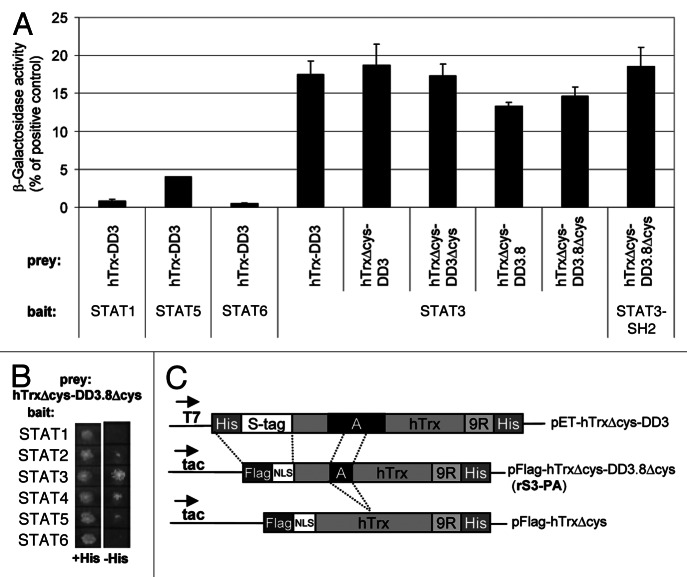
Figure 1. Structure and binding specificity of recombinant peptide aptamer rS3-PA. (A) YTH interaction analysis of the peptide aptamer rS3-PA with members of the STAT family. Yeast cells (Y187) were co-transformed with bait and prey constructs. Interaction of these proteins was quantified in standard β galactosidase assays (displayed in Miller Units). (B) Interaction of bait and prey fusion proteins described in (A) also results in the growth of colonies on plates lacking histidine. (C) Schematic presentation of peptide aptamer DD3 40 amino acids (A) inserted into the hTrx scaffold (upper construct). Modifications performed to generate pFlag-hTrxΔcys-DD3.8Δcys encoding rS3-PA are shown (middle construct). rS3-PA encodes a 20mer peptide fragment of DD3 devoid of cysteines. The peptide aptamer present in rS3-PA corresponds to the sequence N-VRH SAL HMA VGP LSW PAR VS-C. The bait used to select this peptide aptamer comprises the STAT3 sequence 655 to 755.21 Secondary structure prediction programs from the Expasy website (e.g., Porter, NtSurfP, JPred) were used and a comparison of the results suggests that the 20 amino acid peptide aptamer sequence consists largely of a α-helical structure in its N-terminal region and a coiled-coil structure in its C-terminal region. The prediction for the 655–755 amino acid sequence of STAT3 suggests that it is largely composed of a coiled-coil domain with a possible α-helical insert at position 76–84. The pFlag-hTrxΔcys control construct was generated by Rsr II digestion removing the aptamer sequences (lower line). NLS, nuclear localization signal; 9R, nine arginine PTD.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.