Abstract
A growing body of evidence suggests that insulators have a primary role in orchestrating the topological arrangement of higher-order chromatin architecture. Insulator-mediated long-range interactions can influence the epigenetic status of the genome and, in certain contexts, may have important effects on gene expression. Here we discuss higher-order chromatin organization as a unifying mechanism for diverse insulator actions across the genome.
Keywords: chromatin, epigenetics, topological domains, transcription, nuclear organization
Introduction
Genomes of metazoan organisms are packaged in a hierarchy of topological configurations that allow for intricate spatiotemporal regulation of complex nuclear functions such as transcription, replication, recombination, and DNA repair (Misteli, 2007). In G1, for example, the chromatin fiber must be arranged in a manner that is favorable for regulation by a complex cascade of transcription factors and chromatin modifying enzymes. In principle, this architecture should also leave chromosomes poised for reorganization, condensation, decondensation, and re-assembly during each subsequent S-phase and mitosis. Reports of fractal organization of genomes provide a biophysical model for how this folding and unfolding can take place in a rapid and precise manner at a larger scale (Lieberman-Aiden et al., 2009), but the molecular mechanisms that regulate chromatin organization at a finer sub-Mb scale are poorly understood.
Understanding the organizing principles responsible for 3-D folding of chromatin remains an important and unanswered question. Progress has been made possible in recent years by the development of a cadre of Chromosome-Conformation-Capture (3C)-based molecular techniques that allow high resolution mapping of inter- and intra-chromosomal interactions (de Wit and de Laat, 2012; Dekker et al., 2002). This information, coupled with genome-wide maps of the distribution of chromatin binding proteins obtained by ChIP-seq has made it possible to correlate the presence of specific proteins binding to the chromatin fiber with large scale structural features as well as point-to-point looping interactions across the genome.
Here we review evidence suggesting that insulator proteins have a conserved role across metazoans as architectural proteins that orchestrate chromatin organization. We use observations in mammals and D. melanogaster to discuss molecular mechanisms regulating the myriad of intra- and inter-chromosomal interactions between regulatory elements and insulator proteins across the genome. Mechanisms of chromatin folding are discussed in the context of transcription, but we note that similar principles could apply to other genome functions linked to chromatin organization (e.g. replication and recombination (Gilbert et al., 2010; Jhunjhunwala et al., 2009).
Modularity and distribution of insulators across the genome
CTCF is considered the primary insulator in mammals. The protein is ubiquitously expressed across most mammalian tissues (Wendt et al., 2008) and is required for early mouse development (Fedoriw et al., 2004). Homozygous CTCF deletion results in early embryonic lethality (Heath et al., 2008; Splinter et al., 2006) and conditional knock-down in mouse oocytes leads to mitotic defects upon fertilization and delayed progression to the blastocyst stage (Fedoriw et al., 2004; Wan et al., 2008). In somatic cells, conditional knock-out confirmed additional important roles for CTCF in cell-cycle progression, apoptosis, and differentiation (Heath et al., 2008; Soshnikova et al., 2010; Splinter et al., 2006). Thus, CTCF has a pervasive role across most cell types during mammalian development.
In addition to ubiquitous expression patterns, CTCF binding sites are also widely distributed across mammalian genomes. In the first ChIP-chip analysis of CTCF, Ren and colleagues reported ~14,000 occupied sites in human IMR90 fibroblasts with a genomic distribution of 46% intergenic, 22% intronic, 12% exonic, and 20% within 2.5 kb upstream of TSSs (Kim et al., 2007). Subsequent ChIP-seq analysis by Zhao and colleagues revealed ~19,000–29,000 occupied sites in CD4+, HeLa, and Jurkat cells, with genomic distributions of 49–56% intergenic, 3–4% exonic, and 32–33% intronic, and 7–15% at TSSs (Barski et al., 2007; Cuddapah et al., 2009; Jothi et al., 2008). Most recently, Stamatoyannopoulos and colleagues found an average of 55,000 CTCF sites per cell type when comparing 19 different cell lines, whereas Chen et al. reported ~66,800 occupied sites from each of 38 different cell types from the ENCODE project (Chen et al., 2012; Wang et al., 2012). Advances in molecular and computational ChIP-seq technologies are the most probable explanation for the marked increase in sensitivity for genome-wide occupied CTCF sites.
In Drosophila, there are five subclasses of insulator sequences (reviewed in (Gurudatta and Corces, 2009)). Each subclass is defined by common accessory proteins and a unique DNA binding protein, including: Suppressor of Hairy-wing [Su(Hw)], Drosophila CTCF (dCTCF), Boundary Element Associated Factor of 32 kDa (BEAF-32), GAGA Binding Factor (GAF) and Zeste-white 5 (Zw5). The biological significance of five different insulators is unclear. However, because some subclasses are only found in Drosophila, a leading hypothesis is that multiple insulators might be critical for precise regulation of compact genomes with much less distance between genes than in vertebrates. For example, the BEAF-32 insulator has been recently linked to the independent regulation of adjacent genes transcribed in opposite orientations (Yang et al., 2012).
Drosophila insulator proteins are ubiquitously expressed, contain DNA binding domains, and null mutations (with the exception of Su(Hw)) result in lethality (Gurudatta and Corces, 2009). Genome-wide analyses indicated that Drosophila insulators bind to 3,000–6,000 sites and show distinct distributions with respect to genomic features (Bartkuhn et al., 2009; Bushey et al., 2009; Negre et al., 2010; Wood et al., 2011). BEAF-32 is preferentially located in promoter regions, whereas Su(Hw) is biased toward intergenic regions. By contrast, dCTCF follows an intermediate distribution similar to that of CTCF in mammals. Distinct distributions of different sub-classes of insulators may be important for predicting their function(s) in genome regulation.
Growing evidence also suggests a widespread role for TFIIIC as the most evolutionarily conserved insulator (Van Bortle and Corces, 2012). Protists, fungi and plants lack CTCF (Heger et al., 2012) and may rely on TFIIIC to carry out aspects of insulator function. For example, tDNA genes containing TFIIIC binding sites show insulator activity in S. saccharomyces, S. pombe and mammals (Raab et al., 2012). In addition, TFIIIC sites unrelated to tDNA genes have been mapped across the mouse genome and appear to correlate with CTCF, suggesting that these two insulators may cooperate at specific genomic sites (Moqtaderi et al., 2010). Interestingly, both cohesin and condensin interact with TFIIIC and are required for its function (D’Ambrosio et al., 2008), suggesting that this insulator might show mechanistic similarities to CTCF (discussed below).
Controversy surrounding insulator mechanisms of action
Insulators have been linked to a vast range of genomic functions, including: activation, repression, enhancer blocking insulation, barrier insulation, promoter proximal pausing, alternative splicing, and protection from DNA methylation. The molecular mechanisms for how insulators confer these pleiotropic effects across the genome remain poorly understood. It has been suggested that mammalian CTCF serves different purposes based on the binding to divergent consensus sequences and the subsequent recruitment of different binding partners and post-translational modifications (Ohlsson et al., 2010). In the case of Drosophila, an influential idea is that the different insulator subclasses may be responsible for performing distinct functions. Thus, an important unresolved question is whether insulators are true multivalent factors with the ability to perform many contrasting functions, or if there is a single unifying mechanism that can explain these divergent roles.
Mammalian CTCF and 3-D chromatin architecture
A body of locus-specific and genome-wide evidence now points to a primary role for mammalian CTCF in genome organization. Prior to the availability of deep sequencing technologies, several clues had already emerged supporting this role. First, mass spectroscopy analysis of Flag-tagged CTCF purified from HeLa cells revealed that CTCF can form both homodimers and multimers in vivo (Yusufzai et al., 2004). Second, yeast two-hybrid experiments showed that CTCF has the capacity to bind other CTCF molecules in vitro (Yusufzai et al., 2004). Third, CTCF molecules bound to probes encoding divergent CTCF consensus sequences also dimerized in vitro (Pant et al., 2004). Finally, GST pull-down assays revealed that the C-terminus of CTCF binds to the 11-zinc-finger domain of CTCF in vitro (Pant et al., 2004). Together, these data provided the initial biochemical evidence in mammalian systems to support a role for CTCF in long-range looping interactions.
More recently, 3C-based methods have been leveraged to analyze higher-order chromatin architecture at kb-resolution. Independent studies at the mouse β-globin, H19/Igf2, and MHC class II genomic loci demonstrated that CTCF sites are involved in forming long-range interactions between specific genomic elements (Kurukuti et al., 2006; Li et al., 2008; Majumder et al., 2008; Murrell et al., 2004; Splinter et al., 2006; Yoon et al., 2007). Since then, 3C has been used to identify insulator-mediated contacts at many mammalian genomic loci, including, but not limited to: human β-globin (Hou et al., 2010), human apolipoprotein (Mishiro et al., 2009), human kcnq5 (Ren et al., 2012), human/mouse HoxA (Ferraiuolo et al., 2010; Kim et al., 2011), human insulin (Xu et al., 2012), and human Interferon-γ (Hadjur et al., 2009). In most of these studies, global RNAi-mediated knock-down of CTCF resulted in marked reduction of 3C signal, providing direct evidence that CTCF is required for at least some subset of long-range interactions.
Handoko et al. used an independent technique termed ChIA-PET to find a sub-population of CTCF-mediated chromatin interactions genome-wide (Handoko et al., 2011). The authors identified 1,816 high-confidence 3-D interactions (1,480 intrachromosomal and 336 interchromosomal) connected by CTCF in mouse ES cells. CTCF siRNA in ES cells showed reduced interaction of specific inter- and intra-chromosomal contacts selected for validation, suggesting that CTCF is essential for the formation of specific long-range interactions. Most recently, Dekker and colleagues reported that CTCF is highly enriched in long-range interactions between transcription start sites (TSSs) and distal regulatory elements throughout ENCODE pilot regions spanning 1% of the human genome (Sanyal et al., 2012).
Insights into the mechanisms governing insulator-mediated genome organization came with the discovery that CTCF co-localizes with cohesin at thousands of sites across mammalian genomes (Parelho et al., 2008; Rubio et al., 2008; Stedman et al., 2008; Wendt et al., 2008). Indeed, 50–80% of CTCF occupied sites overlap with cohesin, depending on the cell line. Because cohesin proteins are traditionally thought to function in sister chromatid cohesion, an influential model suggests that CTCF recruits cohesins to DNA and then, in turn, cohesin subunits form a ring-like structure that stabilizes higher-order organization of chromatin during interphase (Gause et al., 2008). Direct evidence from knockdown experiments is so far consistent with this model, as RNAi for cohesin subunits results in disruption of long-range looping interactions at several distinct loci (Hadjur et al., 2009; Hou et al., 2010; Mishiro et al., 2009; Nativio et al., 2009). Although it is clear that both cohesin-dependent and cohesin-independent CTCF sites can form long-range interactions, a critical unresolved question would be why some sites require cohesin and why some sites do not. Altogether, these data provide unequivocal evidence that mammalian CTCF is involved in and essential for higher-order chromatin organization genome-wide.
Drosophila insulators and 3-D chromatin architecture
Direct and indirect evidence is also consistent with a role for Drosophila insulators in mediating 3-D chromatin interactions. The scs and scs’ insulators flank the hsp70 genes and bind Zw5 and BEAF proteins, respectively. These insulators are separated by 15 kb on the linear genome, but show high interaction frequency by 3C in Drosophila embryos (Blanton et al., 2003). Co-IP and GST pull-down assays indicated that Zw5 and BEAF proteins can bind directly to each other in vitro and in vivo. These results support the idea that direct hetero-dimerization by insulator DNA binding proteins could be one mechanism driving scs-scs’ looping interactions.
Another leading idea is that insulators form long-range interactions by recruiting co-factors such as Centrosomal Protein of 190 kDa (CP190) and/or Modifier of mdg4 [Mod(mdg4)] to the DNA-binding proteins. Yeast two-hybrid experiments indicate that Mod(mdg4) proteins can form direct heterodimers with Su(Hw) and also homodimerize with each other (Gause et al., 2001; Ghosh et al., 2001). Furthermore, Co-IP, yeast two-hybrid, and affinity chromatography experiments suggested that CP190 can bind directly to CTCF as well as Su(Hw) and Mod(mdg4) insulator proteins (Gerasimova et al., 2007; Mohan et al., 2007; Pai et al., 2004). Different subclasses of insulators group together in Drosophila nuclei in subnuclear structures termed ‘insulator bodies’. CP190 is essential for the formation of these bodies, suggesting that CP190 homodimerization may be a key mechanism in the formation of a 3-D interaction network in Drosophila nuclei.
Contrary to vertebrates, Drosophila insulators do not appear to rely on cohesin to establish or maintain interactions with other sequences in the genome (Dorsett, 2009). We favor a model in which CP190 or Mod(mdg4) play a similar functional role to that of cohesin, as both proteins contain BTB domains that may be involved in mediating inter-insulator interactions between independent genomic loci. In addition to CP190 and Mod(mdg4), several other proteins such as Chromator and L(3)mbt have been recently shown to interact or co-localize with Drosophila insulator proteins, but their possible role in chromatin organization has not been studied in detail (Gan et al., 2011). Importantly, a subset of so-called “aligned insulator elements” contain clustered occupied sites for CP190, Su(Hw), BEAF-32 and/or dCTCF in close proximity to each other within 100–300 bp-sized elements across the genome (Van Bortle et al., 2012). The presence of multiple insulator proteins clustered together within a small genomic element may give these sequences a unique role in 3-D chromatin organization compared to sites that only bind single insulators (discussed below).
Mechanisms of insulator-mediated chromatin organization
Insulators as boundaries of higher-order topological domains
Four independent studies have reported the discovery of highly self-interacting genomic units termed topologically associating domains (TADs) (Dixon et al., 2012; Hou et al., 2012; Nora et al., 2012; Sexton et al., 2012). Genomic sequences within TADs have a high frequency of 3-D interactions with each other compared to sequences in adjacent TADs. Between TADs are distinct boundaries where chromatin interactions switch their directionality from an upstream bias (interactions within the current TAD) to a downstream bias (interactions within the adjacent TAD).
What are the mechanisms that causally demarcate the boundaries of TADs? In mammals, Ren and colleagues found that >75% of all TAD boundaries contain CTCF occupied sites. Specifically, ~28% of all boundaries contain CTCF plus active housekeeping genes, ~20% contain CTCF and other genes, ~28% contain CTCF alone without genes, whereas ~9% have only genes and ~15% do not display a particular mark (Dixon et al., 2012). In Drosophila, boundaries are enriched for BEAF-32, CTCF, and CP190, but not Su(Hw) (Sexton et al., 2012). There also appears to be an enrichment for aligned insulator elements containing binding sites for two or more insulator DNA binding proteins plus CP190, whereas single insulator sites are enriched inside TADs (Hou et al., 2012). On the basis of this finding, we hypothesize that clusters of mammalian CTCF and its numerous binding partners (reviewed in detail (Herold et al., 2012; Ohlsson et al., 2010)) might also create tandemly aligned sites that contribute to the formation of boundaries between TADs in mammals.
It is also important to note that factors other than insulators also contribute to the formation of TAD boundaries. Indeed, in Drosophila and mammals, boundaries are enriched in active genes in addition to insulators, suggesting that high levels of transcription may be essential in the establishment and/or maintenance of topological domains. TAD boundaries are also enriched in tRNA genes and Alu/B1 and B2 SINE elements (Dixon et al., 2012), which contain binding sites for TFIIIC (Lunyak and Atallah, 2011). Because both CTCF and TFIIIC interact with cohesin and condensins (D’Ambrosio et al., 2008), we speculate that TFIIIC alone or in combination with CTCF might have a causal role in genome organization at TAD boundaries.
Together these data suggest that insulators (with or without active transcription) may be important causal factors in the organization of topological transitions from one TAD to another. Several important questions related to the mechanisms of TAD boundary function remain to be answered: (1) Is CTCF necessary and/or sufficient for functional boundary formation? (2) What additional mechanisms (e.g. transcription) causally affect the specificity of CTCF action at TAD borders genome-wide? (3) Are CTCF and active chromatin modifications a cause or consequence of boundary formation? and (4) How are insulators at TAD boundaries vs. internal to TADs mechanistically distinguished?
Insulators as borders of functional chromatin domains
Evidence that CTCF organizes 3-D chromatin topology genome-wide raises important questions related to the putative role for CTCF as a barrier insulator. Barrier insulators were originally defined by their ability to protect transgenes from position effect silencing (defined in Figure 1A). Initial studies in vertebrates reported > 1 kb-sized genomic sequences containing CTCF binding sites that exhibited barrier activity in transgene assays (Cho et al., 2005; Filippova et al., 2005; Pikaart et al., 1998). Interestingly, specific CTCF binding sites could be deleted without affecting the barrier activity of a ~275 bp-sized chicken HS4 insulator sequence (Recillas-Targa et al., 2002). On the basis of this observation, it was first proposed that CTCF is not essential for barrier function and primarily worked through enhancer blocking mechanisms (discussed below).
Figure 1. Experimental paradigms for enhancer blocking and barrier insulation.
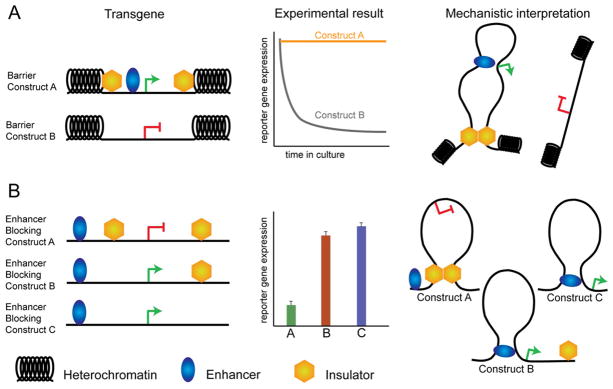
(A) To test barrier insulator activity, a transgene flanked by insulators is randomly integrated into the genome. Multiple integration sites are considered to control for position-effect variegation. If sequences are true barrier insulators, reporter expression over time in culture should remain constant, whereas a control transgene that does not contain insulators should eventually be silenced by encroachment of heterochromatin. (B) To test enhancer blocking insulator activity, transgene constructs are designed by placing a putative insulator sequence in various positions with respect to an enhancer driving a reporter gene. The degree of ‘insulation’ (or ability to abrogate the enhancer) is assayed as the level of reporter gene expression after transient transfection, or integration of the vector, into target cells. To rule out effects of position-independent silencing, results are compared to control constructs in which insulators are placed adjacent to, but not in between, linked enhancer-promoter sequences. Limitations of these assays to be considered during data interpretation, include: the spacing between elements which does not mimic the endogenous locus, the integration of the reporter into multiple ectopic genomic locations, and the often use of heterologous enhancer-promoter sequences that also do not represent the genomic context of insulator sequences.
More recently, the role for insulators in barrier function was brought back into question with a genome-wide query (Cuddapah et al., 2009). Cuddupah et al. identified 30,000+ domains of the repressive chromatin modification H3K27me3 that ranged in size from 5 to 25 kb across the human genome. A search for proteins binding at the borders on either side of these H3K27me3 domains revealed 1,606 and 793 CTCF occupied sites in CD4+ and HeLa cells, respectively. Although this result is widely cited as supporting a genome-wide role for CTCF in barrier function, a closer look suggests a different interpretation. Only a very small fraction of H3K27me3 domain borders contain CTCF: 1578/39,900 (~4%) in CD4+ cells and 771/32,704 (~2.4%) in HeLa cells. Furthermore, only a very small number of total CTCF binding sites (4–6%) are associated with these chromatin borders. We interpret this result to support the original hypothesis (Recillas-Targa et al., 2002) that there must be other mechanisms, in addition to or independent from CTCF, that are essential for demarcating most of H3K27me3-marked repressive domains.
What is known about the mechanism(s) by which the small fraction of CTCF sites at borders contribute to the establishment of functional domains of heterochromatin? Cuddapah et al. observed that the majority of their identified repressive domains were cell type-specific (Cuddapah et al., 2009). When considering only borders bound by CTCF, only 5–11% were constant between cells types and also have constant CTCF occupancy, whereas 23–40% were cell type-specific and mirrored by cell type-specific binding of CTCF. By contrast, 55–66% of borders bound by CTCF displayed constant occupancy of CTCF but cell type-specific boundary function. From a mechanistic perspective, these observations suggest that CTCF occupancy is not likely the critical factor for barrier function.
Two recent studies in Drosophila explored the mechanistic role of insulators in maintaining borders for domains of silencing histone modifications (Schwartz et al., 2012; Van Bortle et al., 2012). Pirrotta and colleagues analyzed H3K27me3 domains genome-wide and reported that half of these domains showed a gradual decline, while half (~100) have edges that represented sharp transitions that could be characterized as borders. Computational analysis of the ~100 sharp borders demonstrated that ~33% display insulator binding and actively transcribed genes, whereas 44% have only actively transcribed genes and ~20% have only insulators. For borders marked only by insulator proteins, 75% showed slight spreading of the H3K27me3 into the surrounding regions after insulator knockdown. For the remaining borders that contain actively transcribed genes (with or without insulators), there was neglibible effect on H3K27me3 spreading after insulator knockdown. van Bortle et al. also explored H3K27me3 domains in Drosophila Kc cells with a focus on the role for insulators in demarcating domains. The authors reported that only 2% of independent CTCF sites without any other insulators were present within 5 kb of H3K27me3 borders, whereas 8% of aligned insulator sites (e.g. CTCF in combination with BEAF-32 and/or Su(Hw)) were located within 5 kb of H3K27me3 borders. In this study, down-regulation of Drosophila insulators by RNAi, individually or in combination, resulted in a decrease of H3K27me3 levels within the domain, but no clear spreading of the modification. These results suggest that the role for insulators may not be to maintain borders of repressive chromatin domains but rather to maintain the level of silencing by alternative mechanisms. One possibility is that insulators are involved in the recruitment of Polycomb (Pc)/H3K27me3 domains to Pc bodies, where clustering of these domains is necessary for the maintenance of silencing (Pirrotta and Li, 2012).
If insulators are not the primary contributor to barrier activity, then what other factors play a role? Studies with the chicken HS4 insulator provided the first mechanistic clues. While deletion of the CTCF site had no effect, deletion of either VEZF1 or USF1/USF2 binding sites markedly disrupted barrier activity (Dickson et al., 2010; Recillas-Targa et al., 2002; West et al., 2004). Intriguingly, the HS4 insulator contains both CTCF and high levels of active chromatin modifications H3K4me2, H3K9ac, and H3K14ac. USF proteins recruit H3K4-specific methyltransferase SET7/9 and the H3-specific acetyltransferase PCAF to HS4. Furthermore, deletion of the USF binding site resulted in loss of active histone modifications in addition to loss of barrier activity (Litt et al., 2001; West et al., 2004). Importantly, although positive histone modifications were necessary, they were not sufficient for insulator barrier activity, because deletion of VEGZF1 sites also resulted in loss of barrier insulation without disrupting chromatin modifications (Dickson et al., 2010). These data are consistent with the idea that functional borders may be modular and require several different epigenetic mechanisms in addition to CTCF.
Together results suggest that barriers are complex genomic elements that require combinatorial action of multiple proteins and chromatin modifications with distinct roles. A leading model from these studies is that multiple proteins (in addition to CTCF) bind to barrier insulator sequences to recruit chromatin-modifying enzymes that lay down persistent positive histone modifications to prevent the processive spread of heterochromatin. The primary role for insulators in these modular elements may be to mediate intra- or inter-chromosomal interactions that form the topological basis of barrier function, whereas the other proteins and epigenetic modifications have the causal responsibility of preventing the spread of the repressive chromatin mark.
Blurring the boundaries between borders, barriers, and loops
Given the diversity of insulator actions across the genome, a critical question is wether the mechanisms by which insulators confer barrier function are the same as the mechanisms by which insulators demarcate TAD boundaries. In Drosophila, many TADs contain a heterogeneous mixture of chromatin states, suggesting that TADs do not always correspond to functional chromatin domains defined by specific histone modifications (Hou et al., 2012). In mouse and human, a small fraction of TADs correspond to domains of H3K9me3, whereas a large proportion contains a mixture of chromatin modifications (Dixon et al., 2012). These results suggest that only a subset of TADs might play a causal role in demarcating blocks of repressive chromatin modifications, while a larger subset of TADs have purposes broader than delimiting chromatin domains that have not yet been defined.
In the subset of cases where TADs align with domains of repressive H3K9me3 chromatin modifications: do the modifications themselves dictate the formation of TADs or do TAD boundaries causally mark heterochromatin borders? To directly test these questions, Heard, Dekker and colleagues first knocked down enzymes that catalyze repressive histone modifications in embryonic stem cells. They showed that TADs were not disrupted on the X chromosome after knock-down of H3K27me3 or H3K9me2, suggesting that repressive chromatin modifications are not causal for the creation of TAD boundaries. By contrast, they also reported that a 58 kb deletion at a boundary between 2 TADs on the X-chromosome (depicted in Figure 2A) disrupts topological organization and results in numerous ectopic point-to-point looping interactions between adjacent TADs (depicted in Figure 2B) (Nora et al., 2012). Importantly, a CTCF occupied site that has been shown by Lee and colleagues to have insulator activity in transgene assays binds within the deleted region (Spencer et al., 2011). This provides a powerful clue suggesting that the actual read out of an insulator transgene system can be functionally observed in the genome as a boundary between TADs. Together, these data suggest that information contained at the boundaries is essential for topological organization of TADs. However, it is not yet known if deletion of TAD boundaries also results in spread of heterochromatin marks into an adjacent topological domain, and future genetic experiments will be important to assess if TADs are causal for demarcating blocks of repressive H3K9me3 or H3K27me3 genome-wide.
Figure 2. Higher-order genome organization as a unifying mechanism for insulator function.
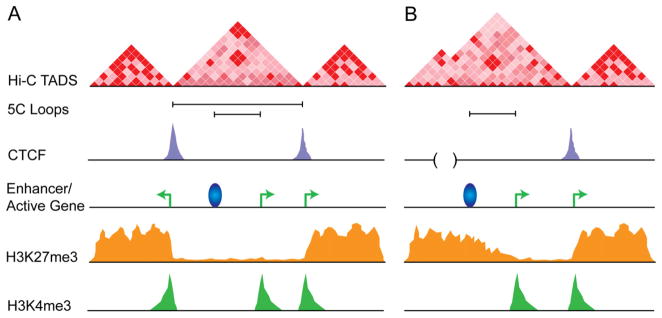
(A) Model for CTCF in mediating TAD boundaries and intra-TAD genomic organization via long-range interactions in wild type cells. (B) Blurring of TAD boundaries after deletion of a 50–80 kb boundary between TADs that contains CTCF and active genes (adapted from (Nora et al., 2012)). In principle, the TAD boundary would also be responsible for demarcating the border of the repressive H3K27me3 mark. Thus, we hypothesize that deletion of the boundary would lead to aberrant heterochromatin spreading. TADs are defined by the genome-wide 3-D mapping technology Hi-C. Counts are directly proportional to the frequency in which genomic fragments interact, with deep red pixels depicting high frequency interactions and light pink pixels depicting low frequency interactions. Point-to-point looping interactions at kb resolution can be mapped by Chromosome-conformation-capture-carbon-copy (5C) and are depicted by black bars connecting two genomic segments. CTCF ChIP-seq track in purple. Active enhancer depicted as a blue ball. Active genes depicted as green arrows. ChIP-seq track for H3K27me3 repressive chromain mark in orange.
Does CTCF demarcate topological units by simply mediating specific long-range interactions between the boundaries on each side of a TAD? Although CTCF is enriched at TAD boundaries, putative boundary elements represent only a small fraction of total CTCF sites. Of the 32,000 CTCF occupied sites genome-wide identified by Ren and colleagues, only 4,800 (15%) are at boundaries, suggesting that the large majority of CTCF sites do not serve a boundary function (Dixon et al., 2012). Thus, we speculate that CTCF sites that are not at boundaries could participate in the formation of additional long-range looping interactions within TADs, between TADs, and possibly between chromosomes. Indirect evidence supporting this idea is underscored by ChIA-PET analysis suggesting that a large proportion of CTCF-mediated loops are smaller than the Mb length scale of TADs (Handoko et al., 2011). Furthermore, an analysis of the epigenetic states of chromatin around and within loops connected by CTCF in mouse ES cells revealed that > 70% of CTCF-mediated looping interactions separate genomic regions enriched for divergent histone modifications. Together these data provide preliminary evidence suggesting that some proportion of CTCF-mediated loops serve to demarcate different chromatin domains. Correlating these looping interactions with TADs would shed further light into the link between TADs and long-range interactions.
Altogether, we favor a model in which classic barrier and EB insulators primarily function to mediate higher-order genome folding, and this 3-D organization can result in a wealth of functional outcomes depending on the genomic context (reviewed in (Phillips and Corces, 2009)). We hypothesize a model in which CTCF is necessary but not sufficient for the formation of TADs through its primary role in long-range looping interactions (depicted in Figure 2A and B). Within this model, an alternative explanation for the enrichment of CTCF at boundaries windowed at 40 kb or greater is that CTCF is actually placed at the edges of each domain, and these occupied sites causally shape each TAD through a hierarchy of long-range point-to-point looping interactions with the other edge of the TAD.
Insulator-Mediated Chromatin Organization and Gene Regulation
Although significant evidence points to a role for CTCF in genome organization, a much more controversial topic is related to whether and how insulator-mediated chromatin organization is linked to gene expression. This question remains unanswered in part due to the complexity of analyzing insulator function at sub-kb resolution in the context of the 3-D topology of an endogenous genomic locus. Emerging rules governing the epigenetic systems linking insulators, chromatin modifications, higher-order architecture, and gene expression are discussed below.
Enhancer blocking is rare in vivo
Insulators are traditionally thought to regulate gene expression through enhancer blocking (EB) mechanisms. The classic definition of an enhancer blocker is the ability to block inappropriate communication between enhancers and promoters in a position-dependent manner (West et al., 2002). Most of our knowledge of EB insulators comes from experiments that rely on transgene constructs (described in Figure 1B). Use of this experimental paradigm has resulted in the identification of hundreds of EB insulator sequences to date (reviewed comprehensively in (Herold et al., 2012; Ohlsson et al., 2010)). However, it has been rare to find sequences that confer EB function in a physiologically relevant system at an endogenous locus in vivo.
Two recent genome-wide analyses have provided new insights that now cause us to re-evaluate the role for EB mechanisms in mammalian systems (Sanyal et al., 2012; Shen et al., 2012). Shen et al. used a computational approach to evaluate EB models. Under the assumption that all CTCF sites serve EB mechanisms, the correlation between the intensity of H3K4me1 signal at enhancers and PolII signal at promoters was calculated only for matched enhancer-promoter pairs within each CTCF-demarcated domain. The resulting Spearman Correlation Coefficient was only slightly higher than that calculated for the random enhancer-promoter pairing control. Furthermore, >35% of enhancer-promoter pairs in CTCF-marked domains were anti-correlated, suggesting that an EB assumption results in a very large number of incorrectly paired regulatory sequences. Consistent with this finding, Sanyal et al. mapped long-range interactions between promoters and distal regulatory elements throughout 30 Mb ENCODE regions and found that > 75% of identified loops pass over one or more CTCF-occupied sites (Sanyal et al., 2012). Together, these studies suggest that enhancer blocking is not a pervasive mechanism for most CTCF sites in mammalian systems.
A critical aspect of assessing the validity of the notion that insulators function through EB mechanisms is to determine whether this mechanism exists in vivo at endogenous genomic loci in a developmentally relevant chromatin environment. The classical example supporting a role for EB insulators is the parent of origin specific expression of Igf2 and H19 genes in mice (reviewed in (Phillips and Corces, 2009)). However, more recent evidence suggests that long-range epigenetic mechanisms could also account for the observed imprinted expression of these genes (discussed below). Similarly, at the mouse β-globin locus, the 3′HS1 and HS5 regulatory elements are gold-standard sequences with demonstrated EB activity in transgene assays (Bulger et al., 2003; Farrell et al., 2002). However, more recent evidence now demonstrates that 3′HS1 and HS5 regulatory elements are connected in 3-D space by long-range looping interactions mediated by CTCF (Splinter et al., 2006). Are these data more consistent with a model in which mammalian CTCF does not function through EB mechanisms or a model in which EB insulation occurs as a consequence of looping interactions at endogenous loci? Interestingly, deletion of the 3′HS1 in vivo did not disrupt expression of β-globin or olfactory genes surrounding the β-globin locus during erythroid differentiation (Splinter et al., 2006). One caveat to this observation is that the undifferentiated erythroid progenitors used in these experiments do not normally express high levels of β-globin (Bulger and Groudine, 2010). Indeed, in a later study by Dean and colleagues, depletion of CTCF in K562 cells resulted in a reduction of β-globin gene expression (Hou et al., 2010). Nevertheless, either deletion of a CTCF binding site or global knock down of CTCF in independent studies did not lead to misregulation of the olfactory genes surrounding the β-globin locus. Together, these data support the idea that CTCF binding at 3′HS1 is necessary for long-range contacts between chromatin around developmentally regulated globin genes, but the function in this specific case is likely not EB insulation.
In contrast to mammalian systems, data consistent with a role for insulators in the proper regulation of gene expression through EB mechanisms in the endogenous context comes from now classical studies of the homeotic genes of the bithorax complex in Drosophila. Spatio-temporal expression of these three genes involves an intricate collection of enhancers, Polycomb response elements (PREs) and insulators (Barges et al., 2000). dCTCF binds to the Fab-8 insulator between the abd-A and Abd-B genes and mutations in CTCF result in abdominal hometic phenotypes due to missexpression of Abd-B (Gerasimova et al., 2007; Mohan et al., 2007). More recent evidence in support for a role of insulators in EB was obtained from analyses of insulator protein binding genome-wide after treatment of Drosophila Kc cells with the steroid hormone ecdysone (Wood et al., 2011). The ecdysone-inducible Eip75B gene encodes at least four different transcripts named Eip75B-RA through Eip75B-RD. A poised but inactive CTCF sites separates the Eip75B-RB promoter from regulatory sequences located within the locus. During the first 3 h after ecdysone induction, the Eip75B-RB RNA is transcribed. After 48 hr, the CTCF insulator becomes activated by recruitment of CP190, resulting in downregulation of the Eip75B-RB promoter. CP190 recruitment occurs in parallel with increased interaction frequency between this site and another CP190 site <100 kb downstream as measured by 3C. RNAi for CP190 reduces the frequency of this 3-D interaction down to wild type levels. We hypothesize that, in this case, the inducible insulator present in the Eip75B locus could possibly inhibit the interaction between enhancers and promoters by creating loops through interactions with adjacent insulators (Wood et al., 2011).
Much more experimental work is necessary to understand EB mechanisms in light of the newly identified TADs. Data so far suggest that canonical EB insulation is not a widespread regulatory mechanism in mammals. However, we cannot yet rule out the possibility that in certain specific cases insulator-mediated looping interactions can prevent inappropriate enhancer-promoter interactions. In the case of Drosophila, studies so far underscore the importance of insulators in the proper regulation of developmental genes. One possibility is that EB mechanisms are much more widespread in Drosophila due to the compact genome and greater need for regulating enhancer specificity. To date, the enhancers that are blocked by fly EB insulators have not yet been identified, so we cannot rule out the possibility that in flies, as in mammals, insulators are simply regulating the higher-order architecture that is critical for proper expression of a genomic locus. Another interesting possibility is that enhancers are not necessarily specific to a single gene and, instead, may activate all promoters within a genomic locus defined by a TAD. In this model, enhancers would have limited ability to sample the genomic space outside of the current TAD, suggesting that the CTCF sites causal for TAD boundary formation might also be the sites involved in EB insulation.
Insulators directly tether promoters to distal regulatory elements
A leading idea is that constitutive insulator sites across cell types may be involved in constitutive long-range interactions that connect the larger architectural framework, whereas the cell type-specific insulator sites have a more important role in regulating gene expression. Initial genome-wide studies observed ~40–70% overlap in CTCF occupied sites between cell types and used this information to support the claim that CTCF binding is largely invariant (Kim et al., 2007). However, because 30–60% can be cell type-specific, we hypothesize that variable CTCF sites play a functionally important role in genome regulation.
In a recent study by Stamatoyannopoulos and colleagues (Wang et al., 2012), CTCF occupancy was mapped by ChIP-seq in 19 different cell types (e.g. 7 immortal cell types and 12 normal cell lines) and showed much more widespread differential occupancy than previously suggested. This result was confirmed in parallel by a study in which CTCF binding was compared across 38 different cell types from the ENCODE project. CTCF overlap between cell types was generally >50%, but with an overlap as high as 80% observed between similar cell lines (Chen et al., 2012). For example, two lymphocyte cell lines (GM12875 vs. GM12873) showed ~80% overlap, while overlap was as low as 25% in unrelated lineages (GM12801 vs. HepG2). Together, these data indicate that cell type-specific CTCF sites are widespread and as important as constitutive CTCF sites.
A clue to the functional significance of cell type-specific CTCF sites comes from the mouse ENCODE project, where ~34,000 and ~41,000 occupied sites were reported in ES cells and MEFs, respectively (Shen et al., 2012). Interestingly, only 50–60% of CTCF sites were found to be common between these cells, and these constitutive sits were biased toward promoters (22% enhancers, 36% promoters, 42% other). By contrast, cell type-specific CTCF sites significantly overlapped with enhancers (50% enhancers, 24% promoters, 26% other). Thus, one possible reason for cell type-specific CTCF sites would be to organize proper chromatin configurations that enable enhancer-promoter interactions instead of blocking them.
Consistent with the idea that CTCF helps tether promoters to distal genomic elements, Handoko et al. reported a small subset of CTCF-mediated loops that function to bring p300-bound enhancers in close spatial proximity to target genes (Handoko et al., 2011). Similarly, Tijan and colleagues reported that TAF3, a component of the basal TFIID transcriptional machinery, binds directly to CTCF in Co-IP assays (Liu et al., 2011). Genome-wide analysis indicated that TAF3 is enriched at core promoters marked by TFIID subunits and H3K4me3, as well as at CTCF sites distal to promoters in ES cells. 3C analysis at Mapk3 and Psmd1 genes indicated that a distal CTCF/TAF3-bound element can form a long-range interaction with a TAF3-bound promoter. This looping interaction might be functional, as a combination of CTCF/TAF3 knock-down reduced expression of these genes, presumably through disruption of loop formation. Although it is not yet clear if distal sites are functional enhancers or some other regulatory element, these data provide evidence that CTCF sites can tether distal regulatory elements to promoters to regulate gene expression.
Finally, evidence also exists for CTCF-mediated loops that tether distal promoters to another promoter. This is perhaps best exemplified at the insulin (INS) locus in human pancreatic β-cells (Xu et al., 2011). The SYT8 gene is important for insulin secretion in response to glucose and it is located 300 kb from the INS gene. Felsenfeld and collaborators used 3C and 4C to show that INS and SYT8 genes physically interact. This interaction is mediated by CTCF and increases in response to glucose. Depletion of CTCF or inactivation of the INS promoter results in a decrease of SYT8 transcription. The results suggest that in addition to enhancer-promoter interactions, CTCF can also help in the coordination of gene expression by mediating long-distance interactions between the promoters of distally separated genes.
A role for CTCF in segregating enhancers during limb patterning
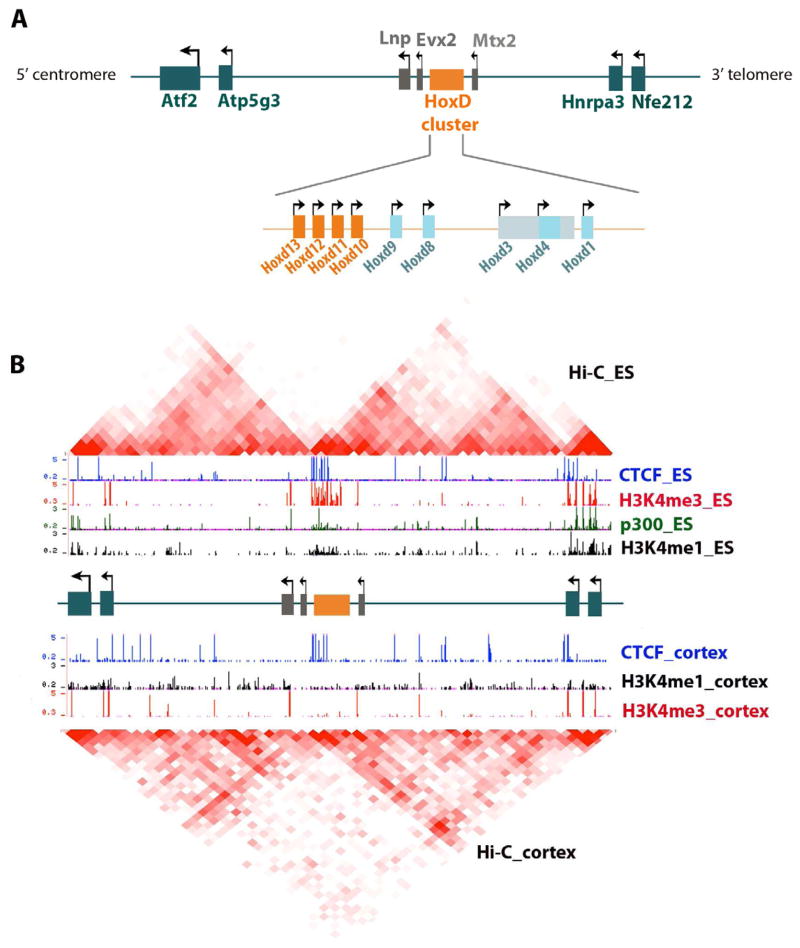
The mouse HoxD locus has 13 genes arranged in descending order (HoxD13 to HoxD1) from 5′ (centromeric) to 3′ (telomeric) on the linear DNA (described in Figure 3A). Precise spatiotemporal expression of different HoxD isoforms within this locus is critical for proper development of proximal and distal segments of the limb bud (Spitz et al., 2005; Tarchini and Duboule, 2006).
Figure 3. Role of CTCF in higher-order chromatin architecture at the mouse HoxD locus.
(A) 2-D organization of the HoxD locus. The HoxD gene cluster is a developmentally regulated locus that must be partitioned into discrete regulatory landscapes. 3′ Hox genes (HoxD9-HoxD1) are activated during early limb bud development via enhancers in a gene desert region on the 3′ side of the cluster toward the telomeres. 5′ Hox genes (HoxD13-HoxD10), as well as adjacent Lnp and Evx2 genes, are activated later in development during patterning of digits, and this wave of transcription is controlled by different enhancers in a gene desert region on the 5′ side of the cluster toward the centromeres. Centromeric enhancers have been well characterized: there is a distal GCR (global control region) 180 kb upstream of HoxD13 that contains multiple enhancers, as well as a proximal enhancer 50 kb upstream from HoxD13. (B) 3-D organization of the HoxD locus. Topological domains identified with HiC analyses by Dixon et al. in ES cells (top) and mouse cortex (bottom) with counts ranging from low (white) to high (deep red). Genome-browser tracks from Dixon et al. are also displayed for CTCF, mark for active genes H3K4me3, and enhancer marks H3K4me1 and p300 in ES cells (Dixon et al., 2012).
Duboule and colleagues hypothesized that CTCF binding sites operate through EB mechanisms to insulate the early HoxD1-9 genes (expressed in the proximal limb bud and regulated by a 3′ gene desert containing early limb telomeric enhancers) from Evx2 and the late HoxD12-10 genes (expressed in the distal limb bud and regulated by a 5′ gene desert containing late limb centromeric enhancers) (Montavon et al., 2011). Consistent with this idea, ChIP-chip analysis (Soshnikova et al., 2010) in distal embryonic limb buds at E10.75 revealed that 7 of the 9 HoxD genes were flanked by CTCF binding sites, whereas there were 4 CTCF sites in the centromeric gene desert and 7 CTCF sites in the telomeric gene desert (Figure 3A).
To analyze the causal role of CTCF at the specific time when an EB mechanism would be needed in development, CTCF alleles were conditionally deleted in distal mouse limb buds at E10.75 with Cre-recombinase under control of the Prx1 promoter. Almost complete knock down of CTCF mRNA and protein expression was achieved and ChIP-chip confirmed a 96% loss of the wild type CTCF occupied sites. At E10.75, mutant and wild type limb buds showed similar sizes and few differences in apoptosis. But, by E11.5, there was extensive apoptosis that ultimately resulted in severe shortening of fore- and hind-limbs. Thus, CTCF knock-down during development results in severe effects on phenotype at organ, tissue, and cellular levels.
Analysis of gene expression at E10.75 after CTCF knock-down revealed 220 downregulated and 177 up-reguated genes with at least 1.5-fold change, with an enrichment for genes involved in apoptotic pathways, oxidative stress pathways, and mitochondrial functions. Importantly, several genes containing CTCF in their TSS, such as Evx2 and HoxD13, were reduced by 4.5- and 2.4-fold, respectively, in CTCF mutants. By contrast, the more telomeric HoxD genes that were not active in the limb bud, such as HoxD8 and HoxD9, showed a 2.4- and 3-fold increase, respectively. Although gene expression was markedly deregulated upon CTCF knock-down, minimal changes in the spatial patterns of Hoxd gene expression were observed. Similar to wild type animals, Evx2 and HoxD13 expression remained localized to the distal limb bud and HoxD9 expression remained localized to the proximal limb bud. Before the discovery of the organization of the 3-D genome into TADs, these results were originally interpreted to suggest that CTCF did not function as a canonical enhancer-blocker, but may function though more general transcription mechanisms.
To consider this result in the context of 3-D genome organization, we compared the HoxD locus with TADs mapped by Hi-C in mouse ES cells and mouse cortex (Dixon et al., 2012). Intriguingly, the HoxD genes and multiple CTCF sites fall directly at the boundary between two TADs (Figure 3B). Gene deserts containing telomeric and centromeric enhancers coincide remarkably well with the interior of each adjacent TAD. This striking co-localization suggests that CTCF also has a role in higher-order organization at the HoxD locus, but further studies will need to explore the causality in this relationship. Because recent reports suggest that insulators often work in combination with active genes to create boundaries between TADs, it is very likely that simple loss of CTCF sites would not be sufficient to disrupt the topological organization leading to abberant enhancer-promoter contacts. We speculate that true de-regulation of the spatial patterns of HoxD gene expression would occur if the TAD domain organization was disrupted by dual knock-down of transcription and CTCF. We also note that CTCF knock-down still results in marked misregulation of several developmental genes responsible for limb patterning as well as many genes encoding many basic cellular processes, ultimately leading to massive apoptosis. Since deregulation is often seen at genes that have CTCF bound to their promoter, we speculate that CTCF knockdown might disrupt long-range interactions that directly connect enhancers to promoters within TADs.
A role for CTCF in alternative promoter selection to generate neural diversity
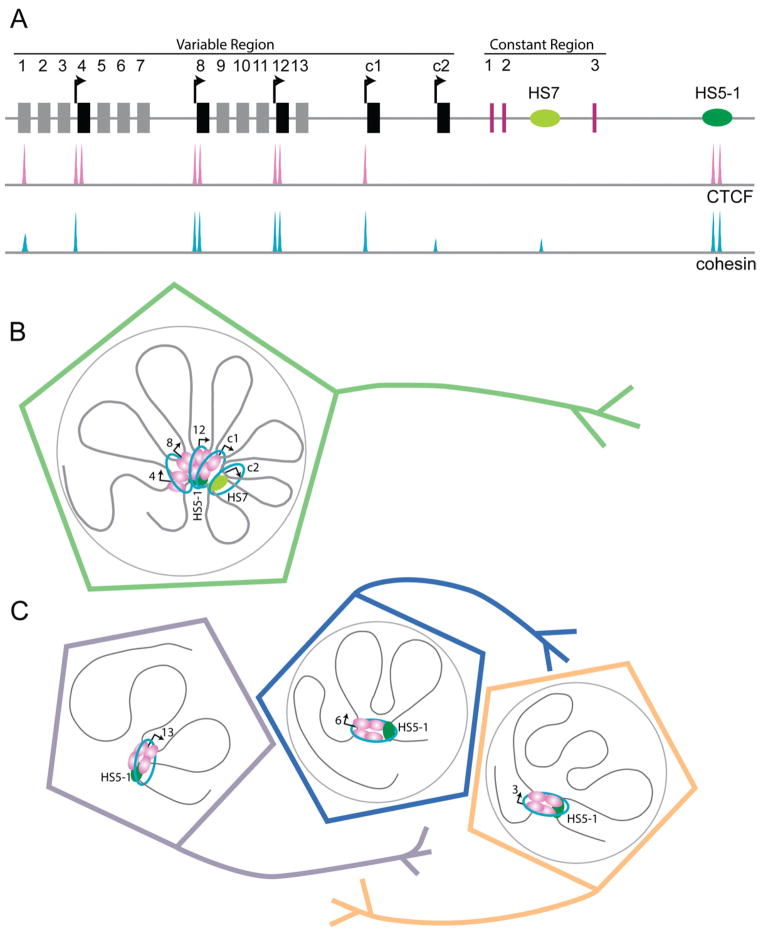
Enormous diversity in neuronal phenotypes is generated during development through combinatorial expression of ~50 protocadherin (Pcdh) isoforms from three primary gene clusters (Pcdhα, Pcdhβ, Pcdhγ). For example, diverse combinations of Pcdhα isoforms are created in humans through stochastic alternative promoter choice from the fifteen variable first exons, followed by alternative splicing of the chosen alternative exons to 3 downstream constant exons (described in Figure 4A).
Figure 4. Role for CTCF in higher-order chromatin architecture at the human Pcdh locus.
Pcdh genes encode a large number of calcium-dependent cell-cell adhesion molecules important in establishing neural diversity. (A) 2-D organization of the human Pcdh locus. Two enhancer elements downstream of the 13 alternative isoforms (1–13) and 2 c-type ubiquitous isoforms (c1, c2) have been identified as necessary for appropriate tissue-specific expression: HS7 in the intron between constant exons 2 and 3 (light green) and HS5-1 downstream of constant exon 3 (dark green) (Ribich et al., 2006). In the diploid human neuroblastoma cell line SK-N-SH, CTCF and cohesin occupied sites have been mapped with ChIP-seq (adapted from (Guo et al., 2012)). (B) Model for the role of enhancer-promoter looping interactions in variable exon expression in the diploid human neuroblastoma cell line SK-H-SH (adapted from (Guo et al., 2012)). (C) Model for neural diversity created by alternative Pcdh isoform expression through looping interactions between alternative promoters and the downstream HS5-1 enhancer.
Maniatis and colleagues hypothesized that CTCF plays a critical role in alternative promoter selection during the generation of mammalian neural diversity. This question has been notoriously difficult to study due to technical challenges mapping protein binding and 3-D chromatin interactions in individual neurons. The authors addressed this issue by using the human diploid neuroblastoma cell line SK-N-SH with stable expression of a select number of specific alternative and ubiquitous Pcdh isoforms (Figure 4A) (Guo et al., 2012). The power of this model system is that it represents an example of a simple expression pattern in a single neuron, thus enabling the study of the mechanisms of neural diversity for one specific clonal scenario.
Using ChIP-seq, the authors discovered that dual CTCF/cohesin sites are bound to the TSS and first exon of α4, α8, and α12 isoforms in SK-N-SH cells. CTCF/cohesin binding correlated with transcription, as these three isoforms were highly expressed compared to the other variable exons. Additionally, a single CTCF/cohesin site was mapped at the c1 isoform and a CTCF-independent cohesin site was mapped at c2 isoform. Both of these c-type ubiquitous isoforms were also expressed at low levels.
In addition to alternative promoters, it was also discovered that the HS7 enhancer contains a CTCF-independent cohesin binding site, whereas the HS5-1 enhancer contains dual CTCF/cohesin occupied sites. (Figure 4A) (Guo et al., 2012). A previous study by the authors in mice had demonstrated that deletion of the CTCF-bound HS5-1 enhancer deregulates expression of specific Pcdhα isoforms and markedly disrupts CTCF binding at the promoters of these genes even though they are separated by up to 250 kb from the enhancer (Kehayova et al., 2011). This result suggested that CTCF-mediated long-range mechanisms may play a role in alternative promoter selection. To test this idea, the authors leveraged 3C to demonstrate that the HS5-1 enhancer forms strong 3-D contacts with α8 and α12 and weaker interactions with α4, αc1, and αc2, whereas interactions with the inactive isoforms were undetected (Figure 4B) (Guo et al., 2012). Similarly, 3C analysis also detected 3-D interactions between the HS7 enhancer and α8, α12, αc1, and αc2, but not with αc4. Importantly, lentiviral shRNA knockdown of CTCF led to reduced expression of several alternative Pcdhα isoforms and disruption of the 3-D interactions at this locus.
Taken together, these data indicate that CTCF binding mirrors alternative promoter expression and that the binding of this protein is required for proper expression of these genes. Because this region was also shown by Ruan and colleagues to have numerous CTCF-mediated looping interactions (Handoko et al., 2011), this supports the idea that CTCF has an important role in creating the unique 3-D configurations favorable for alternative promoter choice and expression of Pcdhα during the generation of neural diversity (Figure 4C).
Insulators influence epigenetic states to regulate alternative splicing
One way that CTCF influences gene expression is through the organization of long-range interactions that influence the epigenetic state of specific genomic loci (reviewed in (Phillips and Corces, 2009)). In a more recent exciting example of the interplay between insulators and epigenetic modifications, CTCF and DNA methylation were recently functionally linked to alternative splicing during lymphocyte development (Shukla et al., 2011). The CD45 gene was used as a model system because exclusion of exons 4, 5, and 6 is correlated with differentiation of peripheral lymphocytes. The long form of CD45 containing exon 4 is expressed early in development and the short form of CD45 lacking all 3 exons is expressed in terminally differentiated lymphocytes. Prior to this study, proteins involved in exon 4 and exon 6 exclusion during terminal differentiation were known, but exon 5 exclusion appeared to be controlled through different unknown mechanisms. Interestingly, Shukla et al. found that CTCF binds specficially to exon 5 and might be casually linked to splicing. CTCF binding correlated with inclusion of exon 5 in the CD45 transcript, whereas disruption of CTCF binding resulted in exclusion/splicing of exon 5 and a shortened CD45 transcript.
What is the mechanism by which CTCF affects splicing? Importantly, exon 5 shows high levels of DNA methylation and is not bound by CTCF at late stages in development when the exon is excluded from the CD45 transcript. Depletion of the DNA maintenace methyltransferase DNMT1 led to reduced DNA methylation, re-acquisition of CTCF binding, and a subsequent increase in inclusion of exon 5 in the CD45 transcripts. Mechanistically, in vitro biochemical studies supported the idea that CTCF facilitates inclusion by promoting transient pausing of polymerase II, while also allowing subsequent pol II elongation after pausing. To our knowledge, this study provided the first evidence linking the epigenetic system of CTCF occupancy and DNA methylation to polymerase pausing and splicing. Independent studies indicate that CTCF may facilitate polymerase II pausing in other genomic contexts (Kang and Lieberman, 2011; Paredes et al., 2012; Wada et al., 2009) and it will be very interesting to see results from more detailed genome-wide analyses of splicing, insulators, epigenetic modifications, and higher-order chromatin organization.
Conclusions
A growing body of evidence now supports the idea that insulators are multi-faceted regulatory sequences that modulate a variety of nuclear processes by mediating long-range interactions between distant sites in the genome. We favor a unifying mechanism for insulators in the formation of inter- and intra-chromosomal interactions, with the underlying consensus and the local chromatin environment at a particular position in the genome providing specificity for protein conformations, binding partners, and post-translational modifications that yield context-dependent effects on gene expression.
Given the global role for insulators in orchestrating genome organization, it is surprising that insulator knock-down appears to have only modest gobal effects on gene expression. It is likely that the small effects on gene expression observed so far are a consequence of only partial knockdown of the proteins. It is equally as likely that insulators are modular elements that work in conjunction with other mechanisms to perform diverse functions. Thus, insulator protein knock-down might have only a partial effect accounted for by redundant mechanisms. Finally, because CTCF’s primary role is in genome organization, we envision that ony a fraction of this organization is important for gene expression and another fraction has primarily topological/architectural roles that may not specifically required for cellular function.
A critical issue in dissecting the role of insulators in nuclear biology is to understand whether genome function is an effector that determines its three-dimensional organization or whether insulator proteins play a structural role to instruct patterns of organization that then allow specific functional outcomes. As is often the case, the answer is likely to be a combination of both options. The role for insulators in defining TADs is often redundant with high gene density and transcription and, therefore, function appears to be an important contributor to the establishment and/or maintenance of topological chromosome domains. At a local level, regulation of enhancer-promoter interactions may first require the establishment of contacts between regulatory sequences by insulators, cohesin and/or mediator, suggesting that function in this case is a consequence of structure. Future studies leveraging deep sequencing in combination with genetic and biochemical perturbation studies should yield valuable insights into the causes and consequences of genome organization.
Acknowledgments
Work in the authors’ laboratory is supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM035463. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.E.P.C. was supported by NIH National Research Service Award 5F32NS065603.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barges S, Mihaly J, Galloni M, Hagstrom K, Muller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes & development. 2003;17:664–675. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol. 2010;339:250–257. doi: 10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Schubeler D, Bender MA, Hamilton J, Farrell CM, Hardison RC, Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol Cell Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes & development. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Tian Y, Shu W, Bo X, Wang S. Comprehensive identification and annotation of cell type-specific and ubiquitous CTCF-binding sites in the human genome. PLoS ONE. 2012;7:e41374. doi: 10.1371/journal.pone.0041374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dickson J, Gowher H, Strogantsev R, Gaszner M, Hair A, Felsenfeld G, West AG. VEZF1 elements mediate protection from DNA methylation. PLoS Genet. 2010;6:e1000804. doi: 10.1371/journal.pgen.1000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D. Cohesin, gene expression and development: lessons from Drosophila. Chromosome Res. 2009;17:185–200. doi: 10.1007/s10577-009-9022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell CM, West AG, Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol Cell Biol. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoriw AM, Stein P, Svoboda P, Schultz RM, Bartolomei MS. Transgenic RNAi reveals essential function for CTCF in H19 gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- Ferraiuolo MA, Rousseau M, Miyamoto C, Shenker S, Wang XQ, Nadler M, Blanchette M, Dostie J. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res. 2010;38:7472–7484. doi: 10.1093/nar/gkq644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN, Cheng MK, Moore JM, Truong JP, Hu YJ, Nguyen DK, Tsuchiya KD, Disteche CM. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Gan M, Moebus S, Eggert H, Saumweber H. The Chriz-Z4 complex recruits JIL-1 to polytene chromosomes, a requirement for interband-specific phosphorylation of H3S10. J Biosci. 2011;36:425–438. doi: 10.1007/s12038-011-9089-y. [DOI] [PubMed] [Google Scholar]
- Gause M, Morcillo P, Dorsett D. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol. 2001;21:4807–4817. doi: 10.1128/MCB.21.14.4807-4817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gause M, Schaaf CA, Dorsett D. Cohesin and CTCF: cooperating to control chromosome conformation? Bioessays. 2008;30:715–718. doi: 10.1002/bies.20787. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. The EMBO journal. 2001;20:2518–2527. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM, Takebayashi SI, Ryba T, Lu J, Pope BD, Wilson KA, Hiratani I. Space and time in the nucleus: developmental control of replication timing and chromosome architecture. Cold Spring Harb Symp Quant Biol. 2010;75:143–153. doi: 10.1101/sqb.2010.75.011. [DOI] [PubMed] [Google Scholar]
- Guo Y, Monahan K, Wu H, Gertz J, Varley KE, Li W, Myers RM, Maniatis T, Wu Q. CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci U S A. 2012;109:21081–21086. doi: 10.1073/pnas.1219280110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurudatta BV, Corces VG. Chromatin insulators: lessons from the fly. Brief Funct Genomic Proteomic. 2009;8:276–282. doi: 10.1093/bfgp/elp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat Genet. 2011;43:630–638. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath H, de Almeida CR, Sleutels F, Dingjan G, van de Nobelen S, Jonkers I, Ling KW, Gribnau J, Renkawitz R, Grosveld F, et al. CTCF regulates cell cycle progression of alphabeta T cells in the thymus. EMBO J. 2008;27:2839–2850. doi: 10.1038/emboj.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P, Marin B, Bartkuhn M, Schierenberg E, Wiehe T. The chromatin insulator CTCF and the emergence of metazoan diversity. Proc Natl Acad Sci U S A. 2012;109:17507–17512. doi: 10.1073/pnas.1111941109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold M, Bartkuhn M, Renkawitz R. CTCF: insights into insulator function during development. Development. 2012;139:1045–1057. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- Hou C, Dale R, Dean A. Cell type specificity of chromatin organization mediated by CTCF and cohesin. Proc Natl Acad Sci U S A. 2010;107:3651–3656. doi: 10.1073/pnas.0912087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C, Li L, Qin ZS, Corces VG. Gene density, transcription, and insulators contribute to the partition of the Drosophila genome into physical domains. Mol Cell. 2012;48:471–484. doi: 10.1016/j.molcel.2012.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138:435–448. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Lieberman PM. Mechanism of glycyrrhizic acid inhibition of Kaposi’s sarcoma-associated herpesvirus: disruption of CTCF-cohesin-mediated RNA polymerase II pausing and sister chromatid cohesion. J Virol. 2011;85:11159–11169. doi: 10.1128/JVI.00720-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehayova P, Monahan K, Chen W, Maniatis T. Regulatory elements required for the activation and repression of the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. 2011;108:17195–17200. doi: 10.1073/pnas.1114357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Cecchini KR, Kim TH. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc Natl Acad Sci U S A. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Hu JF, Qiu X, Ling J, Chen H, Wang S, Hou A, Vu TH, Hoffman AR. CTCF regulates allelic expression of Igf2 by orchestrating a promoter-polycomb repressive complex 2 intrachromosomal loop. Mol Cell Biol. 2008;28:6473–6482. doi: 10.1128/MCB.00204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- Liu Z, Scannell DR, Eisen MB, Tjian R. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell. 2011;146:720–731. doi: 10.1016/j.cell.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Atallah M. Genomic relationship between SINE retrotransposons, Pol III-Pol II transcription, and chromatin organization: the journey from junk to jewel. Biochem Cell Biol. 2011;89:495–504. doi: 10.1139/o11-046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Gomez JA, Chadwick BP, Boss JM. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J Exp Med. 2008;205:785–798. doi: 10.1084/jem.20071843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishiro T, Ishihara K, Hino S, Tsutsumi S, Aburatani H, Shirahige K, Kinoshita Y, Nakao M. Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 2009;28:1234–1245. doi: 10.1038/emboj.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. The EMBO journal. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147:1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, Lobanenkov V, Klenova E. Does CTCF mediate between nuclear organization and gene expression? Bioessays. 2010;32:37–50. doi: 10.1002/bies.200900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Pant V, Kurukuti S, Pugacheva E, Shamsuddin S, Mariano P, Renkawitz R, Klenova E, Lobanenkov V, Ohlsson R. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol Cell Biol. 2004;24:3497–3504. doi: 10.1128/MCB.24.8.3497-3504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes SH, Melgar MF, Sethupathy P. Promoter proximal CTCF binding is associated with an increase in the transcriptional pausing index. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaart MJ, Recillas-Targa F, Felsenfeld G. Loss of transcriptional activity of a transgene is accompanied by DNA methylation and histone deacetylation and is prevented by insulators. Genes Dev. 1998;12:2852–2862. doi: 10.1101/gad.12.18.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Curr Opin Genet Dev. 2012;22:101–109. doi: 10.1016/j.gde.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab JR, Chiu J, Zhu J, Katzman S, Kurukuti S, Wade PA, Haussler D, Kamakaka RT. Human tRNA genes function as chromatin insulators. EMBO J. 2012;31:330–350. doi: 10.1038/emboj.2011.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci U S A. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Wang Y, Shi M, Wang X, Yang Z, Zhao Z. CTCF mediates the cell-type specific spatial organization of the Kcnq5 locus and the local gene regulation. PLoS ONE. 2012;7:e31416. doi: 10.1371/journal.pone.0031416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribich S, Tasic B, Maniatis T. Identification of long-range regulatory elements in the protocadherin-alpha gene cluster. Proc Natl Acad Sci U S A. 2006;103:19719–19724. doi: 10.1073/pnas.0609445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Linder-Basso D, Kharchenko PV, Tolstorukov MY, Kim M, Li HB, Gorchakov AA, Minoda A, Shanower G, Alekseyenko AA, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 2012;22:2188–2198. doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnikova N, Montavon T, Leleu M, Galjart N, Duboule D. Functional analysis of CTCF during mammalian limb development. Dev Cell. 2010;19:819–830. doi: 10.1016/j.devcel.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Spencer RJ, del Rosario BC, Pinter SF, Lessing D, Sadreyev RI, Lee JT. A boundary element between Tsix and Xist binds the chromatin insulator Ctcf and contributes to initiation of X-chromosome inactivation. Genetics. 2011;189:441–454. doi: 10.1534/genetics.111.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz F, Herkenne C, Morris MA, Duboule D. Inversion-induced disruption of the Hoxd cluster leads to the partition of regulatory landscapes. Nat Genet. 2005;37:889–893. doi: 10.1038/ng1597. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes & development. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarchini B, Duboule D. Control of Hoxd genes’ collinearity during early limb development. Dev Cell. 2006;10:93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Van Bortle K, Corces V. tDNA insulators and the emerging role of TFIIIC in genome organization. Transcription. 2012;3:277–284. doi: 10.4161/trns.21579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bortle K, Ramos E, Takenaka N, Yang J, Wahi JE, Corces VG. Drosophila CTCF tandemly aligns with other insulator proteins at the borders of H3K27me3 domains. Genome research. 2012;22:2176–2187. doi: 10.1101/gr.136788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, Komura D, Kitakami J, Oshida N, Papantonis A, et al. A wave of nascent transcription on activated human genes. Proc Natl Acad Sci U S A. 2009;106:18357–18361. doi: 10.1073/pnas.0902573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan LB, Pan H, Hannenhalli S, Cheng Y, Ma J, Fedoriw A, Lobanenkov V, Latham KE, Schultz RM, Bartolomei MS. Maternal depletion of CTCF reveals multiple functions during oocyte and preimplantation embryo development. Development. 2008;135:2729–2738. doi: 10.1242/dev.024539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Maurano MT, Qu H, Varley KE, Gertz J, Pauli F, Lee K, Canfield T, Weaver M, Sandstrom R, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt KS, Yoshida K, Itoh T, Bando M, Koch B, Schirghuber E, Tsutsumi S, Nagae G, Ishihara K, Mishiro T, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Wood AM, Van Bortle K, Ramos E, Takenaka N, Rohrbaugh M, Jones BC, Jones KC, Corces VG. Regulation of chromatin organization and inducible gene expression by a Drosophila insulator. Mol Cell. 2011;44:29–38. doi: 10.1016/j.molcel.2011.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Lefevre GM, Felsenfeld G. Chromatin structure, epigenetic mechanisms and long-range interactions in the human insulin locus. Diabetes Obes Metab. 2012;14(Suppl 3):1–11. doi: 10.1111/j.1463-1326.2012.01645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei G, Chepelev I, Zhao K, Felsenfeld G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat Struct Mol Biol. 2011;18:372–378. doi: 10.1038/nsmb.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ramos E, Corces VG. The BEAF-32 insulator coordinates genome organization and function during the evolution of Drosophila species. Genome research. 2012;22:2199–2207. doi: 10.1101/gr.142125.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon YS, Jeong S, Rong Q, Park KY, Chung JH, Pfeifer K. Analysis of the H19ICR insulator. Mol Cell Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]