Abstract
The sequencing of the complete genome of the nematode Caenorhabditis elegans was a landmark achievement and ushered in a new era of whole-organism, systems analyses of the biology of this powerful model organism. The success of the C. elegans genome sequencing project also inspired communities working on other organisms to approach genome sequencing of their species. The phylum Nematoda is rich and diverse and of interest to a wide range of research fields from basic biology through ecology and parasitic disease. For all these communities, it is now clear that access to genome scale data will be key to advancing understanding, and in the case of parasites, developing new ways to control or cure diseases. The advent of second-generation sequencing technologies, improvements in computing algorithms and infrastructure and growth in bioinformatics and genomics literacy is making the addition of genome sequencing to the research goals of any nematode research program a less daunting prospect. To inspire, promote and coordinate genomic sequencing across the diversity of the phylum, we have launched a community wiki and the 959 Nematode Genomes initiative (www.nematodegenomes.org/). Just as the deciphering of the developmental lineage of the 959 cells of the adult hermaphrodite C. elegans was the gateway to broad advances in biomedical science, we hope that a nematode phylogeny with (at least) 959 sequenced species will underpin further advances in understanding the origins of parasitism, the dynamics of genomic change and the adaptations that have made Nematoda one of the most successful animal phyla.
Keywords: genome, nematode, next-generation sequencing, second-generation sequencing, wiki
Introduction
The phylum Nematoda is fascinating because it is the most ubiquitous, numerous and diverse of all animal phyla, present in just about every ecological niche on our small planet. Nematodes have been indispensable for research programs in developmental biology,1 genome biology,2,3 evolutionary genomics,4 neurobiology,5 aging,6 health7 and parasitology.8
In the last two decades, DNA sequencing technology has evolved dramatically and allowed us to create genome resources for many of these nematodes, which have transformed our understanding of the biology of not just this phylum, but of all organisms.9-12
Raw sequencing costs have dropped five orders of magnitude13 in the past ten years, which means that it is now a viable research goal to obtain genome sequences for all nematodes of interest, rather than just a few model organisms. Inspired by large-scale genome initiatives for other major taxa14-17 we have initiated a push to sequence, in the first instance, 959 nematode genomes. Why (only) 959 genomes? The adult hermaphrodite C. elegans has 959 somatic cells, and one of the first major projects that turned C. elegans from a local curiosity into a key global research organism was the deciphering of the near-invariant developmental cell lineage that gives rise to these adult cells, starting from the fertilised zygote. In an analogous way we hope that a nematode phylogeny (the evolutionary lineage of the extant species) with 959 or more species will be similarly catalytic in driving nematode research programs across the spectrum of basic and applied science. Obviously, as sequencing technologies improve and become more accessible, we will move beyond this initial goal of 959, especially with over 23,000 described species and an estimated one to two million undescribed species in the phylum.
The goal of this article is to describe the current status of nematode genome research, to encourage everyone to sequence their favorite nematode and to share genome sequencing experiences and data. We show how inexpensive it has become to obtain high quality draft genomes and introduce the 959 Nematode Genomes wiki18 as a way to collate and track sequencing projects worldwide.
The Genomes We Have
The worm community has been at the forefront of animal genome sequencing since 1998, when Caenorhabditis elegans was the first metazoan to be fully sequenced.2 The C. elegans genome and its extensive annotation is accessible through the WormBase portal.19 WormBase was one of the first databases to integrate genomic, genetic and phenotypic data, and its curators aim to catalog and link all C. elegans literature and research, including large scale analyses such as modENCODE.20
Since the release of the C. elegans genome, nine other nematode genomes have been published, including six species parasitic in plants and animals (Table 1). Only C. elegans and C. briggsae have been sequenced to “finished” status2 with all sequence data organized into chromosome-sized pieces. The remaining eight are high-quality draft genomes and all ten can be accessed at WormBase19 through graphical genome browsers and via bulk-data downloads.
Table 1. Published nematode genomes.
| Species | Systematic position (Blaxter Clade, Helder Clade*) | Year Published | Technology | Genome Size (Mbp)† | Number of chromosomes or scaffolds in assembly† | Scaffold N50 (kbp)†,‡ | AT content (%)† | Number of genes/ proteins† |
|---|---|---|---|---|---|---|---|---|
|
Caenorhabditis elegans |
V, 9E |
19982 |
Sanger |
100 |
6 chromosomes |
17,494 |
64.6 |
20,461 / 25,244 |
|
Caenorhabditis briggsae |
V, 9E |
20033 |
Sanger |
108 |
6 chromosomes + 5 fragments |
17,485 |
62.6 |
NA / 21,986 |
|
Brugia malayi |
III, 8 |
200752 |
Sanger |
96 |
27,210 |
38 |
69.4 |
18,348 / 21,332 |
|
Meloidogyne hapla |
IV, 11 |
200824 |
Sanger |
53 |
3,452 |
38 |
72.6 |
NA / 13,072 |
|
Meloidogyne incognita |
IV, 11 |
200827 |
Sanger |
82 |
9,538 |
13 |
68.6 |
NA / 21,232 |
|
Pristionchus pacificus |
V, 9B |
200853 |
Sanger |
172 |
18,083 |
1,245 |
57.2 |
NA / 24,217 |
|
Caenorhabditis angaria |
V, 9E |
201033 |
Illumina |
80 |
33,559 |
9 |
63.7 |
22,662 / 26,265 |
|
Trichinella spiralis |
I, 2A |
201136 |
Sanger |
64 |
6,863 |
6,373 |
66.1 |
16,380 / 16,380 |
|
Bursaphelenchus xylophilus |
IV, 10D |
201137 |
Roche 454, Illumina |
75 |
5,527 |
950 |
59.6 |
18,074 / 18,074 |
| Ascaris suum | III, 8 | 201125 | Illumina | 273 | 29,831 | 408 | 62 | 18,542 / 18,542 |
Nuclear genome only, not including mitochondria or endosymbionts, computed from WormBase release WS227 where available or from data URLs in Table 2.
Scaffold N50: Half the assembly is in scaffolds of this size or larger in the nuclear genome.
On the 959 Nematode Genomes wiki, 26 additional genome sequencing projects are listed with publicly available draft assemblies and, in some cases, annotations (Table 2). Seven of these are hosted at WormBase and the rest are available either through the 959 Nematode Genomes website or at sequencing center websites. These draft genomes are expected to have at least 95% of the genes present in multi-gene sized contigs, but the exact ordering and chromosomal location of the contigs is usually not known. Despite these shortcomings, draft data are very useful for comparative and evolutionary genomics or simply for identifying single genes of interest. Early access to these data not only allows researchers to test hypotheses, but, equally importantly, to identify potential problems early in the assembly process. Researchers wishing to publish analyses using pre-publication draft data should contact the sequencing center or lead researchers for permissions (and also to see if better versions of these data are or will soon be available).
Table 2. Nematode species for which published or draft genome data are publicly available.
Note: See www.nematodes.org/nematodegenomes/index.php/Strains_with_Data for an up-to-date list.
Why We Need More: One Nematode Genome Does Not a Phylum Make
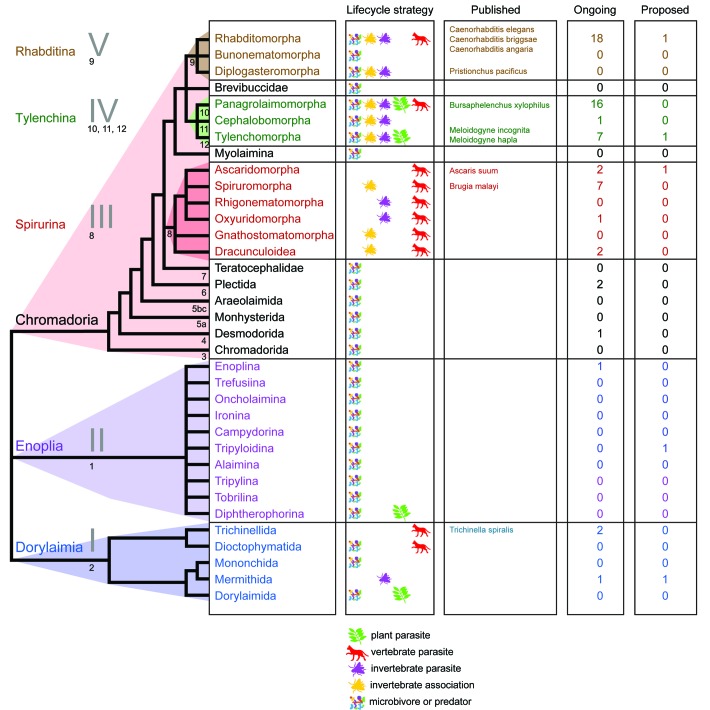
C. elegans is an excellent model nematode and its genome, with its wealth of annotation, is an excellent model genome. However C. elegans cannot be taken to represent all nematode genomes (Fig. 1). We know that C. elegans is quite derived within Nematoda21 and that it lacks many genes shared between other nematodes and other Metazoa.22 Nematode genomes have been sized from 20 Mb to 500 Mb (i.e., one fifth to five times that of C. elegans).23 Sequenced nematode genomes range from Meloidogyne hapla24 at 54 Mb to Ascaris suum25 at 273 Mb. Interesting genomic features in other species include chromatin diminution in Ascaris suum and other ascaridids (i.e., the germline has a larger genome than the soma26), aneuploid triploidy in the Meloidogyne incognita genome27 and the presence of obligate, vertically-transmitted symbiont alphaproteobacterial Wolbachia and their genomes inside the cells of many filarial nematodes.28
Figure 1. Systematic tree of Nematoda indicating current sequenced, in progress or proposed genome sequencing projects. The systematic arrangement of Nematoda is based on De Ley and Blaxter;51 the clades defined by Blaxter et al.21 and van Mengen et al.54 are indicated. For each major group we summarize the trophic ecology (microbivore, predator, fungivore, plant parasite, non-vertebrate parasite or associate, vertebrate parasite) and the number of species for which genome projects are reported in the 959 Nematode Genomes wiki. Figure developed from Blaxter.55
Apart from understanding genome organization and origins, richer sampling of sequenced genomes would allow a better understanding of the phylogeny of Nematoda and the evolutionary dynamics of important traits—such as parasitism of plants and animals—and developmental modes. The most comprehensive molecular phylogenies of Nematoda have been based on a single gene, the ~1600 bp nuclear small subunit rRNA locus,21,29,30 but this single locus is insufficient for robust resolution of the deep divergences in the phylum. Methods for generating large-scale multi-gene phylogenies now exist and can be applied even to draft genomes.
When large scale expressed sequence tag (EST) sequencing was first performed,31 new insights into nematode gene evolution became possible from the partial catalogs of expressed genes.22 More nematode genomes, even draft-quality ones, take those insights several steps further, as they allow analysis of complete gene catalogs. Additionally, whole-genome resources include non-genic regions, such as the regulatory regions upstream of genes, which are often even more conserved than coding regions and may function in developmental regulation.32,33
How to Make More
C. elegans was sequenced over a decade ago using Sanger sequencing. At that time, sequencing the genome to ten-fold depth took a decade and cost roughly $10 M. Once the sequencing was completed, similar resources were required to finish the genome. Sanger sequencing is still considered the gold standard in terms of quality, but because of the high cost and time investment, it is unlikely that there will be any more Sanger-sequenced nematode genomes.
Sequencing the C. elegans genome was based on an array of mapped and ordered large-insert genomic clones, which greatly facilitated assembly. Most genome sequencing today avoids this time-consuming step and uses only whole-genome shotgun sequencing. As a result, current genome projects typically result in draft genomes with multi-gene sized contigs rather than chromosome-sized sequences. The substantial additional effort required to finish a genome is necessary if the goal is to study chromosome organization or long-range regulation. However, many questions about phylogenetics, gene evolution and shared or novel gene functions can be approached using high-quality draft genomes generated at a tiny fraction of the time and cost of a finished genome.
Second-generation sequencing platforms have dramatically reduced costs and increased throughput, with the trade-off of reduced read length compared with Sanger dideoxy reads (Table 3). Shorter reads mean that most genomic repeats are longer than a read, and the only way to attempt to resolve them in a genome assembly is to use pairs of reads sequenced from opposite ends of fragments that are longer than the repeats. Sophisticated assembly programs that use high sequencing depth and multiple insert libraries to get around the problems of sequencing errors and repeats have been developed specifically for second-generation data.34
Table 3. Current sequencing costs, throughput and read lengths.
| Technology | Read length (bases) | Error model | Recommended sequencing depth | Cost per base (£/€/$) | Cost per 100 Mb genome (£/€/$) | Throughput (bases/ day/ instrument) | Time per 100 Mb genome per instrument (days) |
|---|---|---|---|---|---|---|---|
| Sanger dideoxy |
1000–1500 |
Gold standard, accurate base quality, typical error probability 0.0001 |
10 X |
10−3 |
106 |
106 |
103 |
| Roche 454 FLX/FLX+ |
400–1000 |
Homopolymer errors |
20–30 X |
10−5 |
2 × 104 |
5 × 108 |
5 |
| Illumina HiSeq2000 | 100–150 | Typical error probability 0.01, Lower quality toward end of read | 50–100 X | 10−7 | 103 | 1010 | 1 |
Each platform has different read lengths and error profiles that affect their suitability for de novo genome sequencing projects. The Illumina platforms generate reads up to 150 bases and are the workhorses of sequencing projects. Illumina sequencing errors are usually miscalled bases and higher read-depths are recommended to consensus-correct such errors. Roche 454 reads can extend to 750 bases but are more expensive than the shorter-read technologies. In Roche 454 data, sequencing depths higher than 30-fold are not recommended35 because homopolymer errors accumulate and confound assembly algorithms. Life Tech's SOLiD technology generates short (~75 base) reads but is not suitable for de novo genome sequencing because each base is represented by two “colors” (readings) and sequencing errors are difficult to identify in the absence of a reference sequence.
Different combinations of technologies, insert lengths and depths of coverage can be employed to exploit the best characteristics of each and minimise known classes of errors. In particular, paired-end sequencing from a mix of library insert sizes appears to be optimal for de novo assembly, using short-insert (200–700 bp) paired end (PE) libraries complemented by long-insert (1–20 kb) mate pair (MP) libraries. While PE data derive from directly captured genome fragments and are thus largely free of chimaeras, construction of MP libraries involves additional manipulations, including circularisation of long DNAs, that can result in high proportions of chimaeric or aberrantly short virtual inserts. MP data are typically used for scaffolding contigs generated from PE data, which are generated in higher coverage. Deep sequencing of the transcriptome can also yield scaffolding information,33 linking genome sequence contigs that contain exons for a gene that cannot be joined by genome sequence data because of repeats.
In the last year, genome sequences have been published for four nematode species. Each project used different sequencing strategies. The genome of Trichinella spiralis was determined using traditional Sanger dideoxy sequencing,36 with a 33-fold base coverage in the final assembly. Bacterial artificial chromosome clones and multiple-size insert clone libraries were used to scaffold the 64 Mb genome. The Bursaphelenchus xylophilus37 genome was sequenced using Illumina PE and Roche 454 single-end reads for basic contig generation and Roche 454 MP for scaffolding contigs. For Caenorhabditis angaria,33 Illumina PE (from libraries with multiple insert sizes from 200–450 bp) totalling 170-fold coverage were used, and then deep transcriptome data (Illumina RNA-Seq) were used to improve this assembly. This was the first genome project to use RNA-Seq reads to scaffold genomic contigs. Two versions of the A. suum genome have been released. Wang et al.38 generated an assembly using Roche 454 and Illumina data from short insert libraries and mate-pair data from 5.5 kb libraries sequenced using Sanger dideoxy technology as part of an extensive transcriptome sequencing project. Jex et al.25 used a mix of Illumina PE 170 bp and 500 bp PE reads, scaffolded with Illumina MP data from 800 bp, 2 kb, 5 kb and 10 kb libraries. Interestingly, these long-insert MP libraries were generated from DNA that was whole-genome amplified using strand-displacing isothermal amplification, a technology that holds great promise for additional nematode genome projects where starting materials may be limiting.
So which strategy should you use? If you are on a bargain-basement budget and want the most value for money, a single lane of Illumina HiSeq2000 PE (100 bases plus 100 bases) sequencing with multiplexed 300 bp and 600 bp PE libraries can provide a highly usable draft genome. For example, in our laboratory, Caenorhabditis species 5 was recently sequenced using this strategy and resulted in a draft assembly spanning 131 Mb in only 16,384 scaffolds, with more than half the assembly in scaffolds larger than 31 kb (S. Kumar, A. Cutter, M-A. Felix, M. Blaxter, unpublished; see www.nematodes.org/nematodegenomes/index.php/Caenorhabditis_sp._5_DRD-2008_JU800). Roche 454 data are more expensive base for base than Illumina, but usually assemble into longer contigs at the same effective coverage. Mate pair data serve to scaffold the primary contigs generated from single-end Roche 454 or PE Illumina sequencing and significantly improve the assembled fragment lengths. Construction of MP libraries for Illumina or Roche 454 sequencing requires much more and higher quality starting DNA than do PE libraries and MP libraries are more costly to produce. In addition to genomic sequencing, a single lane of Illumina HiSeq2000 RNA-Seq data (100 base PE reads from 300 bp libraries made from RNA pooled from many stages) is highly recommended for aiding assembly and annotation.
The Costly As: Assembly, Annotation and Analysis
The generation of the raw sequence data are rapidly becoming a marginal cost in a genome sequencing program: the relative cost and time taken for assembly, annotation and analysis post-sequencing is much greater. Raw reads need to be quality checked, checked for contaminants and assembled. The assemblies need to be verified and possibly repeated in turn and then annotated to identify genes and other genomic features of interest such as regulatory regions, repeats and transposons. The intricacies of assembly algorithms, assembly strategies and annotation options are beyond the scope of this article, but a wide variety of excellent methods and tools have been published.35,39-46 The most comprehensive recent analysis of assembly strategies for complex eukaryotic genomes was the Assemblathon.34 If all goes well, a nematode genome can, in theory, be shepherded from DNA extraction to an annotated assembly and be ready for further analyses in as little as a month.
Both bioinformatics and sequencing technologies are changing so rapidly that recommendations on strategies may quickly become obsolete. For bioinformatics solutions, the most up-to-date tips and recommendations will probably come from low-latency sources such as conference presentations, blog posts, forums, crowdsourced Q & A sites and collaborative wikis such as 959 Nematode Genomes (as described below). For sequencing, the two emerging wet laboratory technologies that could dramatically change how we sequence nematodes are whole genome amplification and single-molecule sequencing.
Whole-genome amplification (WGA) has been used to generate sufficient quantities of DNA from tissues of single A. suum for MP libraries.25 This opens the prospect of using WGA on single nematode specimens, though the mass of DNA input from A. suum used by the BGI team (200 ng) is much more than is present in most individual nematodes (one C. elegans adult contains ~200 pg). Proof that amplification does not overly bias sequencing coverage or generate chimaeras that mislead assembly algorithms would be a major advance. Sequencing from single nematodes will reduce the assembly issues arising from extremes of heterozygosity observed in wild populations and will allow researchers to select specimens directly from environmental samples.
The promise of single-molecule sequencing is the generation of ultra-long reads (several kilobases) from templates that have undergone a minimum of in vitro manipulation. It is well recognized in second-generation sequencing that the several PCR steps involved can exclude some regions of a genome from sequencing and positively bias sequencing to regions that have GC content closer to 50%. Ultra-long reads could span repetitive regions and thus ease assembly. PacBio SMRT47 is the first single-molecule technology to be released commercially and can produce reads over 2 kb, but has relatively low throughput and an accuracy far lower than second-generation sequencers. Another single-molecule technology is from Oxford Nanopore,48 which promises high-quality, high-throughput reads with no theoretical length limit. However, the company has not yet released any data or metrics on error rates, read lengths, throughput or costs, so all we can say is that the technology will change genome sequencing if it works.
Keeping Track Using the 959 Nematode Genomes Wiki
We set up the 959 Nematode Genomes wiki (959NG wiki) at www.nematodegenomes.org to keep track of genomes being sequenced and published.18 As second-generation sequencing becomes more accessible, we anticipate that several hundred nematodes will be sequenced in the next few years. Although the INSDC databases (GenBank/ENA/DDBJ) are the first sources that most of us turn to when looking for sequences from or related to our organism of interest, genomes are often deposited there only at the time of publication, and this can yield the impression that no project is underway. We hope the 959NG wiki will enable genomic resources to be shared pre-publication, avoid duplication of effort, allow new genomes to be proposed and forge collaborations between researchers interested in the same species or clade.
Web-based databases for tracking genome sequencing projects are not a new idea and we know of at least four (diArk,49 Genomes OnLine Database (GOLD),50 The International Sequencing Consortium (www.intlgenome.org) and Genome News Network (www.genomenewsnetwork.org). However, only the first two are currently maintained and all four rely on centrally updating the database whenever a new genome is proposed or released. As 959NG is a wiki where anyone can sign in to add or edit information, we anticipate that the site will stay up to date, and because it is specific to nematodes, it is more likely to be of use to the nematode community.
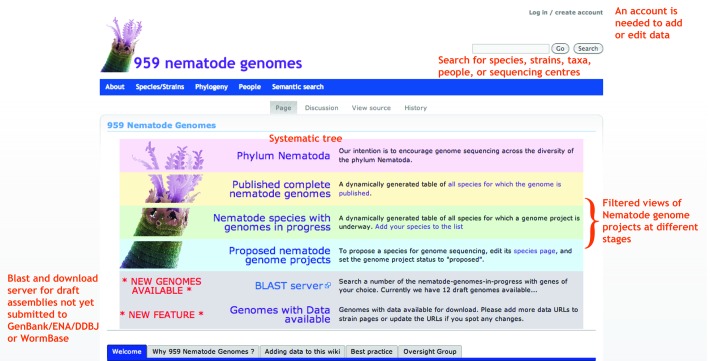
The homepage of the wiki has links to all the important parts of the site (Fig. 2). The 959NG wiki is organized taxonomically using the systematics proposed by De Ley and Blaxter51 (Fig. 1). We also use the clades defined by Blaxter et al.21 and Holterman et al.29 and derive other systematic information from the NCBI taxonomy. The tree is editable, so if new evidence is found for resolution of any paraphyletic nodes or rearrangements, additional nodes can be added or taxa reassigned simply by changing the parent taxon for that set of taxa. For any taxon (class, order, family, genus, etc.), the wiki lists all the species and strains that have active or proposed genome projects. For published projects we encourage addition of PubMed IDs for publications and links to genome browsers and data repositories. For species where a genome project is “ongoing” we associate the project with a strain of the species, to permit more than one independent project to be registered. Again, genome project leaders are encouraged to add links to project web pages and data access portals.
Figure 2. The 959 Nematode Genomes wiki home page.
One goal of the 959NG wiki is to reduce the “activation energy” for starting a new genome project. Embarking on a genome scale endeavor can be daunting, but we hope that the 959NG wiki will promote collaboration on genomes of interest. Individual researchers can “propose” a species (and strain) for genome sequencing and register interest in species that have been proposed. By making interests known, it is more likely that fruitful collaborations will ensue. We know of two multi-center projects, both now mature, where the proponents first met on the 959NG wiki.
Finding current genome data can be frustrating. We therefore provide a list of available data portals for genomes sequenced and in sequencing. These include genome browsers (such as those provided by WormBase) as well as data download sources. The 9595NG wiki also includes a standard BLAST search portal that allows researchers with specific (gene-centered) interests in one or many genomes to query available published and pre-publication draft genomes.
The 959NG wiki is built on the MediaWiki platform (the same tools that run Wikipedia) and uses the Semantic MediaWiki (SMW) extension. Each page about a strain, species, taxon, researcher or sequencing center has semantic properties associated with it that can be queried in new ways to extract inferred relationships and new properties can be added to any page without changing any database schemas. For example, we plan to add lifecycle strategies (as shown in Fig. 1) to taxon pages to enable queries such as “List all ongoing genome projects for plant parasitic nematodes with genomes smaller than 100 Mb.” Other query examples and details of how SMW is an appropriate technology for such sites can be found in Kumar, Schiffer and Blaxter.18
Join the 959 Nematode Genomes Initiative
The 959 Nematode Genomes initiative (and the 959NG wiki) is open to all and we encourage all interested to join. Anyone can view the wiki (and free registration gives editing rights). The 959NG wiki will only be as good as (and as up to date as) the information we, collectively, enter. In particular, we would encourage registration of interest in ongoing and proposed genomes and the active proposal of additional nematode genomes for sequencing. As the community of researchers producing and consuming new nematode genomes grows, the synergy of combining skills and discoveries in data generation, assembly and annotation will become more evident and will facilitate the generation of new genomes. The availability of large numbers of phylogenetically diverse genomes will also—we hope—inspire a new breed of nematode genomics researchers not wedded to any one species but hungry for data across the phylum and thus eager to collaborate in the analyses of new genomes.
The 959NG wiki will evolve as the community evolves. The snapshot presented here (Tables 1 and 2) will soon be out of date. The open architecture of the SMW system will allow us to add additional concepts and linking data between genomes and thus the wiki should also be able to nucleate and serve special interest groups where the core themes are not simply systematic, but rather other shared phenotypes (reproductive mode, parasitism) or specific gene sets or systems. By identifying colleagues with shared interests, joint funding to generate nematode genome data will be more easily sourced. The collective experience embodied in the 959NG wiki will also mean that the costs (in both consumables and human effort) of de novo sequencing a genome will continue to drop and multi-genome projects will become even more attractive to funding agencies and more rewarding for the nematode genomes community.
Acknowledgments
We acknowledge the input of Philipp Schiffer in the initiation of the 959NG wiki. We thank all our colleagues around the world who are sequencing nematode genomes and adding information to the wiki. At the University of Edinburgh, we especially want to thank Karim Gharbi and colleagues at the GenePool Genomics and Bioinformatics Facility for their expertise and advice on de novo genome sequencing and the members of the Blaxter Lab for their constant support and advice on bioinformatics analyses. We would also like to thank Dan Lawson at the European Bioinformatics Institute for inspiring us by setting up arthropodgenomes.org using the SMW platform. The SMW community was very helpful in answering questions about customising the platform. S. K. is funded by a School of Biological Sciences PhD studentship and the Overseas Research Student Awards Scheme at the University of Edinburgh; G. Kuar is funded by MRC funding to the GenePool, Edinburgh (G00900740) and the EU Project “Enhanced protective Immunity Against Filariasis” focused research project (SICA; contract number 242131); G. Koutsovoulos is funded by a BBSRC School of Biological Sciences University of Edinburgh PhD studentship.
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/19046
References
- 1.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 2.The C elegans Genome Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–8. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 3.Stein LD, Bao Z, Blasiar D, Blumenthal T, Brent MR, Chen N, et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:e45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cutter AD, Dey A, Murray R. Evolution of the Caenorhabditis elegans Genome. Mol Biol Evol. 2009;26:1199–234. doi: 10.1093/molbev/msp048. [DOI] [PubMed] [Google Scholar]
- 5.White JG, Southgate E, Thomson JN, Brenner S. The Structure of the Nervous System of the Nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 6.Crittenden SL, Eckmann C, Wang L, Bernstein D, Wickens M, Kimble J. Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philos Trans R Soc Lond B Biol Sci. 2003;358:1359–62. doi: 10.1098/rstb.2003.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang MC, O'Rourke E, Ruvkun G. Fat Metabolism Links Germline Stem Cells and Longevity in C. elegans. Science. 2008;322:957–60. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooker S. Estimating the global distribution and disease burden of intestinal nematode infections: adding up the numbers–a review. Int J Parasitol. 2010;40:1137–44. doi: 10.1016/j.ijpara.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 10.Hengartner MO, Ellis R, Horvitz R. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature. 1992;356:494–9. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 11.Horvitz HR, Sternberg P. Multiple intercellular signalling systems control the development of the Caenorhabditis elegans vulva. Nature. 1991;351:535–41. doi: 10.1038/351535a0. [DOI] [PubMed] [Google Scholar]
- 12.Blaxter M.Nematoda: Genes, Genomes and the Evolution of Parasitism. Advances in Parasitology 2003; 54:101-95 [DOI] [PubMed] [Google Scholar]
- 13.National Human Genome Research Institute DNA Sequencing Costs. genome.gov, 2011
- 14.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Genome 10K Community of Scientists Genome 10K: A Proposal to Obtain Whole-Genome Sequence for 10 000 Vertebrate Species. J Hered. 2009;100:659–74. doi: 10.1093/jhered/esp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson G, Hackett K, Purcell-Miramontes M, Brown S, Evans J, Goldsmith M, et al. Creating a buzz about insect genomes. Science 2011; 331 [DOI] [PubMed] [Google Scholar]
- 17.Cao J, Schneeberger K, Ossowski S, Gunther T, Bender S, Fitz J, et al. Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet. 2011;43:956–63. doi: 10.1038/ng.911. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S, Schiffer P, Blaxter M.959 Nematode Genomes: a semantic wiki for coordinating sequencing projects. Nucleic Acids Research 2011 [DOI] [PMC free article] [PubMed]
- 19.Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen W, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–7. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerstein MB, Lu Z, Van Nostrand E, Cheng C, Arshinoff B, Liu T, et al. Integrative Analysis of the Caenorhabditis elegans Genome by the modENCODE Project. Science. 2010;330:1775–87. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blaxter ML, De Ley P, Garey J, Liu L, Scheldeman P, Vierstraete A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–5. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 22.Wasmuth J, Schmid R, Hedley A, Blaxter M. On the extent and origins of genic novelty in the phylum Nematoda. PLoS Negl Trop Dis. 2008;2:e258. doi: 10.1371/journal.pntd.0000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gregory TR, Nicol J, Tamm H, Kullman B, Kullman K, Leitch I, et al. Eukaryotic genome size databases. Nucleic Acids Res. 2007;35:D332–8. doi: 10.1093/nar/gkl828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opperman CH, Bird D, Williamson V, Rokhsar D, Burke M, Cohn J, et al. Sequence and genetic map of Meloidogyne hapla: A compact nematode genome for plant parasitism. Proc Natl Acad Sci USA. 2008;105:14802–7. doi: 10.1073/pnas.0805946105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jex A, Liu S, Li B, Young N, Hall R, Li Y, et al. Ascaris suum draft genome. Nature 2011 [DOI] [PubMed]
- 26.Müller F, Bernard V, Tobler H. Chromatin diminution in nematodes. Bioessays. 1996;18:133–8. doi: 10.1002/bies.950180209. [DOI] [PubMed] [Google Scholar]
- 27.Abad P, Gouzy J, Aury J-M, Castagnone-Sereno P, Danchin E, Deleury E, et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat Biotechnol. 2008;26:909–15. doi: 10.1038/nbt.1482. [DOI] [PubMed] [Google Scholar]
- 28.Fenn K. Are filarial nematode Wolbachia obligate mutualist symbionts? Trends Ecol Evol. 2004;19:163–6. doi: 10.1016/j.tree.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Holterman M, van der Wurff A, van den Elsen S, van Megen H, Bongers T, Holovachov O, et al. Phylum-Wide Analysis of SSU rDNA Reveals Deep Phylogenetic Relationships among Nematodes and Accelerated Evolution toward Crown Clades. Mol Biol Evol. 2006;23:1792–800. doi: 10.1093/molbev/msl044. [DOI] [PubMed] [Google Scholar]
- 30.van Megen H, van den Elsen S, Holterman M, Karssen G, Mooyman P, Bongers T, et al. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology. 2009;11:927–50. doi: 10.1163/156854109X456862. [DOI] [Google Scholar]
- 31.Parkinson J, Mitreva M, Whitton C, Thomson M, Daub J, Martin J, et al. A transcriptomic analysis of the phylum Nematoda. Nat Genet. 2004;36:1259–67. doi: 10.1038/ng1472. [DOI] [PubMed] [Google Scholar]
- 32.Vavouri T, Walter K, Gilks W, Lehner B, Elgar G. Parallel evolution of conserved non-coding elements that target a common set of developmental regulatory genes from worms to humans. Genome Biol. 2007;8:R15. doi: 10.1186/gb-2007-8-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mortazavi A, Schwarz E, Williams B, Schaeffer L, Antoshechkin I, Wold B, et al. Scaffolding a Caenorhabditis nematode genome with RNA-seq. Genome Res. 2010;20:1740–7. doi: 10.1101/gr.111021.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Earl D, Bradnam K, St John J, Darling A, Lin D, Faas J, et al. Assemblathon 1: A competitive assessment of de novo short read assembly methods. Genome Research 2011 [DOI] [PMC free article] [PubMed]
- 35.Finotello F, Lavezzo E, Fontana P, Peruzzo D, Albiero A, Barzon L, et al. Comparative analysis of algorithms for whole-genome assembly of pyrosequencing data. Briefings in Bioinformatics 2011 [DOI] [PubMed]
- 36.Mitreva M, Jasmer D, Zarlenga D, Wang Z, Abubucker S, Martin J, et al. The draft genome of the parasitic nematode Trichinella spiralis. Nat Genet. 2011;43:228–35. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kikuchi T, Cotton J, Dalzell J, Hasegawa K, Kanzaki N, McVeigh P, et al. Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011;7:e1002219. doi: 10.1371/journal.ppat.1002219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Czech B, Crunk A, Wallace A, Mitreva M, Hannon G, et al. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011;21:1462–77. doi: 10.1101/gr.121426.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaisson MJ, Brinza D, Pevzner P. De novo fragment assembly with short mate-paired reads: Does the read length matter? Genome Res. 2009;19:336–46. doi: 10.1101/gr.079053.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nat Methods. 2009;6:S6–12. doi: 10.1038/nmeth.1376. [DOI] [PubMed] [Google Scholar]
- 41.Pop M. Genome assembly reborn: recent computational challenges. Brief Bioinform. 2009;10:354–66. doi: 10.1093/bib/bbp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alkan C, Sajjadian S, Eichler E. Limitations of next-generation genome sequence assembly. Nat Methods. 2011;8:61–5. doi: 10.1038/nmeth.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Picardi E, Pesole G. Computational methods for ab initio and comparative gene finding. Methods Mol Biol. 2010;609:269–84. doi: 10.1007/978-1-60327-241-4_16. [DOI] [PubMed] [Google Scholar]
- 44.Miller JR, Koren S, Sutton G. Assembly algorithms for next-generation sequencing data. Genomics. 2010;95:315–27. doi: 10.1016/j.ygeno.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin Y, Li J, Shen H, Zhang L, Papasian C, Deng HW. Comparative studies of de novo assembly tools for next-generation sequencing technologies. Bioinformatics. 2011;27:2031–7. doi: 10.1093/bioinformatics/btr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671–82. doi: 10.1038/nrg3068. [DOI] [PubMed] [Google Scholar]
- 47.Korlach J, Bjornson K, Chaudhuri B, Cicero R, Flusberg B, Gray J, et al. Real-time DNA sequencing from single polymerase molecules. Methods Enzymol. 2010;472:431–55. doi: 10.1016/S0076-6879(10)72001-2. [DOI] [PubMed] [Google Scholar]
- 48.Maglia G, Heron A, Stoddart D, Japrung D, Bayley H. Analysis of single nucleic acid molecules with protein nanopores. Methods Enzymol. 2010;475:591–623. doi: 10.1016/S0076-6879(10)75022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammesfahr B, Odronitz F, Hellkamp M, Kollmar M. diArk 2.0 provides detailed analyses of the ever increasing eukaryotic genome sequencing data. BMC Research Notes. 2011;4:338. doi: 10.1186/1756-0500-4-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz V, et al. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38:D346–54. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Ley P, Blaxter M. Systematic position and phylogeny. In: Lee DL, ed. The biology of nematodes: Taylor & Francis, 2002:1-30. [Google Scholar]
- 52.Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, et al. Draft genome of the filarial nematode parasite Brugia malayi. Science. 2007;317:1756–60. doi: 10.1126/science.1145406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dieterich C, Clifton S, Schuster L, Chinwalla A, Delehaunty K, Dinkelacker I, et al. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet. 2008;40:1193–8. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Megen H, van den Elsen S, Holterman M, Karssen G, Mooyman P, Bongers T, et al. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology. 2009;11:927–50. doi: 10.1163/156854109X456862. [DOI] [Google Scholar]
- 55.Blaxter M. Nematodes: the worm and its relatives. PLoS Biol. 2011;9:e1001050. doi: 10.1371/journal.pbio.1001050. [DOI] [PMC free article] [PubMed] [Google Scholar]