Abstract
Aromatic-aromatic hydrogen bonds are important in many areas of chemistry, biology and materials science. In this study we have analyzed the roles played by the π-π interactions in interleukins (ILs) and tumor necrosis factor (TNF) proteins. Majority of π-π interacting residues are conserved in ILs and TNF proteins. The accessible surface area calculations in these proteins reveal that these interactions might be important in stabilizing the inner core regions of these proteins. In addition to π-π interactions, the aromatic residues also form π-networks in ILs and TNF proteins. The results obtained in the present study indicate that π-π interactions and π-π networks play important roles in the structural stability of ILs and TNF proteins.
Keywords: π-π interactions, TNF proteins, ILs, structure, stability, π-network
Background
Noncovalent interactions are ubiquitous in chemistry and are a main source of stability for many molecular complexes in nanoscience, materials chemistry and biochemistry [1–3]. Weak interactions although small in magnitude, being numerous, contribute to the overall stability of protein molecules [4, 5]. Protein structures are stabilized with various non-covalent interactions such as hydrophobic, electrostatic, hydrogen bonds and Vander Waals interactions. In addition, the π-π interaction is an important non-covalent binding interaction in structural biology. Interactions between aromatic residues (Phe, Tyr, Trp and His) contribute to the stability of native fold [6]. Interactions among side chains of protein residues and protein subunits are primarily responsible for the structure, stability and function of proteins. The π-π interactions between aromatic rings are recognized to play an important role in structural stability in protein and DNA [6–8] and form recognition motifs in proteins and enzymes [9, 10]. Cytokines are proteins that regulate and mediate immunity, inflammation, and hematopoiesis. ILs and TNF are immunoregulatory, proinflammatory cytokines that strongly synergize in numerous biological functions, both in vitro and in vivo. IL-1α is produced mainly by mononuclear phagocytes but also by many other types of cells in response to inflammatory stimuli [11]. It has a wide range of biological activities on many target cell types, including B cells, T cells and monocytes [12, 13]. TNF-α is known to regulate growth and differentiation of a variety of immune cells. It is secreted by activated macrophages, monocytes, and many other cells, including B cells, T cells and fibroblasts [13–15]. TNF-α, in particular, has received much attention and cardiac-specific over expression of this cytokine promotes a phenotype mimicking several features of clinical heart failure including cardiac hypertrophy, ventricular dilatation, fibrosis and several biochemical and cellular defects [16]. Recent data indicate that a complex TNF-α mediated regulation of matrix metalloproteinases (MMP) and tissue inhibitors of metalloproteinases (TIMP) may significantly contribute to myocardial remodeling [17, 18]. In autoimmune diseases, the pro-inflammatory cytokines are typically considered to be pathogenic, whereas the anti-inflammatory cytokines are regarded to be protective in nature [19–21]. Our group has already analyzed the cation-π interactions in ILs and TNF proteins [22]. There is no systematic analysis of π-π interactions in ILs and TNF proteins. Hence, we investigated the role of π-π interactions in ILs and TNF proteins by bioinformatics approaches.
Methodology
Data Set:
We have selected five ILs and two TNF proteins from PDB (Protein data bank) [23] for our investigation, the details of which are given in Table 1 & Table 2 (see supplementary material). Criteria that were followed for selecting the data set are (i) the three dimensional structures of these proteins have been solved with ≤3.0 Å resolution and (ii) the sequence identity among the proteins in the dataset was less than 40%.
π-π interactions:
The stand alone program NCI (Non-canonical interactions) server [24] was used to identify π-π interactions in a protein. NCI used the geometric criteria of separation 4-6.5 Å between the centroids of aromatic ring of interacting residues to predict π-π interactions. All significant π-π interactions, where the aromatic residues (Phe, Tyr & Trp) donate π electron density with other acceptor aromatic residues were identified. We did not consider C-H..π interactions for our analysis in the present study. Because in C-H..π interactions, the donor residues were not always π-residues. The π-π interaction types were represented by a two letter code S and S5. S represented the side-chain (6 membered aromatic ring) of amino acids Phe, Tyr and Trp. S5 represented the side-chain (5 membered ring) in Trp. Eventhough His has a 5 membered aromatic ring, it was not considered as the protonation state of histidine depended on the pH and local environment of residues. It may act as either cation or π residue and can change the preferred relative orientations of the π-π interaction. π-π interactions were classified into three types namely (π-π -SS) side-chain to sidechain π-π interactions, (π-π -SS5) side-chain to side-chain five member aromatic ring π-π interactions and (π-π -S5S) sidechain five member aromatic ring to side-chain π-π interactions.
Location of π-π interaction forming residues based on secondary structure and solvent accessibility:
Secondary structure and solvent accessibility are very important to understand the biochemical activity of proteins. A systematic analysis of each π-π interaction forming residue was performed based on their location in different secondary structures and solvent accessibility of ILs and TNF proteins. We obtained the information about secondary structures from PDB [23] and identified solvent accessibility of the proteins using the program ASAView [25].
Conservation score:
Conservation score is a useful parameter for the identification of conserved residues in a protein sequence based on the phylogenetic relations between homologous sequences. For computing conservation score, we used Consurf [26] in which the following methodology was adopted. 1) The amino acid sequence was extracted from the PDB file; 2) Search for close homologous sequences using BLAST (or PSI-BLAST) [27, 28]. The sequences were clustered and highly similar sequences were removed using CD-HIT [29]; 3) Multiple sequence alignment of the homologous sequences was constructed using CLUSTALW [30]; 4) The MSA was then used to build a phylogenetic tree using the neighbor-joining algorithm as implemented in the Rate4Site program [31]. Position-specific conservation scores were computed using the empirical Bayesian [32] or ML algorithms [31]; 5) The continuous conservation scores were divided into a discrete scale of nine grades. Grade 1 contained the most variable positions, grade 5 contained intermediately conserved positions, and grade 9 contained the most conserved positions [26, 33].
Computation of stabilization center:
Stabilization centers (SCs) were clusters of residues that were involved in medium or long range interactions [34]. Computer simulations and analysis of experimentally determined real protein structures indicate that long range interactions play the dominant role in stabilizing the native structures [35–37]. Two residues were considered to be in long-range interaction if they were separated by at least ten residues in the sequence and at least one of their heavy atom contact distances was less than the sum of the van der Waals radii of the two atoms plus 1.0 Å. Residues can be considered part of stabilization centers if they were involved in medium or long range interactions and if two supporting residues can be selected from both of their flanking tetra peptides, which together with the central residues form at least seven out of the nine possible contacts. The interactions of stabilization center residues hardly influence the formation of the various secondary structure elements and the distribution of the stabilization center residues was rather uneven among the secondary structure element [34]. We used the SCide for predicting stabilization centers among π-π interacting residues [38].
Sequential separation between residues that are involved in π-π interactions:
The sequential distance between the π-π interacting residues was computed using standard methodology [39–42]. The short, medium and long range interactions have been classified according to the distance of separation between the residues along the polypeptide chain. The contribution from <±4 were treated as short-range contacts, greater than ±4 to less than ±10 as medium-range contacts and greater than ±10 were treated as long-range contacts.
Discussion
Preference of aromatic residues for forming π-π interaction in interleukins and TNF Proteins:
We have analyzed the frequency of occurrence of aromatic amino acid residues which are involved in π-π interactions. There are 25 π-π interactions in ILs and 6 interactions in TNF proteins in the whole data set. It is interesting to observe that, there is an average of one significant π-π interaction for almost 2 aromatic aminoacid residues in ILs and 5 aromatic aminoacid residues in TNF proteins. We observe that in ILs, the contribution of Phe residue is higher than Tyr and Trp residues. The occurrence of Phe, Tyr and Trp residues involved in π-π interactions are 48%, 30% and 22% respectively. Considering the benzene in Phe residues, the greater electro negativity of sp2 C relative to H, produces substantial C--H+ dipole. The C-H dipole accounts well for π-π interaction in phenylalanine [43]. In TNF proteins, Tyr residues have the highest contribution than other two aromatic residues for π-π interactions. The occurrence of Phe, Tyr and Trp residues involved in π-π interactions are 25%, 58% and 17% respectively. In tyrosine, the hydroxyl group in the ortho position on the benzene ring, increase the π-stacking by withdrawing π-electron density from the substituted benzene; reduce the electrostatic repulsion with other benzene [44]. Also the ability of hydroxyl group of tyrosine to act as H-bond donor considerably potentiates the binding ability of phenolic ring [45]. These results are depicted in (Figure 1). From these results we believe that, Phe residues in ILs and Tyr in TNF proteins are more important than other aromatic residues for π-π interactions. In general, Trp is the least occurring aminoacid in any protein [46]. This is in accord with our result that Trp have the lowest occurrence in both ILs and TNF proteins. These observations are comparable to the earlier reports on CH- π hydrogen bonds in interleukins [47].
Figure 1.
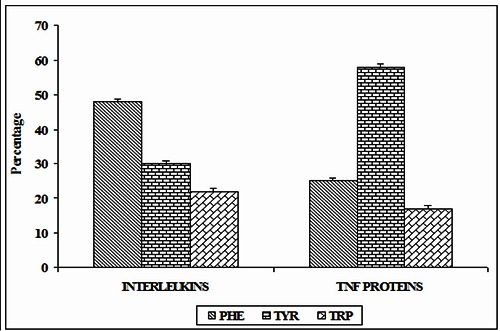
Preference of π-π interacting residues in ILs and TNF proteins.
Secondary structure preferences:
The systematic analysis of secondary structure preference of each amino acid, which participated in different types of π-π interactions are performed. The preferences of secondary structure correlate with the chemical structure and stereochemistry of the amino acids [48]. Secondary structure types were assigned using letters: H for helix, T for turn, and S for strand. The secondary structure preference of each of the amino acids involved in π-π interactions are obtained from PDB. In ILs, among the π-π interacting Phe residues, 42, 46 and 12 % of residues prefer to be in helix, strand and turn conformations respectively. 60% of Tyr residues are in strand conformation and 7, 33 % of residues are in helix and turn conformations. Similarly, 55% of Trp residues are preferred to be in strand, only 27 and 18% Trp residues are in helix and turn conformation. We observe that in ILs, the π-π interacting Phe, Tyr and Trp residues predominantly prefer the strand conformation. These results are depicted in (Figure 2A). We find that in TNF proteins, none of the π-π interacting residues are in helix conformation. All the Phe and Trp residues are in β- strand. Among the Tyr residues, 86% of residues are preferred to be strand and 14% are in turn conformation. These results are depicted in (Figure 2B). In both ILs and TNF proteins, the π-π interacting aromatic residues prefer strand conformation. These results are consistent with the conformational preferences of amino acids in globular proteins [48].
Figure 2.
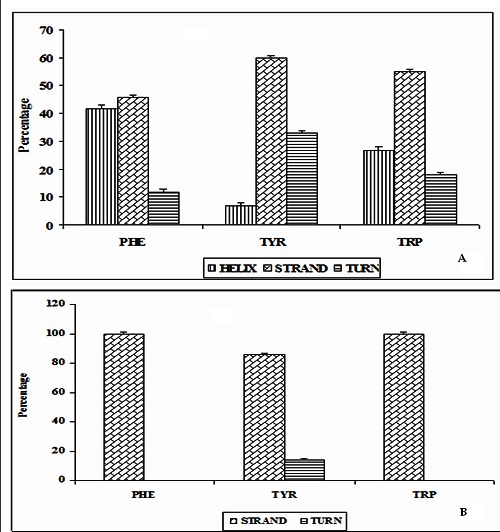
Secondary structure preferences of proteins A) Structural preferences of π-π interacting residues in ILs; B) Structural perferences of residues in TNF Proteins.
Conservation score of interacting residues:
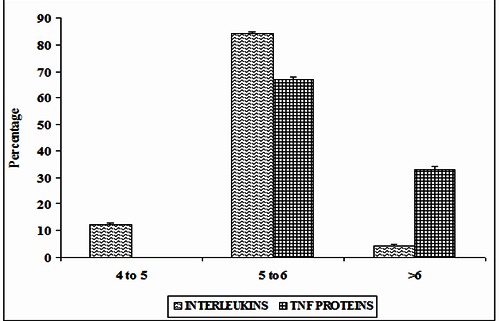
The level of evolutionary conservation is used often as an indicator of importance of the position in maintaining the protein's structure and/or function [26, 33]. Conservation score of ≥ 6 is the cutoff value used to identify the stabilizing residues. In ILs, 90% of residues have the conservation score ≥6. In TNF proteins, 83% of amino acid residues involved in π-π interactions have the conservation score ≥6. These results are shown in (Figure 3A). Thus from these observations, we are able to infer that most of the amino acid residues involved in π- π interactions are evolutionary conserved in both ILs and TNF proteins . Hence, we believe that π-π interacting residues have an additional role in maintaining the structure and function of ILs and TNF proteins. The conservation grade of π-π interacting residues in ILs [PDB ID 1I16] using PyMol is shown in (Figure 3B).
Figure 3.
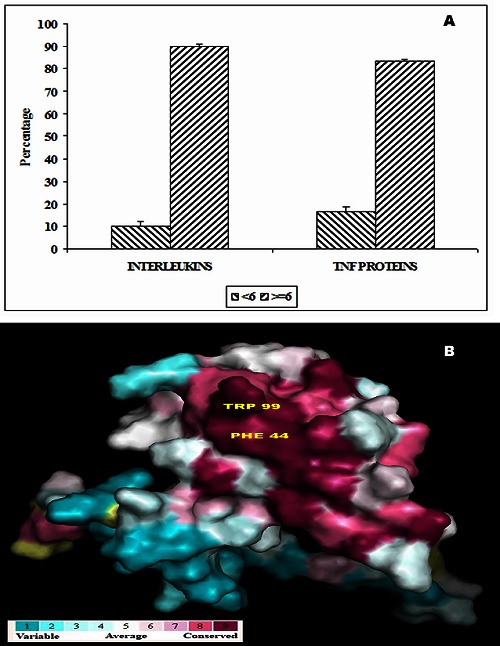
A) Conservation score of π-π interacting residues in ILs and TNF proteins; B) Conservation pattern of ILS [PDB ID 1116] using PyMol.
Stabilization centers:
The unit of SC (Stabilization center) is one pair of interacting residues that are far enough in the primary structure and whose interactions are also supported by other interactions formed by residues located in their vicinity in the primary structure [38]. We have computed the stabilization center [40] for all π-π interaction forming residues in both ILs and TNF proteins. 32% of π-π interacting residues in ILs and 50% of π-π interacting residues in TNF proteins have one or more stabilization centers. The percentage of stabilization centers is more in the case of TNF proteins than ILs. From these results we infer that, the role of π-π interacting residues might be significant in the structural stability of TNF proteins through π-π interactions. All these SC residues might contribute additional stability to ILs and TNF proteins in addition to their participation in π-π interactions. These results are shown in (Figure 4).
Figure 4.
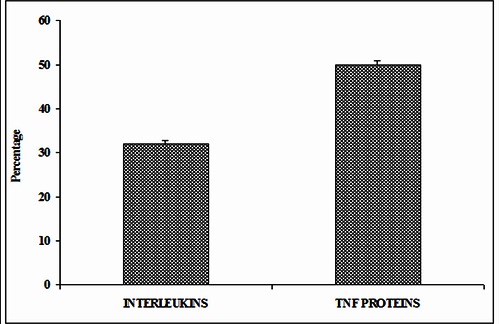
Stabilization centers of π-π interacting residues in ILs and TNF proteins
π-π interacting residues in different ASA ranges:
The solvent accessible surface area (ASA) is computed using the program ASAview to estimate the solvent accessibility of the residues involved in π-π interactions. In ILs, considering the Phe residues, 92% of residues are in solvent buried region whereas each 4% of residues are in partially buried and exposed regions. 86% of Tyr is buried and each 7% of residues are in partially buried and exposed regions. Among the Trp residues, 73% of residues are in buried region and 27% in partially buried regions. In TNF proteins, all the Trp and Phe residues are in solvent buried region. 71% and 29% of Tyr residues are in buried and partially buried regions. It is interesting to note that none of the π-π interacting residues are exposed to solvent. In both ILs and TNF proteins, it is observed that all π-π interacting residues are preferred to be in the buried region. Hence, π-π interacting residues stabilize the inner core regions in these proteins. These results are shown in (Figure 5A & 5B).
Figure 5.
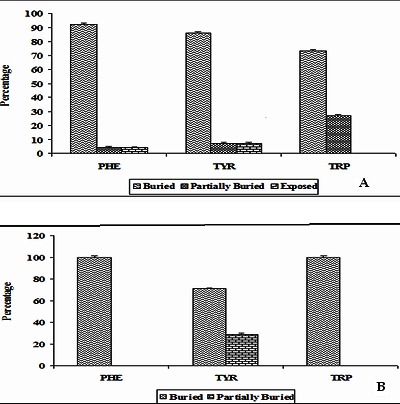
A) Solvent accessibility patterns of π-π interacting residues in ILs; B) Solvent accessibility patterns of π-π interacting residues in TNF proteins.
Residue pairs involved in π-π interactions and π-networks:
There are six possibilities of π-π interacting pairs namely Phe- Phe, Phe-Tyr, Phe-Trp, Tyr-Tyr, Tyr-Trp and Trp-Trp pairs. When homo-pairs of aromatic side chains are considered, the highest percentage of interactions is observed between Phe-Phe residues. 20% of interactions are Phe-Phe interactions, 8% are Tyr-Tyr interactions and there are no Trp-Trp interactions. The percentages of interacting pairs (F-Y, F-W, and Y-W) towards π- π interactions in ILs are 28, 28, and 16% respectively. In TNF proteins, we find four types of interacting pairs, Phe-Tyr, Tyr- Tyr, Phe-Trp and Tyr-Trp interactions. The percentages of interacting pairs (F-Y, Y-Y, F-W, Y-W) towards π-π interactions in TNF proteins are 33, 17, 33 and 17% respectively. We have analyzed the distance between the centroids of π-π interacting residues. The π-π interacting pairs in ILs and TNF proteins are most favorable in the distance range of 5-6 Å as shown in (Figure 6). We have also analyzed the π-networks in ILs and TNF proteins. There are π-networks in ILs and TNF proteins. These π-networks might add more stability and play an important role in understanding the 3D structure of proteins. The 3π, 4π and 7π - networks in the data set of ILs and TNF proteins are shown in (Figure 7A). The PyMol view of Trp-Phe interacting pair in ILs [PDB ID 1I16] and linear 7π network in ILs [PDB ID 1F45] are shown in (Figure 7B & 7C).
Figure 6.
Preferential distance (in A) of π-π interacting residues in ILS and TNF proteins
Figure 7.
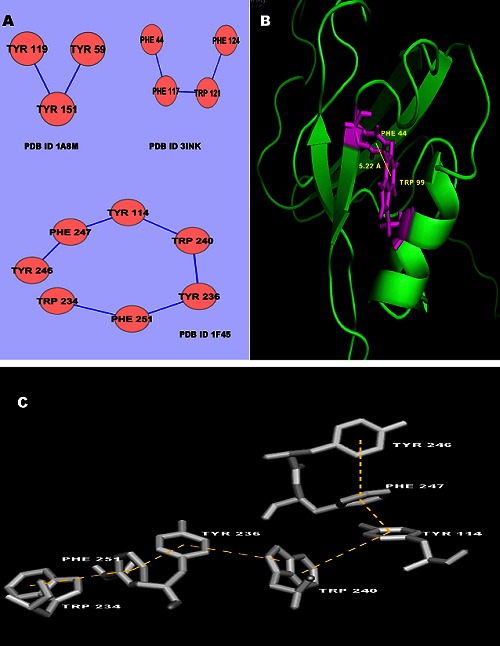
A) 3π, 4π and 7π- -networks in ILS and TNF proteins; B) PyMol view of Trp-Phe interacting pairs in ILs [PDB ID 1116]; C) The PyMol view of 7π network in ILs [PDB ID 1F45].
Sequential separation of interacting pairs:
Long range interactions play an active role in the stability of protein molecules. Short, medium and long range interactions as a function of percentage of interaction in ILs and TNF proteins are studied. It is interesting to note that, 68% of π-π interactions in ILs are long range interactions and 8, 24% of interactions are short-range interactions and medium-range interactions respectively. It is observed that in TNF proteins, 67% of π-π interactions are long range interactions whereas 33% are medium range interactions. Long range π-π interactions are the predominant type of interactions in ILs and TNF proteins and hence π-π interactions contribute significantly to the tertiary structure stability of these proteins. These results are shown in (Figure 8).
Figure 8.
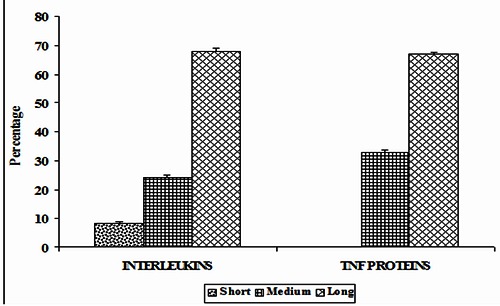
Sequential distances of interacting pairs in ILs and TNF proteins.
Conclusion
We have analyzed the role of π-π interactions in ILs and TNF proteins and made a comparative study. In ILs, Phe residues have maximum contribution whereas, it is Tyr in TNF proteins to form π-π interactions. The occurrence of Trp is very low in both ILs and TNF. Several π-π interactions are observed between the residues in β-strand. Since, most of the π-π interacting residues have the tendency of being buried in the interior of ILs and TNF proteins, these interactions might be important in stabilizing the inner core regions of these proteins. It is interesting to note that in both ILs and TNF proteins, none of the π-π interacting Trp residues are at the surface. Our investigations on sequential distance between the interacting pairs suggest that most of the π-π interacting residues are in long-range contacts and hence, these interacting residues might play an important role in global structural stability of these proteins. Majority of π-π interacting residues are evolutionary conserved in both ILs and TNF proteins. Conserved residues are comparatively high in case of ILs than TNF proteins. Considerable percentage of π-π interacting residues had one or more stabilization centers and these residues might contribute significantly to the stability to ILs and TNF proteins. Most of the π-π interacting pairs prefer to be in distance range of 5-6 Å in ILs and TNF proteins. There is also a significant number of π- networks in both ILs and TNF proteins. On the whole the results obtained from this study will be very helpful in further understanding the structural stability and functions of ILs and TNF proteins.
Supplementary material
Acknowledgments
Dr. Anand Anbarasu gratefully acknowledges the Indian Council of Medical Research [ICMR] Government of India agent for the research grant IRIS ID: 2011-03260. P. Lavanya thanks ICMR for the research fellowship from the ICMR grant IRIS ID: 2011- 03260 and would like to thank the management of VIT University for providing the necessary facilities to carry out this research project. We also thank Dr. M. Madan Babu, MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB22QH, UK, for helping us with the NCI server.
Footnotes
Citation:Sivasakthi et al, Bioinformation 9(8): 432-439 (2013)
References
- 1.CG Claessens, Stoddart JF. J Phys Org Chem. 1997;10:254. [Google Scholar]
- 2.Fyfe MCT, Stoddart JF. Acc Chem Res. 1997;30:393. [Google Scholar]
- 3.Kannan N, Vishveshwara S. Protein Eng. 2000;13:753. doi: 10.1093/protein/13.11.753. [DOI] [PubMed] [Google Scholar]
- 4.Burley SK, Petsko GA. Adv Protein Chem. 1988;39:125. doi: 10.1016/s0065-3233(08)60376-9. [DOI] [PubMed] [Google Scholar]
- 5.Samanta U, et al. Proteins. 2000;38:288. [PubMed] [Google Scholar]
- 6.Burley SK, Petsko GA. Science. 1985;229:23. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 7.Hunter CA, Sanders JKM. J Am Chem. Soc. 1990;112:5525. [Google Scholar]
- 8.Chakrabarti P, Samanta U. J Mol. Biol. 1995;251:9. doi: 10.1006/jmbi.1995.0411. [DOI] [PubMed] [Google Scholar]
- 9.Samanta U, Chakrabarti P. Protein Eng. 2001;14:7. doi: 10.1093/protein/14.1.7. [DOI] [PubMed] [Google Scholar]
- 10.Macias MJ, et al. FEBS Lett. 2002;513:30. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrakis MG, et al. Oncol Rep. 2000;7:1327. doi: 10.3892/or.7.6.1327. [DOI] [PubMed] [Google Scholar]
- 12.Alexandrakis MG. Anticancer Res. 1999;19:3607. [PubMed] [Google Scholar]
- 13.Antony VB, et al. Am Rev Resp Dis. 1992;145:1236. doi: 10.1164/ajrccm/145.5.1236. [DOI] [PubMed] [Google Scholar]
- 14.Antony VB, et al. J Immunol. 1993;151:7216. [PubMed] [Google Scholar]
- 15.Van Deuren M, et al. J Pathol. 1992;168:349. doi: 10.1002/path.1711680403. [DOI] [PubMed] [Google Scholar]
- 16.Mann DL. Circ Res. 2002;91:988. doi: 10.1161/01.res.0000043825.01705.1b. [DOI] [PubMed] [Google Scholar]
- 17.Bradham WS, et al. Am J Physiol Heart Circ Physiol. 2002;282:1288. doi: 10.1152/ajpheart.00526.2001. [DOI] [PubMed] [Google Scholar]
- 18.Sivasubramanian N, et al. Circulation. 2001;104:826. doi: 10.1161/hc3401.093154. [DOI] [PubMed] [Google Scholar]
- 19.Feldmann M, Maini RN. Annu Rev Immunol. 2001;19:163. doi: 10.1146/annurev.immunol.19.1.163. [DOI] [PubMed] [Google Scholar]
- 20.OShea JJ, et al. Nat Rev Immunol. 2002;2:37. [Google Scholar]
- 21.Trinchieri G. Nat Rev Immuno. 2003;3:133. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 22.Anbarasu A, et al. Cytokine. 2006;35:263. doi: 10.1016/j.cyto.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Berman HM, et al. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babu MM. Nucl Acids Res. 2003;31:3345. doi: 10.1093/nar/gkg528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shandar A, et al. Bioinformatics. 2004;5:51. [Google Scholar]
- 26.Glaser F, et al. Bioinformatics. 2003;19:163. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 27.Altschul SF, et al. FEBS J. 2005;272:5101. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, et al. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Godzik A. Bioinformatics. 2006;22:1658. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 30.Thompson JD, et al. Nucl Acids Res. 1994;22:4673. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pupko T. Bioinformatics. 2002;18:71. doi: 10.1093/bioinformatics/18.8.1116. [DOI] [PubMed] [Google Scholar]
- 32.Mayrose I, et al. Mol Biol Evol T. 2004;21:1781. doi: 10.1093/molbev/msh194. [DOI] [PubMed] [Google Scholar]
- 33.Landau M, et al. Nucleic Acids Res. 2005;33:299. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dosztanyi ZS, et al. J Mol Biol. 1997;272:597. doi: 10.1006/jmbi.1997.1242. [DOI] [PubMed] [Google Scholar]
- 35.Abkevich V, et al. J Mol Biol. 1995;252:460. doi: 10.1006/jmbi.1995.0511. [DOI] [PubMed] [Google Scholar]
- 36.Mirny LA, Shakhnovich E. J Mol Biol. 1996;264:1164. doi: 10.1006/jmbi.1996.0704. [DOI] [PubMed] [Google Scholar]
- 37.Bahar I, Jernigan R. J Mol Biol. 1997;266:195. doi: 10.1006/jmbi.1996.0758. [DOI] [PubMed] [Google Scholar]
- 38.Dosztányi Z, et al. Bioinformatics. 2003;19:899. doi: 10.1093/bioinformatics/btg110. [DOI] [PubMed] [Google Scholar]
- 39.Gromiha MM, Selvaraj S. J Biol Phys. 1997;23:151. doi: 10.1023/A:1004981409616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gromiha MM, et al. Int J Biol Macromol. 2004;34:203. doi: 10.1016/j.ijbiomac.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Selvaraj S, Gromiha MM. Biophys J. 2003;84:1919. doi: 10.1016/s0006-3495(03)75000-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anbarasu A, Sethumadhavan R. J Theor Biol. 2007;247:346. doi: 10.1016/j.jtbi.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 43.Hunter CA, et al. J mol biol. 1991;218:837. doi: 10.1016/0022-2836(91)90271-7. [DOI] [PubMed] [Google Scholar]
- 44.Cozzi F, et al. J Am Chem Soc. 1992;114:5729. [Google Scholar]
- 45.Mecozzi S, et al. Proc Natl Acad Sci USA. 1996;93:10566. doi: 10.1073/pnas.93.20.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gallivan JP, Dougherty DA. Proc Natl Acad Sci USA. 1999;96:9459. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anand S, et al. In Silico Biology. 2008;8:261. [PubMed] [Google Scholar]
- 48.Levitt M, et al. Biochemistry. 1978;17:4277. doi: 10.1021/bi00613a026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


