncam levels are altered by graded-controlled cortical impact injury. Neural Plast. 2012;2012:378307. doi: 10.1155/2012/378307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli V, Kujala P, Klumperman J, Goldstein LS. Sunday driver links axonal transport to damage signaling. J Cell Biol. 2005;168:775–787. doi: 10.1083/jcb.200410136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah PS, Ramshaw HS, Thomas PQ, Toyo-Oka K, Xu X, Martin S, Coyle P, Guthridge MA, Stomski F, van den Buuse M, Wynshaw-Boris A, Lopez AF, Schwarz QP. Neurodevelopmental and neuropsychiatric behaviour defects arise from 14-3-3zeta deficiency. Mol Psychiatry. 2012;17:451–466. doi: 10.1038/mp.2011.158. [DOI] [PubMed] [Google Scholar]
- Chen L, Wang Z, Ghosh-Roy A, Hubert T, Yan D, O'Rourke S, Bowerman B, Wu Z, Jin Y, Chisholm AD. Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron. 2011;71:1043–1057. doi: 10.1016/j.neuron.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Levedakou EN, Millen KJ, Wollmann RL, Soliven B, Popko B. Proprioceptive sensory neuropathy in mice with a mutation in the cytoplasmic dynein heavy chain 1 gene. J Neurosci. 2007;27:14515–14524. doi: 10.1523/JNEUROSCI.4338-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier-Larsen ES, Wallace KE, Pennise CR, Holzbaur EL. Lysosomal proliferation and distal degeneration in motor neurons expressing the G59S mutation in the p150Glued subunit of dynactin. Human Mol Genet. 2008;17:1946–1955. doi: 10.1093/hmg/ddn092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosker KE, Courchesne SL, Segal RA. Action in the axon: Generation and transport of signaling endosomes. Curr Opin Neurobiol. 2008;18:270–275. doi: 10.1016/j.conb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai JG, Murakami K. Constitutively and autonomously active protein kinase c associated with 14-3-3 zeta in the rodent brain. J Neurochem. 2003;84:23–34. doi: 10.1046/j.1471-4159.2003.01254.x. [DOI] [PubMed] [Google Scholar]
- Dillman JF, 3rd, Pfister KK. Differential phosphorylation in vivo of cytoplasmic dynein associated with anterogradely moving organelles. J Cell Biol. 1994;127:1671–1681. doi: 10.1083/jcb.127.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encalada SE, Szpankowski L, Xia CH, Goldstein LS. Stable kinesin and dynein assemblies drive the axonal transport of mammalian prion protein vesicles. Cell. 2011;144:551–565. doi: 10.1016/j.cell.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer MJ, Hulihan MM, Kachergus JM, Dachsel JC, Stoessl AJ, Grantier LL, Calne S, Calne DB, Lechevalier B, Chapon F, Tsuboi Y, Yamada T, Gutmann L, Elibol B, Bhatia KP, Wider C, Vilariño-Güell C, Ross OA, Brown LA, Castanedes-Casey M, Dickson DW, Wszolek ZK. DCTN1 mutations in Perry syndrome. Nat Genet. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Roy A, Goncharov A, Jin Y, Chisholm AD. Kinesin-13 and tubulin posttranslational modifications regulate microtubule growth in axon regeneration. Dev Cell. 2012;23:716–728. doi: 10.1016/j.devcel.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth S, Wierenga CJ, Bradke F. Plasticity of polarization: Changing dendrites into axons in neurons integrated in neuronal circuits. Curr Biol. 2008;18:992–1000. doi: 10.1016/j.cub.2008.06.026. [DOI] [PubMed] [Google Scholar]
- Ha J, Lo KW, Myers KR, Carr TM, Humsi MK, Rasoul BA, Segal RA, Pfister KK. A neuron-specific cytoplasmic dynein isoform preferentially transports trkb signaling endosomes. J Cell Biol. 2008;181:1027–1039. doi: 10.1083/jcb.200803150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habura A, Tikhonenko I, Chisholm RL, Koonce MP. Interaction mapping of a dynein heavy chain. Identification of dimerization and intermediate-chain binding domains. J Biol Chem. 1999;274:15447–15453. doi: 10.1074/jbc.274.22.15447. [DOI] [PubMed] [Google Scholar]
- Harada A, Takei Y, Kanai Y, Tanaka Y, Nonaka S, Hirokawa N. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol. 1998;141:51–59. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, Ori-McKenney KM, Scoto M, Tuck EP, Bell S, Ma D, Masi S, Allred P, Al-Lozi M, Reilly MM, Miller LJ, Jani-Acsadi A, Pestronk A, Shy ME, Muntoni F, Vallee RB, Baloh RH. Mutations in the tail domain of dync1h1 cause dominant spinal muscular atrophy. Neurology. 2012;78:1714–1720. doi: 10.1212/WNL.0b013e3182556c05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerssen HM, Pazyra MF, Segal RA. Dynein motors transport activated trks to promote survival of target-dependent neurons. Nat Neurosci. 2004;7:596–604. doi: 10.1038/nn1242. [DOI] [PubMed] [Google Scholar]
- Höök P, Vallee R. Dynein dynamics. Nat Struct Mol Biol. 2012;19:467–469. doi: 10.1038/nsmb.2290. [DOI] [PubMed] [Google Scholar]
- Ilieva HS, Yamanaka K, Malkmus S, Kakinohana O, Yaksh T, Marsala M, Cleveland DW. Mutant dynein (loa) triggers proprioceptive axon loss that extends survival only in the sod1 als model with highest motor neuron death. Proc Natl Acad Sci USA. 2008;105:12599–12604. doi: 10.1073/pnas.0805422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglin XH, Poirier K, Saillour Y, Buhler E, Tian G, Bahi-Buisson N, Fallet-Bianco C, Phan-Dinh-Tuy F, Kong XP, Bomont P, Castelnau-Ptakhine L, Odent S, Loget P, Kossorotoff M, Snoeck I, Plessis G, Parent P, Beldjord C, Cardoso C, Represa A, Flint J, Keays DA, Cowan NJ, Chelly J. Mutations in the beta-tubulin gene tubb2b result in asymmetrical polymicrogyria. Nat Genet. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J Biol Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- Karki S, Holzbaur EL. Cytoplasmic dynein and dynactin in cell division and intracellular transport. Curr Opin Cell Biol. 1999;11:45–53. doi: 10.1016/s0955-0674(99)80006-4. [DOI] [PubMed] [Google Scholar]
- Keays DA. Neuronal migration: Unraveling the molecular pathway with humans, mice, and a fungus. Mamm Genome. 2007;18:425–430. doi: 10.1007/s00335-007-9034-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, Cleak J, Oliver PL, Fray M, Harvey RJ, Molnar Z, Pinon MC, Dear N, Valdar W, Brown SD, Davies KE, Rawlins JN, Cowan NJ, Nolan P, Chelly J, Flint J. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell. 2007;128:45–57. doi: 10.1016/j.cell.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Ling SC, Rogers GC, Kural C, Selvin PR, Rogers SL, Gelfand VI. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J Cell Biol. 2007;176:641–651. doi: 10.1083/jcb.200608128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Brown CL, Maier KC, Quintyne NJ, Schroer TA. Analysis of the dynein-dynactin interaction in vitro and in vivo. Mol Biol Cell. 2003;14:5089–5097. doi: 10.1091/mbc.E03-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini AR, Collins CA. Modulation of cytoplasmic dynein ATPase activity by the accessory subunits. Cell Motil Cytoskeleton. 2001;48:52–60. doi: 10.1002/1097-0169(200101)48:1<52::AID-CM5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Kono K, Saeki Y, Yoshida S, Tanaka K, Pellman D. Proteasomal degradation resolves competition between cell polarization and cellular wound healing. Cell. 2012;150:151–164. doi: 10.1016/j.cell.2012.05.030. [DOI] [PubMed] [Google Scholar]
- Kuta A, Deng W, Morsi El-Kadi A, Banks GT, Hafezparast M, Pfister KK, Fisher EM. Mouse cytoplasmic dynein intermediate chains: Identification of new isoforms, alternative splicing and tissue distribution of transcripts. PloS one. 2010;5:e11682. doi: 10.1371/journal.pone.0011682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C, Lin X, Chandran J, Shim H, Yang WJ, Cai H. The G59S mutation in p150(glued) causes dysfunction of dynactin in mice. J Neurosci. 2007;27:13982–13990. doi: 10.1523/JNEUROSCI.4226-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts D, Robberecht W, Carmeliet P. Heterogeneity in motoneuron disease. Trends Neurosci. 2007;30:536–544. doi: 10.1016/j.tins.2007.07.002. [DOI] [PubMed] [Google Scholar]
- LaMonte BH, Wallace KE, Holloway BA, Shelly SS, Ascano J, Tokito M, Van Winkle T, Howland DS, Holzbaur EL. Disruption of dynein/dynactin inhibits axonal transport in motor neurons causing late-onset progressive degeneration. Neuron. 2002;34:715–727. doi: 10.1016/s0896-6273(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Lloyd TE, Machamer J, O'Hara K, Kim JH, Collins SE, Wong MY, Sahin B, Imlach W, Yang Y, Levitan ES, McCabe BD, Kolodkin AL. The p150(glued) cap-gly domain regulates initiation of retrograde transport at synaptic termini. Neuron. 2012;74:344–360. doi: 10.1016/j.neuron.2012.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KW, Kan HM, Pfister KK. Identification of a novel region of the cytoplasmic dynein intermediate chain important for dimerization in the absence of the light chains. J Biol Chem. 2006;281:9552–9559. doi: 10.1074/jbc.M511721200. [DOI] [PubMed] [Google Scholar]
- Maday S, Wallace KE, Holzbaur EL. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012;196:407–417. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus SM, Plevock KM, St Germain BJ, Punch JJ, Meaden CW, Lee WL. Quantitative analysis of Pac1/LIS1-mediated dynein targeting: Implications for regulation of dynein activity in budding yeast. Cytoskeleton (Hoboken) 2011;68:157–174. doi: 10.1002/cm.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matta A, Siu KM, Ralhan R. 14-3-3 zeta as novel molecular target for cancer therapy. Expert opinion on therapeutic targets. 2012 doi: 10.1517/14728222.2012.668185. In press. [DOI] [PubMed] [Google Scholar]
- McKenney RJ, Weil SJ, Scherer J, Vallee RB. Mutually exclusive cytoplasmic dynein regulation by nude-lis1 and dynactin. J Biol Chem. 2011;286:39615–39622. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DJ, Blasier KR, Jeffery ED, Ross MW, Pullikuth AK, Suo D, Park J, Smiley WR, Lo KW, Shabanowitz J, Deppmann CD, Trinidad JC, Hunt DF, Catling AD, Pfister KK. Trk activation of the erk1/2 kinase pathway stimulates intermediate chain phosphorylation and recruits cytoplasmic dynein to signaling endosomes for retrograde axonal transport. J Neurosci. 2012;32:15495–15510. doi: 10.1523/JNEUROSCI.5599-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Cooper JA. Coordinating mitosis with cell polarity: Molecular motors at the cell cortex. Semin Cell Dev Biol. 2010;21:283–289. doi: 10.1016/j.semcdb.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Sept D, Cooper JA. Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration. Proc Natl Acad Sci U S A. 2009;106:5147–5152. doi: 10.1073/pnas.0810828106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris-Rosendahl DJ, Najm J, Lachmeijer AM, Sztriha L, Martins M, Kuechler A, Haug V, Zeschnigk C, Martin P, Santos M, Vasconcelos C, Omran H, Kraus U, Van der Knaap MS, Schuierer G, Kutsche K, Uyanik G. Refining the phenotype of alpha-1a tubulin (tuba1a) mutation in patients with classical lissencephaly. Clin Genet. 2008;74:425–433. doi: 10.1111/j.1399-0004.2008.01093.x. [DOI] [PubMed] [Google Scholar]
- Moughamian AJ, Holzbaur EL. Dynactin is required for transport initiation from the distal axon. Neuron. 2012;74:331–343. doi: 10.1016/j.neuron.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, Prudlo J, Peraus G, Hanemann CO, Stumm G, Ludolph AC. Point mutations of the p150 subunit of dynactin (dctn1) gene in als. Neurology. 2004;63:724–726. doi: 10.1212/01.wnl.0000134608.83927.b1. [DOI] [PubMed] [Google Scholar]
- Neal CL, Xu J, Li P, Mori S, Yang J, Neal NN, Zhou X, Wyszomierski SL, Yu D. Overexpression of 14-3-3zeta in cancer cells activates pi3k via binding the p85 regulatory subunit. Oncogene. 2012;31:897–906. doi: 10.1038/onc.2011.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey JP, Smith DS. A Cdk5-dependent switch regulates Lis1/Ndel1/dynein-driven organelle transport in adult axons. J Neurosci. 2011;31:17207–17219. doi: 10.1523/JNEUROSCI.4108-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuhov SI, Fujiki K, Nix P, Kanao S, Bastiani M, Matsumoto K, Hisamoto N. Endocannabinoid-goalpha signalling inhibits axon regeneration in caenorhabditis elegans by antagonizing gqalpha-pkc-jnk signalling. Nat Commun. 2012;3:1136. doi: 10.1038/ncomms2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penney DP, Powers JM, Frank M, Willis C, Churukian C. Analysis and testing of biological stains--the biological stain commission procedures. Biotech Histochem. 2002;77:237–275. doi: 10.1080/bih.77.5-6.237.275. [DOI] [PubMed] [Google Scholar]
- Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated map kinase in injured nerve. Neuron. 2005;45:715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- Perlson E, Jeong GB, Ross JL, Dixit R, Wallace KE, Kalb RG, Holzbaur EL. A switch in retrograde signaling from survival to stress in rapid-onset neurodegeneration. J Neurosci. 2009;29:9903–9917. doi: 10.1523/JNEUROSCI.0813-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlson E, Maday S, Fu MM, Moughamian AJ, Holzbaur EL. Retrograde axonal transport: Pathways to cell death? Trends Neurosci. 2010;33:335–344. doi: 10.1016/j.tins.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Fisher EM, Gibbons IR, Hays TS, Holzbaur EL, McIntosh JR, Porter ME, Schroer TA, Vaughan KT, Witman GB, King SM, Vallee RB. Cytoplasmic dynein nomenclature. J Cell Biol. 2005;171:411–413. doi: 10.1083/jcb.200508078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister KK, Salata MW, Dillman JF, 3rd, Vaughan KT, Vallee RB, Torre E, Lye RJ. Differential expression and phosphorylation of the 74-kda intermediate chains of cytoplasmic dynein in cultured neurons and glia. J Biol Chem. 1996;271:1687–1694. doi: 10.1074/jbc.271.3.1687. [DOI] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Keays DA, Francis F, Saillour Y, Bahi N, Manouvrier S, Fallet-Bianco C, Pasquier L, Toutain A, Tuy FP, Bienvenu T, Joriot S, Odent S, Ville D, Desguerre I, Goldenberg A, Moutard ML, Fryns JP, van Esch H, Harvey RJ, Siebold C, Flint J, Beldjord C, Chelly J. Large spectrum of lissencephaly and pachygyria phenotypes resulting from de novo missense mutations in tubulin alpha 1a (tuba1a) Hum Mutat. 2007;28:1055–1064. doi: 10.1002/humu.20572. [DOI] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH, Jr, Ludlow CL, Fischbeck KH. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- Puls I, Oh SJ, Sumner CJ, Wallace KE, Floeter MK, Mann EA, Kennedy WR, Wendelschafer-Crabb G, Vortmeyer A, Powers R, Finnegan K, Holzbaur EL, Fischbeck KH, Ludlow CL. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann Neurol. 2005;57:687–694. doi: 10.1002/ana.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reck-Peterson SL, Vale RD, Gennerich A. Motile properties of cytoplasmic dynein. In: Hirose K, Amos L, editors. Handbook of dynein. Pan Stanford Publishing; 2011. [Google Scholar]
- Reiner O, Sapoznik S, Sapir T. Lissencephaly 1 linking to multiple diseases: Mental retardation, neurodegeneration, schizophrenia, male sterility, and more. Neuromolecular Med. 2006;8:547–565. doi: 10.1385/NMM:8:4:547. [DOI] [PubMed] [Google Scholar]
- Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- Schroer TA. Dynactin. Annu Rev Cell Dev Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- Sheng ZH, Cai Q. Mitochondrial transport in neurons: Impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci. 2012;13:77–93. doi: 10.1038/nrn3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siman R, Giovannone N, Toraskar N, Frangos S, Stein SC, Levine JM, Kumar MA. Evidence that a panel of neurodegeneration biomarkers predicts vasospasm, infarction, and outcome in aneurysmal subarachnoid hemorrhage. PloS one. 2011;6:e28938. doi: 10.1371/journal.pone.0028938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siman R, McIntosh TK, Soltesz KM, Chen Z, Neumar RW, Roberts VL. Proteins released from degenerating neurons are surrogate markers for acute brain damage. Neurobiol Dis. 2004;16:311–320. doi: 10.1016/j.nbd.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Steinacker P, Aitken A, Otto M. 14-3-3 proteins in neurodegeneration. Semin Cell Dev Biol. 2011;22:696–704. doi: 10.1016/j.semcdb.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Sweet ES, Firestein BL. Neuronal polarization: Old cells can learn new tricks. Curr Biol. 2008;18:R661–R663. doi: 10.1016/j.cub.2008.06.057. [DOI] [PubMed] [Google Scholar]
- Tischfield MA, Baris HN, Wu C, Rudolph G, Van Maldergem L, He W, Chan WM, Andrews C, Demer JL, Robertson RL, Mackey DA, Ruddle JB, Bird TD, Gottlob I, Pieh C, Traboulsi EI, Pomeroy SL, Hunter DG, Soul JS, Newlin A, Sabol LJ, Doherty EJ, de Uzcategui CE, de Uzcategui N, Collins ML, Sener EC, Wabbels B, Hellebrand H, Meitinger T, de Berardinis T, Magli A, Schiavi C, Pastore-Trossello M, Koc F, Wong AM, Levin AV, Geraghty MT, Descartes M, Flaherty M, Jamieson RV, Moller HU, Meuthen I, Callen DF, Kerwin J, Lindsay S, Meindl A, Gupta ML, Jr, Pellman D, Engle EC. Human tubb3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokito MK, Howland DS, Lee VM, Holzbaur EL. Functionally distinct isoforms of dynactin are expressed in human neurons. Mol Biol Cell. 1996;7:1167–1180. doi: 10.1091/mbc.7.8.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trokter M, Mucke N, Surrey T. Reconstitution of the human cytoplasmic dynein complex. Proc Natl Acad Sci U S A. 2012;109:20895–20900. doi: 10.1073/pnas.1210573110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LH, Gleeson JG. Nucleokinesis in neuronal migration. Neuron. 2005;46:383–388. doi: 10.1016/j.neuron.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Twelvetrees AE, Yuen EY, Arancibia-Carcamo IL, MacAskill AF, Rostaing P, Lumb MJ, Humbert S, Triller A, Saudou F, Yan Z, Kittler JT. Delivery of gabaars to synapses is mediated by hap1-kif5 and disrupted by mutant huntingtin. Neuron. 2010;65:53–65. doi: 10.1016/j.neuron.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan SH, Gee MA, Vallee RB. Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J Biol Chem. 2000;275:32769–32774. doi: 10.1074/jbc.M001537200. [DOI] [PubMed] [Google Scholar]
- Vallee RB, McKenney RJ, Ori-McKenney KM. Multiple modes of cytoplasmic dynein regulation. Nat Cell Biol. 2012;14:224–230. doi: 10.1038/ncb2420. [DOI] [PubMed] [Google Scholar]
- Vaughan KT. Roles of Cytoplasmic Dynein during Mitosis. In: King SM, editor. Dyneins: Structure, Biology and Disease. Elsevier; 2011. [Google Scholar]
- Vaughan KT, Vallee RB. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150glued. J Cell Biol. 1995;131:1507–1516. doi: 10.1083/jcb.131.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villapol S, Yaszemski AK, Logan TT, Sanchez-Lemus E, Saavedra JM, Symes AJ. Candesartan, an angiotensin ii at(1)-receptor blocker and ppar-gamma agonist, reduces lesion volume and improves motor and memory function after traumatic brain injury in mice. Neuropsychopharmacology. 2012;37:2817–2829. doi: 10.1038/npp.2012.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Hastings R, Caswell R, Xie W, Paszkiewicz K, Antoniadi T, Williams M, King C, Greenhalgh L, Newbury-Ecob R, Ellard S. Exome sequencing identifies a dync1h1 mutation in a large pedigree with dominant axonal charcot-marie-tooth disease. Am J Hum Genet. 2011;89:308–312. doi: 10.1016/j.ajhg.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynshaw-Boris A. Lissencephaly and lis1: Insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- Yang L, Ukil L, Osmani A, Nahm F, Davies J, De Souza CP, Dou X, Perez-Balaguer A, Osmani SA. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in aspergillus nidulans. Eukaryot Cell. 2004;3:1359–1362. doi: 10.1128/EC.3.5.1359-1362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Cao W, Zhang L, Zhang W, Zhang X, Lin H. Targeting 14-3**3zeta in cancer therapy. Cancer Gene Ther. 2012;19:153–159. doi: 10.1038/cgt.2011.85. [DOI] [PubMed] [Google Scholar]
- Yeh TY, Quintyne NJ, Scipioni BR, Eckley DM, Schroer TA. Dynactin's pointed-end complex is a cargo-targeting module. Mol Biol Cell. 2012;23:3827–3837. doi: 10.1091/mbc.E12-07-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JH, Katagiri Y, Susarla B, Figge D, Symes AJ, Geller HM. Alterations in sulfated chondroitin glycosaminoglycans following controlled cortical impact injury in mice. J Comp Neurol. 2012;520:3295–3313. doi: 10.1002/cne.23156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JY, Ori-McKenney KM, McKenney RJ, Vershinin M, Gross SP, Vallee RB. High-resolution imaging reveals indirect coordination of opposite motors and a role for lis1 in high-load axonal transport. J Cell Biol. 2011;195:193–201. doi: 10.1083/jcb.201104076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Wang Z, Tchantchou F, Chiu CT, Zhang Y, Chuang DM. Lithium ameliorates neurodegeneration, suppresses neuroinflammation, and improves behavioral performance in a mouse model of traumatic brain injury. J Neurotrauma. 2012;29:362–374. doi: 10.1089/neu.2011.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yao X, Fischer L, Abenza JF, Penalva MA, Xiang X. The p25 subunit of the dynactin complex is required for dynein-early endosome interaction. J Cell Biol. 2011;193:1245–1255. doi: 10.1083/jcb.201011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang F, Cao J, Shen Y, Huang Q, Bao L, Zhu X. Nudel promotes axonal lysosome clearance and endo-lysosome formation via dynein-mediated transport. Traffic. 2009;10:1337–1349. doi: 10.1111/j.1600-0854.2009.00945.x. [DOI] [PubMed] [Google Scholar]
- Zhou B, Cai Q, Xie Y, Sheng ZH. Snapin recruits dynein to bdnf-trkb signaling endosomes for retrograde axonal transport and is essential for dendrite growth of cortical neurons. Cell Rep. 2012;2:42–51. doi: 10.1016/j.celrep.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.