Abstract
Bioinformatic and direct cloning approaches have led to the identification of over 100 novel miRNAs expressed in DNA viruses, although the function of the majority of these small regulatory RNA molecules is unclear. Recently, a number of reports have now identified potential targets of viral miRNAs, including cellular and viral genes as well as an ortholog of an important immune-regulatory cellular miRNA. In this review, we will cover the identification and characterization of miRNAs expressed in the herpesvirus family and discuss the potential significance of their role in viral infection.
Keywords: Herpesvirus, microRNA, miRNA, HCMV, HSV, KSHV, EBV, Virus
Introduction
Herpesviruses belong to a large family of enveloped, double-stranded DNA viruses disseminated throughout nature, which maintain persistent or latent infections during the lifetime of the host [1]. Membership to the herpesviridae family is determined by virion morphology, and gene expression involves a complex pattern of transcriptional regulation during acute infection and during the establishment, maintenance and reactivation from latency [1]. Herpesviruses are divided into three groups (alpha, beta and gamma) and members of all three groups encode miRNAs, indicating that herpesviruses have utilized RNAi throughout their evolution. Bioinformatic and direct cloning approaches have led to the identification of over 100 novel miRNAs expressed in DNA viruses [2], although the function of the majority of these small regulatory RNA molecules is unclear. Recently, a number of reports have now identified the potential targets of viral miRNAs, including cellular and viral genes as well as an ortholog of an important immune-regulatory cellular miRNA [3–8]. In this review, we will cover the identification and characterization of miRNAs expressed in the herpesvirus family and discuss the potential significance of their role in viral infection.
Alpha-herpesviruses
Herpes simplex virus
Herpes simplex virus is a member of the alpha herpesvirus subfamily and is neurotropic in nature, initially infecting the mucosal epithelia before establishing a latent infection in the sensory neurons of the trigeminal ganglia. Reactivation of HSV-1 is associated with the occurrence of the common cold sore around the mouth, but can cause more severe infections in immunecompromised patients. Unlike HCMV, a number of animal models of infection are available, as HSV-1 infects a range of different species. A number of well-established in vitro models of latency also exist. Furthermore, HSV replicates rapidly in vitro, unlike EBV and KSHV, allowing for the analysis of the lytic stage of viral replication. These features make HSV an attractive model for the study of viral miRNAs. Of particular interest is the expression of a latency-specific, large non-coding RNA known as the latency-associated transcript (LAT). Expressed exclusively within infected neurons, LATs are the only transcript thought to be expressed by HSV-1 during latency [9]. Although no function has been definitively shown for LAT, it has been suggested to be involved in the restriction of viral trans-activating proteins and in blocking cellular apoptosis in response to infection, both by unknown mechanisms [10, 11] (Fig. 1).
Fig. 1.
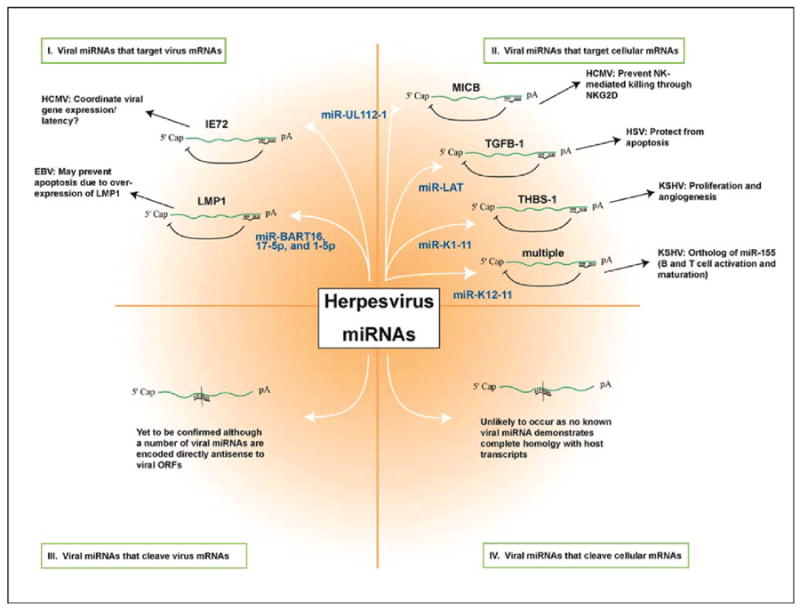
Proposed functions of herpesvirus miRNAs. Recent reports have identified a number of potential targets for viral miRNAs. In HSV-1,miR-LAT was reported to block apoptosis by targeting the cellular transcripts encoding TGFβ and SMAD3 [4]. HCMV miR-UL112-1 has been reported to target both the viral transactivator IE72 and the cellular immune ligand MICB [3,7]. In KSHV multiple miRNAs were found to target THBS-1. miR-K12-11 of KSHV was also identified as an ortholog of the important immunoregulatory miRNA miR-155 [6,8]. In EBV, miRNAs from the BART region were found to efficiently target expression of the oncogenic latency gene LMP1 [5]
To date, two reports have described the identification of miRNAs expressed by HSV-1 close to or within the LAT [4, 12]. The initial study using a bioinformatics approach predicted 13 pre-miRNAs located throughout the HSV-1genome [12]. Although northern blot analysis identified three bands corresponding to the predicted pre-miRNAs, only one mature miRNA, located upstream of the minor LAT transcript, was detected. Using a more directed approach concentrating on the LAT region of the virus, another group characterized an additional miRNA (miR-LAT) encoded within exon 1 of minor LAT. The authors demonstrated that the expression of the LAT transcript alone could block apoptosis and that this activity was dependent on the cellular RNAi pathway. Additional studies by this group suggested that miR-LAT blocks apoptosis through targeting translation of the cellular genes, TGF-β1 and SMAD3, through conserved sequences within the 3′UTRs [4].
Marek’s disease virus
Two cloning studies used a more comprehensive approach to identify miRNAs expressed by chicken alpha herpesvirus Marek’s disease virus (MDV) 1 and 2 [13, 14]. In the first study, eight miRNAs were identified in MDV-1 clustered within the LAT transcript and either side of the viral oncogene, meq [13]. In MDV-2, 17 miRNAs were identified. Although there was no sequence homology between the miRNAs of MDV-1 and 2, the miRNAs were located in similar genomic regions of the viruses [14]. The miRNAs were detected in the infected fibroblast cells as well as in the viral-induced tumor tissue from infected chickens. As yet, there is no information on the potential function of these miRNAs, although identification of multiple miRNAs encoded by MDV would suggest that additional miRNAs are yet to be discovered in HSV.
Beta-herpesviruses
Human cytomegalovirus
A number of studies, including our own, have identified viral miRNAs encoded by the beta-herpesvirus, human cytomegalovirus (HCMV) [15, 16]. HCMV infects a large percentage of the population and, although generally benign, this virus is considered a significant pathogen in immunosuppressed patients, either undergoing immunosuppressive therapy following solid organ or bone marrow transplantation, or with AIDS [17–24]. In addition, HCMV is one of the leading viral causes of birth defects [25]. Primary HCMV infection is followed by a lifelong persistence of the virus in a latent state, although a low level of persistent infection has not been ruled out as a relevant mechanism of viral maintenance.
A combination of both direct cloning and bioinformatics approaches were used to identify miRNAs encoded by fibroblasts lytically infected with HCMV. A total of 11 HCMV-encoded miRNAs have been identified to date and include miR-UL22A-1, miR-UL36-1, miR-UL70-1, miR-UL112-1, miR-UL148D-1, miR-US4-1, miR-US5-1, miR-US5-2, miR-US25-1, miR-US25-2, and miR-US33-1 (15,16, 26). Unlike the miRNAs encoded by the alpha- and gamma-herpesviruses, which are found clustered within the regions of the genome associated with the latent gene expression, the HCMV-encoded miRNAs are located throughout the viral genome [16, 26]. Many are located in the intergenic regions, while miR-UL36-1 is found within the intronic region of UL36. Four of the identified miRNAs(UL70-1, UL112-1, UL148D, and US33-1) are within the predicted coding regions of HCMV and, as all four are directly antisense to the ORFs of the annotated genes, they are predicted to cleave the expressed transcripts. However, studies performed in our lab have shown that the UL114 transcript, which encodes the viral uracil DNA glycosylase is resistant to miR-UL112-1-mediated cleavage, even though both RNAs are present during acute infection(unpublished observation).
However, two recent studies have identified the potential target transcripts of miR-UL112-1. Interestingly it appears that miR-UL112-1 is capable of targeting both cellular and viral transcripts [7, 27].
In a study by our lab, a comparative bioinformatics approach was used to identify the potential transcripts regulated by the HCMV-encoded miRNAs [27]. Of particular interest is a cluster of targets identified for miR-UL112-1 within the major immediate early (MIE) region of HCMV. These target sites were within the 3′UTR sequences of UL120, UL121 and UL123 (IE72). The MIE genes are known to express a number of regulatory proteins that coordinate viral gene expression. Disruption of one of the miR-UL112-1 targets results in a significant attenuation of viral replication following low multiplicity infections [28]. All three target sites effectively down-regulated luciferase expression when cloned downstream of the reporter gene in the presence of miR-UL112-1. In addition, expression of miR-UL112-1 with a vector containing the major immediate early region significantly reduced the levels of IE72 expression. Since IE72 is an important transactivator that is required for efficient viral replication, we hypothesized that aberrant expression of miR-UL112-1 may result in viral attenuation. HCMV DNA replication in cells transfected with synthetic miR-UL112-1 RNA duplex was inhibited up to fivefold, indicating that expression of miR-UL112-1 has the potential to attenuate the acute replication of HCMV. This may have important implications for latency control of HCMV.
Recently, authors Stern-Ginossar et al. [7] demonstrated that miR-UL112-1 targets the major histocompatibility complex class-I related chain B (MICB), a cellular gene involved in host immune responses to viral infection. Using target prediction algorithms based on the identification of repeated target sites within cellular 3′UTR sequences(RepTar; cRepTar), a potential target site for miR-UL112-1 was identified within the 3′UTR of MICB. MICB is a stress-induced ligand of NKG2D, a receptor present on NK cells. Activation of NK cells through the association of MICB with NKG2D can lead to cell killing. Preventing this interaction and the subsequent killing of infected cells would clearly benefit the virus. Deletion of miR-UL112-1 led to an increase in MICB expression and susceptibility to NK cell-mediated killing. Interestingly, HCMV also encodes the UL16 protein, which was previously shown to inhibit MICB signaling by intracellular sequestration, suggesting a level of redundancy in the targeting of MICB[15].
Murine cytomegalovirus
Murine cytomegalovirus (MCMV) belongs to the beta-herpesvirus family and has long served as an important small animal model for HCMV infection and pathogenesis. Two recent studies have identified multiple viral miRNAs encoded by MCMV [29, 30]. Using a combination of bioinformatic and small RNA cloning approaches, authors Buck et al. and Dolken et al. identified and characterized MCMV miRNAs expressed in the lytically infected mouse fibroblasts. A total of 18 MCMV miRNAs were described, originating from at least 12 pre-miRNAs. Similar to HCMV, the MCMV-encoded miRNAs are expressed early during the lytic phase of replication and are found located throughout the viral genome individually or in clusters. Nine of the miRNAs are located in predicted 3′UTRs, seven in the coding sequences of predicted genes (m01, M55, M87, M88, m107 and m108) and two are located 5′ of the predicted ORFs (m01, −14nt and M95, > −1, 100nt). Two particular clusters of miRNAs in proximity to m21, m22, M23, m107 and m108 include members that are derived from complementary strands. None of the identified MCMV miRNAs show significant homology to any of the HCMV-encoded miRNAs, with only the MCMV-encoded miRNAs in the M23 region localized to a similar region as HCMV-miRNAs.
Northern blot analysis in combination with cycloheximide or foscarnet (PAA) treatment was used to determine the expression kinetics during MCMV infection. While mature MCMV miRNAs were detected as early as 4–8hpi, consistent with early kinetics, mature MCMV miRNAs were not detected in cells treated with cyclohexamide, indicating that synthesis of viral proteins may be required for their production. Interestingly, in a number of cases, expression of MCMV pre-miRNA species were not blocked by cycloheximide, despite efficient inhibition of the mature miRNAs that they encode. This pattern of expression is similar to the HCMV miRNA miR-UL36-1: both the HCMV UL36 transcript and UL36 pre-miRNA are expressed with immediate early kinetics, but the mature miR-UL36-1 is not expressed on cycloheximide treatment [16]. However, prolonged cycloheximide treatment may deplete cells of factors required to process pre-miRNAs into mature forms. Whether production of mature miRNAs requires synthesis of viral proteins, or prolonged cycloheximide treatment depletes factors necessary to process mature miRNAs, remains unknown. mir-m108-1-3p, mir-m108-1-5p.2 and mir-m108-1 were sensitive to treatment with PAA and, therefore, are considered late. There are no reported targets of MCMV-encoded miRNAs. However, the use of MCMV as a model infection of β-herpesviruses will enable crucial in vivo studies that are not possible with HCMV.
Gamma-herpesviruses
Epstein–Barr virus
Viral miRNAs were first shown to exist following the cloning of small RNAs from a B cell line latently infected with Epstein–Barr virus (EBV) [31]. EBV was initially identified as the etiological agent responsible for the lymphoproliferative disease, Burkitt’s lymphoma, and is the prototypic member of the gamma herpesvirus subfamily. In addition to Burkitt’s lymphoma, EBV is associated with a number of lymphoproliferative diseases in humans including Castle-man’s disease, Hodgkin’s disease and primary effusion lymphoma (PEL). Following the initial infection of the mucosal epithelia, EBV establishes a latent infection in B lymphocytes and has a powerful transforming ability in these cells. EBV was found to encode five miRNAs clustered within two genomic regions. miR-BHRF1-1, 2 and 3 are located within the untranslated region (UTR) of BHRF1, an anti-apoptosis Bcl-2 homologue. miR-BHRF1-1 is located within the 5′UTR with miR-BHRF1–2 and 3 encoded within the 3′UTR. miR-BART-1 and 2 are located within intronic regions of the BART family of transcripts, which are extensively spliced. The BART family of transcripts has been suggested to encode a number of proteins, although it remains to be convincingly demonstrated that these proteins are expressed during viral infection [32]. Subsequent investigation identified a further 22 EBV miRNAs not identified in the original study, due to a large deletion in the B95-8 strain used for the initial cloning strategy [33, 34].
Three miRNAs encoded within the BART region, miR-BART16, miR-BART17-5p and miR-BART1-5p, have since been shown to target sequences within the 3′UTR of the viral latent membrane protein 1 (LMP1) following the transient transfection of an LMP1 expression plasmid and various BART-derived miRNAs [5]. LMP1 is an oncogenic protein expressed in the majority of EBV-associated malignancies and is thought to induce transformation through the activation of a number of cellular signaling pathways, including NFκB, JNK, JAK/STAT, p38/MAP and Ras/MAPK [35]. The authors demonstrated a correlative link between levels of BART miRNAs and LMP1, but have not, as yet, demonstrated a direct effect on LMP1 levels by BART miRNAs in infected cells. It is currently unclear as to the functional relevance of LMP1 targeting by BART-derived miRNAs, although overexpression of LMP1 can lead to the triggering of apoptosis. The authors suggest that the regulation of LMP1 by BART miRNAs may dampen LMP1 expression levels to avoid triggering cell death. Indeed, expression of BART cluster 1 miRNAs reduced the sensitivity of LMP1-expressing cells to cisplatin-induced apoptosis. As the majority of BART miRNAs have been deleted in the B95-8 strain, it would be of interest to deter-mine whether cells infected with B95-8 produce higher levels of LMP1 than strains with intact BART regions of the genome.
Kaposi’s sarcoma-associated herpesvirus
Cloning studies by multiple groups identified viral miRNAs encoded by a second human gamma herpesvirus, Karposi’s sarcoma-associated herpesvirus (KSHV) [26, 36, 37]. Similar to EBV, KSHV infection is associated with a number of tumors, most notably Kaposi’s sarcoma, a skin malignancy common amongst AIDS patients and older men of Mediterranean descent [38]. In common with all herpesviruses, KSHV has a complex viral replication cycle involving initial acute replication followed by life-long latent infection with the ability to reactivate [38]. In three independent studies, a total of 11 distinct miRNAs were cloned from 2B-cell lines derived from the latently infected lymphomas. Of the 11 miRNAs, 10 are encoded within an approximately 4 kb region, previously thought to be non-coding, between open reading frames (ORF) K12 (kaposin) and ORF71. A further miRNA resides within the coding region of the Kaposin gene. Like EBV, KSHV latent infection is characterized by limited gene expression. Many of the genes expressed during latent infection flank the genomic region encoding the viral miRNAs. Detailed mapping of the latent transcripts in two independent studies have subsequently identified three overlapping transcripts driven by two separate promoters that have the potential to encode 10 of the 11 miRNAs within a single intron [39, 40]. All ten miRNAs were detected following the transient transfection of the genomic region encoding the transcripts, strongly suggesting that all ten miRNAs are in fact encoded for within the identified intron [40]. The three spliced transcripts are also predicted to encode the kaposin ORF that contains the additional miRNA. As the transcript would be cleaved to release the pre-miRNA stem loop, processing of this miRNA by Drosha would theoretically preclude the expression of Kaposin protein, leading to a novel form of gene regulation. However, as both miRNA and Kaposin protein are detected during latent infection, sufficient Kaposin transcript apparently escapes Drosha processing to allow translation of the Kaposin mRNA. Regardless of the precise mechanism, the close association of the viral miRNAs with the expression of Kaposin does suggest the possibility of a functional association. The Kaposin gene encodes for a number of different proteins, including Kaposin B, which was shown to bind and activate MAPK-associated protein kinase 2 (MK2) [41]. Activation of MK2 by Kaposin B leads to an increase in cytokine expression in KSHV-infected cells by blocking the destabilizing effect of AU-rich elements (ARE’s) within the 3′UTRs of cytokine mRNAs. Whether the miRNAs possess any functional relevance to this mechanism, or to one of the Kaposin gene products, awaits further investigation. Ten miRNAs have also been identified in the related Rhesus monkey rhadinovirus (RRV) using a cloning approach [42]. Although the miRNAs expressed by RRV show no sequence homology to those encoded by KSHV, they are encoded within the same genomic region as the miRNAs of KSHV.
Two recent reports from Rolf Renne’s group have suggested cellular targets for miRNAs expressed by KSHV [6,8]. In the first study, Samols et al. [8] constructed stable cell lines expressing ten KSHV miRNAs derived from the single intronic transcript described above. To identify the potential cellular targets of the KSHV miRNAs, global transcript levels were determined by microarray analysis and compared to negative control cell lines. Previous studies have shown that transcript degradation can occur from miRNA targeting, even when the target sequence is not fully complementary. Therefore, microarray analysis has proven to be an effective tool for identifying potential target transcripts of miRNAs. In this study, the authors found 65 genes downregulated in the miRNA-expressing cells, including genes involved in proliferation, immune modulation, angiogenesis and apoptosis. More detailed analysis demonstrated that one of the identified genes, thrombospondin-1 (THBS-1), was targeted by all ten of the KSHV miRNAs examined through multiple binding sites within the 3′UTR. Surprisingly, despite the high level of redundancy in targeting, inhibition of any one of the KSHV miRNAs led to derepression of the target transcript. THBS-1 has anti-proliferative and anti-angiogenic properties and would therefore represent an attractive target for a prooncogenic virus such as KSHV.
In a second study, Skalsky et al. [6], discovered that the seed sequence of miRK12-11 of KSHV exactly matched that of the cellular miRNA miR-155. This was of particular interest as overexpression of miR-155 has been associated with a number of B cell lymphomas and solid organ tumors, as well as the development of B cell lymphomas intransgenic miR-155 overexpressing mice [43, 44]. It is currently unclear as to the role of miR-155 in the development of tumors, but expression of miR-155 has been shown to drive B cell germinal center development and is required for normal B and T cell activation and maturation. Skalsky et al. suggest that miR-K12-11 may mimic the regulatory functions of miR-155 to the advantage of the virus, possibly by inducing survival and proliferation of the infected cells. Although microarray data demonstrate similar regulation of transcripts by miR-155 and miR-K12-11, functional data showing rescue of miR-155 deletion phenotypes by miR-K12-11 would be particularly compelling. The use of cellular miRNA orthologs by viruses is an attractive concept, as the virus would be able to highjack the target sequences already evolved by the host, thereby avoiding potential problems caused by sequence polymorphisms that could lead to the disruption of viral targeting of the cellular transcript. It is therefore somewhat surprising that more viral miRNAs do not show sequence similarities with known cellular miRNAs. One possible explanation is that more cellular orthologs of viral miRNAs do exist, but have not yet been discovered. As additional cellular and viral miRNAs are identified, further examples of virally expressed orthologs of cellular miRNAs may be revealed.
Murine herpesvirus-68
A third member of the gamma herpesviruses, and a distant relative of EBV and KSHV, has been shown to express miRNAs during latent infection [26]. Murine herpesvirus-68 (MHV-68) infects and establishes a latent infection within B cells of the germinal centers of the spleen. Following cDNA cloning of small RNAs from a stably infected B cell line, nine miRNAs were identified, again clustered within a genomic region of the virus known to encode latency-associated transcripts. Of particular interest is the existence of eight virally encoded tRNA-like transcripts (vtRNA), co-linear with the identified miRNAs. Northern blot analysis and secondary RNA structure predictions suggest that these miRNAs are encoded within two terminal stem loops downstream of each vtRNA transcript. These sequences are also predicted to encode a further six potential miRNAs. However, these miRNAs have so far not been detected. Each of the vtRNAs is driven by an RNA polymerase III promoter, and would therefore be predicted to drive expression of the associated miRNAs. This situation is unique to viruses, with all other cellular miRNA transcripts identified being driven by RNA polymerase II transcription. In vivo analysis of mouse tissues following MHV-68 infection has identified the expression of vtRNAs during both lytic and latent infections [45].Although no function has been identified for either the MHV-68 miRNAs or the vtRNAs, vtRNAs 1–4 (and their associated miRNAs) can be deleted from the virus with no obvious effect on the ability of the virus to establish or reactivate from latency, suggesting either a subtle role for these miRNAs or a certain level of redundancy in their actions [46].
Conclusion
From the recent studies discussed above, it is likely that herpesviruses make use of miRNAs for a variety of aspects including inhibiting apoptosis, immune avoidance, cellular proliferation and control of viral gene expression and replication. Targets of herpesvirus miRNAs include both cellular and viral transcripts. Interestingly, miRUL112-1 of HCMV has been shown to regulate both multiple viral genes as well as a cellular gene [7, 27]. In addition to the identification of target genes, a viral ortholog of an important cellular miRNA has also been discovered in KSHV that may prove to contribute to the oncogenic nature of the virus [6]. It is perhaps unsurprising that herpesviruses make extensive use of small RNA regulators of gene expression. The long-term nature of herpesvirus infections requires strict regulation of viral genes and multiple strategies to avoid both the innate and adaptive host defense mechanisms. miRNAs have the ability to regulate multiple genes, while their small size and RNA nature avoids triggering potentially detrimental host defense responses. They are also economic in terms of genomic space, as they require little coding sequence, an aspect attractive for even large viruses whose genome size is restrictive. Although a number of studies have now been published on targets and possible functions for viral miRNAs, a great deal more work will be required to truly understand the functional role of viral miRNAs. One of the major challenges ahead will be to convincingly show a phenotypic effect of viral miRNAs in the context of a physiologically relevant infection.
References
- 1.Roizman B, Pellet E. Fields virology. Vol. 2. Raven press; Philadelphia: 2001. The family Herpesviridae: a brief introduction; pp. 2381–2397. [Google Scholar]
- 2.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grey F, Meyers H, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3:e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2006;442:82–85. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- 5.Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci USA. 2007;104:16164–16169. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi’s Sarcoma-associated Herpesvirus Encodes an Ortholog of miR-155. J Virol. 2007;81(23):12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Marg-alit H, Mandelboim O. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 2007;3:e65. doi: 10.1371/journal.ppat.0030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preston CM. Repression of viral transcription during herpessimplex virus latency. J Gen Virol. 2000;81:1–19. doi: 10.1099/0022-1317-81-1-1. [DOI] [PubMed] [Google Scholar]
- 10.Branco FJ, Fraser NW. Herpes simplex virus type 1 latency-associated transcript expression protects trigeminal ganglion neurons from apoptosis. J Virol. 2005;79:9019–9025. doi: 10.1128/JVI.79.14.9019-9025.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson RL, Sawtell NM. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J Vi-rol. 2001;75:6660–6675. doi: 10.1128/JVI.75.14.6660-6675.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui C, GriYths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of herpessimplex virus 1-encoded microRNAs. J Virol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burnside J, Bernberg E, Anderson A, Lu C, Meyers BC, Green PJ, Jain N, Isaacs G, Morgan RW. Marek’s disease virus en-codes MicroRNAs that map to meq and the latency-associated transcript. J Virol. 2006;80:8778–8786. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Sewer A, Zavolan M, Nair V. Marek’s disease virus type 2 (MDV-2)-encoded MicroRNAs show no sequence conservation with those encoded by MDV-1. J Virol. 2007;81:7164–7170. doi: 10.1128/JVI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC, Cosman D. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2Dligands, protecting against natural killer cell cytotoxicity. J ExpMed. 2003;197:1427–1439. doi: 10.1084/jem.20022059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grey F, Antoniewicz A, Allen E, Saugstad J, McShea A, Carring-ton JC, Nelson J. Identification and characterization of human cytomegalovirus-encoded microRNAs. J Virol. 2005;79:12095–12099. doi: 10.1128/JVI.79.18.12095-12099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson JA, Gnann JW, Jr, Ghazal P. Regulation and tissue-specific expression of human cytomegalovirus. Curr Top Micro-biol Immunol. 1990;154:75–100. doi: 10.1007/978-3-642-74980-3_4. [DOI] [PubMed] [Google Scholar]
- 18.Boehler A, SchaVner A, Salomon F, Keusch G. Cytomegalovirus disease of late onset following renal transplantation: a potentially fatal entity. Scand J Infect Dis. 1994;26:369–373. doi: 10.3109/00365549409008606. [DOI] [PubMed] [Google Scholar]
- 19.Ballard RA, Drew WL, Hufnagle KG, Riedel PA. Acquired cytomegalovirus infection in preterm infants. Am J Dis Child. 1979;133:482–485. doi: 10.1001/archpedi.1979.02130050026005. [DOI] [PubMed] [Google Scholar]
- 20.Adler SP. Transfusion-associated cytomegalovirus infections. Rev Infect Dis. 1983;5:977–993. doi: 10.1093/clinids/5.6.977. [DOI] [PubMed] [Google Scholar]
- 21.Einhorn L, Ost A. Cytomegalovirus infection of human blood cells. J Infect Dis. 1984;149:207–214. doi: 10.1093/infdis/149.2.207. [DOI] [PubMed] [Google Scholar]
- 22.Macher AM, Reichert CM, Straus SE, Longo DL, Parrillo J, Lane HC, Fauci AS, Rook AH, Manischewitz JF, Quinnan GV., Jr Death in the AIDS patient: role of cytomegalovirus. N Engl J Med. 1983;309:1454. doi: 10.1056/NEJM198312083092312. [DOI] [PubMed] [Google Scholar]
- 23.Neiman P, Wasserman PB, Wentworth BB, Kao GF, Lerner KG, Storb R, Buckner CD, Clift RA, Fefer A, Fass L, Glucksberg H, Thomas ED. Interstitial pneumonia and cytomegalovirus infection as complications of human marrow transplantation. Transplantation. 1973;15:478–485. [PubMed] [Google Scholar]
- 24.Tegtmeier GE. The use of cytomegalovirus-screened blood in neonates. Transfusion. 1988;28:201–203. doi: 10.1046/j.1537-2995.1988.28388219141.x. [DOI] [PubMed] [Google Scholar]
- 25.Melnick JL, Adam E, DeBakey ME. Cytomegalovirus and atherosclerosis. Arch Immunol Ther Exp (Warsz) 1996;44:297–302. [PubMed] [Google Scholar]
- 26.PfeVer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 27.Grey F, Meyers HL, White EA, Spector DH, Nelson J. A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication. PLoS Pathog. 2007;3(11):e163. doi: 10.1371/journal.ppat.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mocarski ES, Kemble GW, Lyle JM, Greaves RF. A deletion mutant in the human cytomegalovirus gene encoding IE1 (491aa) is replication defective due to a failure in auto regulation. Proc Natl Acad Sci USA. 1996;93:11321–11326. doi: 10.1073/pnas.93.21.11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buck AH, Santoyo-Lopez J, Robertson KA, Kumar DS, Reczko M, Ghazal P. Discrete clusters of viral encoded miRNAs are associated with complementary strands of the genome and the 7.2-kb stable intron in murine cytomegalovirus. J Virol. 2007;81(23):13761–13770. doi: 10.1128/JVI.01290-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolken L, Perot J, Cognat V, Alioua A, John M, Soutschek J, Ruzsics Z, Koszinowski U, Voinnet O, PfeVer S. Mouse cytomegalovirus microRNAs dominate the cellular small RNAs profile during lytic infection and show features of post-transcriptional regulation. J Virol. 2007 doi: 10.1128/JVI.01313-07. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.PfeVer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 32.Smith PR, de Jesus O, Turner D, Hollyoake M, Karstegl CE, Griffin BE, Karran L, Wang Y, Hayward SD, Farrell PJ. Structure and coding content of CST (BART) family RNAs of Ep-stein–Barr virus. J Virol. 2000;74:3082–3092. doi: 10.1128/jvi.74.7.3082-3092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai X, Schafer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein–Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoSPathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:733–750. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young LS, Rickinson AB. Epstein–Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 36.Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore P, Chang Y. Fields virology. Vol. 2. Raven press; Philadelphia: 2001. Kaposi’s sacoma-associated Herpesvirus; pp. 2803–2831. [Google Scholar]
- 39.Pearce M, Matsumura S, Wilson AC. Transcripts encoding K12, v-FLIP, v-cyclin, and the microRNA cluster of Kaposi’s sarcoma-associated herpesvirus originate from a common promoter. J Virol. 2005;79:14457–14464. doi: 10.1128/JVI.79.22.14457-14464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai X, Cullen BR. Transcriptional origin of Kaposi’s sarcoma-associated herpesvirus microRNAs. J Virol. 2006;80:2234–2242. doi: 10.1128/JVI.80.5.2234-2242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCormick C, Ganem D. The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science. 2005;307:739–741. doi: 10.1126/science.1105779. [DOI] [PubMed] [Google Scholar]
- 42.Schafer A, Cai X, Bilello JP, Desrosiers RC, Cullen BR. Cloning and analysis of microRNAs encoded by the primate gamma-herpesvirus rhesus monkey rhadinovirus. Virology. 2007;364:21–27. doi: 10.1016/j.virol.2007.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 44.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowden RJ, Simas JP, Davis AJ, Efstathiou S. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are ex-pressed during latency. J Gen Virol. 1997;78(Pt 7):1675–1687. doi: 10.1099/0022-1317-78-7-1675. [DOI] [PubMed] [Google Scholar]
- 46.Simas JP, Bowden RJ, Paige V, Efstathiou S. Four tRNA-like sequences and a serpin homologue encoded by murine gammaherpesvirus 68 are dispensable for lytic replication in vitro and latency in vivo. J Gen Virol. 1998;79(Pt 1):149–153. doi: 10.1099/0022-1317-79-1-149. [DOI] [PubMed] [Google Scholar]


