Abstract
A recent study by Greer et al. in the nematode C. elegans has shown transgenerational epigenetic inheritance of longevity in the descendants of worms deficient for subunits of a complex responsible for histone H3 lysine 4 trimethylation (H3K4me3). In this commentary, we discuss the implications of this epigenetic memory of longevity and the potential mechanisms underlying this phenomenon. The transgenerational inheritance of longevity could result from heritable depletion of H3K4me3 at particular aging-regulating gene loci that would only be progressively replenished. The epigenetic memory of longevity could also be explained by the transgenerational transmission of other molecules, for example other proteins or non-coding RNAs. The discovery of an epigenetic memory of longevity in worms raises the intriguing possibility that environmental cues modulating longevity in ancestors might affect subsequent generations in a non-Mendelian manner. Another remaining intriguing question is whether transgenerational inheritance of longevity also exists in other species, including mammals.
Keywords: aging, epigenetics, H3K4me3, longevity, transgenerational inheritance
Introduction
Longevity is a complex phenotype, with both genetic and environmental components. The heritability of longevity in humans is estimated to be approximately 30%,1,2 suggesting a relatively strong genetic component for such an integrative trait. Characterization of the genetic component of longevity has greatly benefited from studies in invertebrates, in particular the nematode Caenorhabditis elegans. For example, studies in worms have revealed the importance of insulin-FoxO signaling,3 a pathway that has subsequently been found to regulate longevity in a conserved manner from worms to humans.4 Even within the heritable component of longevity, however, disentangling the role of genetics from that of environment is far from simple.1 A recent report of transgenerational epigenetic inheritance of longevity in C. elegans5 has provided support for the possibility that part of the heritability of longevity could also be epigenetic.
Epigenetics is the study of phenotypic changes that do not involve modifications of the DNA sequence.6 This term is often used to refer to changes in chromatin states by DNA methylation or histone post-translational modifications, including histone acetylation or methylation. However, in its stricto sensu definition, epigenetics represent mitotic or meiotic heritable changes in phenotypes.7 Epigenetic heritability most frequently refers to mitotic heritability—how marks are inherited in dividing cells. Epigenetic meiotic heritability was long thought to be exceedingly rare, especially because the epigenome is usually considered to be mostly erased and reprogrammed during meiosis and early development.8 However, accumulating evidence of transgenerational epigenetic inheritance of several traits suggests that there may be more selective maintenance and erasure of epigenetic marks between generations than initially thought.9-12
Transgenerational Inheritance of Longevity in C. elegans
A recent study indicates that deficiencies in members of the COMPASS complex can induce transgenerational inheritance of longevity.5 The COMPASS complex is conserved from yeast to humans and is responsible for depositing the H3K4 trimethylation (H3K4me3) histone mark,13 a mark that is usually present at the promoters of actively transcribed genes.14 In C. elegans, deficiencies in individual members of the COMPASS complex (composed of ASH-2, WDR-5 and methyltransferase SET-2) were shown to prolong the lifespan of fertile worms in the parental generation.15 Accordingly, overexpression of the counteracting RBR-2 H3K4me3 demethylase also extends worm lifespan,15 suggesting that an excess of the H3K4me3 is detrimental to worm longevity. Lifespan extension due to deficiencies in this COMPASS complex depends on the presence of a mature adult germline.15 This is in contrast with other chromatin modifiers (e.g., SET-9, SET-15 or UTX-1), which modulate worm lifespan independently of the presence of a functional germline.15-17 The observation that the COMPASS H3K4me3 complex specifically regulates lifespan by acting in the germline raised the question of whether deficiencies in this complex could still extend the lifespan in subsequent generations even when descendants were no longer deficient for members of the complex. Surprisingly, genetically wild-type descendants from ancestors with a mutation in the COMPASS complex still display extended lifespan up until the third generation5 (Table 1). Thus, the lifespan of worms can be influenced by the COMPASS activity status of their great-great-grandparents. This inheritance of the longevity phenotype without changes in the DNA sequence over several generations is a striking example of transgenerational epigenetic inheritance.
Table 1. Transgenerational effects of COMPASS deficiency on lifespan and gene expression.
| Generation* | COMPASS-status (genotype) | Lifespan | Global H3K4me3 | Transcriptome |
|---|---|---|---|---|
| P0 |
wild-type (+/+) |
Normal |
Normal |
Normal |
| P0 |
deficient (mutant/mutant) |
Extended |
Down |
Differential regulation of 7,820 genes |
| F1 |
Heterozygotes (mutant/+) |
n.d. |
n.d. |
n.d. |
| F2 |
wild-type (+/+) |
n.d. |
n.d. |
n.d. |
| F3 |
wild-type (+/+) |
Extended |
Normal |
n.d. |
| F4 |
wild-type (+/+) |
Extended |
Normal |
Differential regulation of 1,740 genes |
| F5 | wild-type (+/+) | Normal | Normal | Normal |
See reference 5 for the setup of the genetic cross schemes; n.d.: not determined.
Specificity of the Transgenerational Inheritance of Longevity
Interestingly, the lifespan extension in the offspring generations relies on similar pathways as that of the parental generation.5 Indeed, deficiency in the RBR-2 demethylase rescues the transgenerational inheritance of longevity phenotype due to deficiencies in members of the COMPASS complex in the parental generation. Moreover, the longevity of wild-type descendants of COMPASS complex mutants remains dependent on the presence of a functional adult germline and on the ability of the worms to produce fertilized eggs.5 These observations suggest that this transgenerational inheritance of longevity is unlikely to result from a non-specific “hybrid vigor” effect of crossing worms, or from another extraneous mutation that might still have been present in the original mutant strains after extensive outcrossing.
Many independent pathways have been found to modulate lifespan in C. elegans,18,19 including the much studied insulin-like signaling pathway and the mitochondria pathways. However, the transgenerational inheritance of longevity appears to be rather specific to the ASH-2/SET-2/WDR-5 complex, as manipulation of these other longevity regulators only led to increased lifespan in the parental generation.5 Importantly, other chromatin modifiers that regulate longevity, but do not affect H3K4me3 or do not act in the germline (e.g., UTX-1, SET-9 and SET-15), do not show similar transgenerational inheritance of longevity. Conversely, other phenotypes depending on H3K4 methylation status have been found to be inherited or amplified across generations in C. elegans, such as germline mortality of worms that are mutated for LSD-1, an H3K4me2 demethylase8,20 or progressive sterility at 25°C of worms carrying the severe set-2(bn129) mutation.21 The progressive sterility phenotype is not observed in set-2(ok952) mutants, which were looked at in the study that identified COMPASS members as regulators of longevity.15,21 Interestingly, the set-2(bn129) mutation is predicted to be a complete null allele (deleting the methyltransferase domain of the SET-2 protein), whereas the set-2(ok952) mutation corresponds to an in-frame deletion, which would more likely be hypomorphic. This is consistent with the contrasting global H3K4me3 levels in both mutants, with total loss of the mark in set-2(bn129) mutants and partial retention of the mark in set-2(ok952) mutants.21 All in all, these results point to H3K4 methylation regulators as specific contributors to the epigenetic memory of acquired traits in C. elegans.
Transgenerational Inheritance of Longevity Correlates with Partial Inheritance of Transcriptome Patterns
What are the mechanisms underlying transgenerational epigenetic inheritance of longevity? There does not seem to be a pure maternal component (i.e., cytoplasmic factors inherited from the oocyte) in the origin of the epigenetic memory of lifespan.5 Indeed, while most of the experiments testing transgenerational inheritance of lifespan were done by crossing hermaphrodite worms deficient in members of the COMPASS complex to wild-type males (Table 1), the lifespan of descendants resulting from a reverse mating scheme (where the males were deficient for a member of the COMPASS complex) was also extended.
The epigenetic memory of longevity was not due to a global heritable dearth in H3K4me3 levels. Global levels of H3K4me3 were restored to wild-type levels in the long-lived genetically wild-type descendants.5,15 These observations raise the possibility that the transgenerational inheritance of the longevity phenotype by deficiencies in the COMPASS complex results from a more targeted epigenetic aberration, that can only be erased and successfully reprogrammed within the span of four generations.
The activity of the COMPASS complex is required for appropriate regulation of gene expression.13,22 Moreover, inheritance of active transcriptional states in daughter cells was found to require trimethylation of H3K4 by the orthologs of ASH-2 and SET-2.23 Thus, Greer and colleagues tested whether COMPASS deficiency in the parental generation might lead to the misregulation of specific subset of genes which could be transgenerationally inherited. Indeed, a subset of WDR-5-regulated genes were still misexpressed in the long-lived descendants at the F4 generation, but returned to normal at the F5 generation (Table 1).5 Just as for the longevity phenotype, the transgenerationally inherited gene misexpression profile returned to normal at the next (F5) generation, suggesting that these phenotypes may be directly linked. This observation points toward the responsibility of a few loci with potentially heritable changes in aberrant H3K4me3 marking across the long-lived generations. Among WDR-5 misregulated genes whose misexpression was transgenerationally inherited, about a third have been previously established as expressed in the worm germline by SAGE,24 and could thus be good candidates to mediate the transgenerational inheritance of the longevity phenotype.5
Remaining Questions
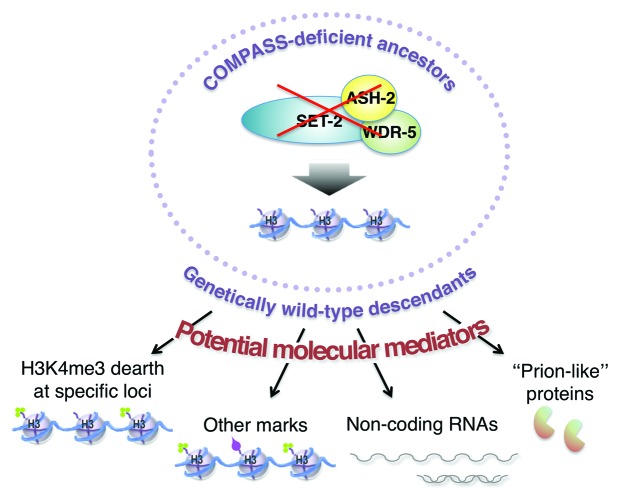
The study by Greer and colleagues has provided the first example of transgenerational epigenetic inheritance of longevity. This epigenetic transmission is compatible with an incomplete reprogramming of chromatin marks in the germline,8 which would take three generations to be finally reset to normal. Much remains to be explored regarding the molecular mechanisms underlying the transgenerational inheritance of longevity induced by deficiencies in the COMPASS complex. The most immediate suspect for mediating epigenetic memory of lifespan would be a dearth of H3K4me3 itself (Fig. 1). However, because there is no global decrease of H3K4me3 in subsequent generations, the dearth of H3K4me3 responsible for the phenotype would have to be restricted to a limited subset of important target loci or to specific cells in the germline. The H3K4me3 dearth would be replenished, but only progressively, at each generation, until the chromatin state near the key mediator genes responsible for the longevity phenotype could be completely reset. Because lifespan is completely restored to a wild-type range between the F4 and F5 generation, one can postulate the existence of a “threshold of H3K4me3” that would be reached in the germline of the F5 generation and prevent further epigenetic transmission of the longevity phenotype.
Figure 1. Potential molecular mechanisms underlying the transgenerational inheritance of longevity. COMPASS deficiency in the ancestral generation could lead to transgenerational epigenetic inheritance of longevity in descendants by several non-mutually exclusive mechanisms: directly by H3K4me3 dearth at specific loci regulating aging, by proxy through other marks (e.g., H3K4me2, H3K36me3 or H3K9me3), by the production and transmission of non-coding RNAs (e.g., siRNAs, lncRNAs, or piRNAs) or even by the production of inheritable proteins with prion-like qualities.
Another possibility is that other histone marks could relay the effect of H3K4me3 on this epigenetic memory of longevity (Fig. 1). Alterations in the H3K4me2 mark profile have been shown to be heritable due to incomplete erasure, which leads to the progressive loss of germline immortality.20 Additionally, levels of the H3K36me3 mark, a modification that is usually associated with actively transcribed genes,14 have recently been found to transmit the epigenetic memory of parental germline gene expression to the next generation.25,26 Another potential culprit could be the H3K9me3 repressive mark, which has been found to act as a transgenerational signal promoting pericentric heterochromatin formation in mouse early embryos.27 Aberrant and transmissible patterns of these “relaying” marks could also affect the expression of specific genes.
Alternatively, coding or non-coding RNA molecules could be crucial in mediating the epigenetic memory in response to COMPASS deficiency (Fig. 1). Non-coding RNAs could serve as guides to re-establish chromatin states in the descendants.28 Indeed RNA transmission was found to play a role in the transgenerational inheritance of some traits in plants and mammals, including purple color in maize and pigmentation patterns or cardiac hypertrophy in mice.29-31 Non-coding RNAs of the piwi-interacting RNA (piRNA) class can be inherited and play a key role in the germline-silencing of transposon activity during chromatin remodeling or translational silencing.32,33 Thus, another potential mechanistic explanation for the transgenerational inheritance of longevity phenotype of COMPASS-deficient worms might be the existence of aberrant non-coding or piRNA complexes transmitted to the progeny. Though this phenomenon has not been reported yet, non-coding micro-RNAs (i.e., miRNA), which are involved in the regulation of cellular processes in C. elegans,34 may also similarly be transgenerationally inherited.
Another potential mechanism for the transgenerational inheritance of longevity could lie in the transmission of small interfering RNAs (siRNA). Indeed, heritable silencing of genes has been observed in C. elegans as a response to double-stranded RNA treatment, with the phenotypic effects of silencing lasting up to the F3 or F4 generations.35 Moreover, a recent study in C. elegans has shown that small-interfering RNAs derived from a viral genome post-infection can be transmitted to the uninfected progeny of worms in a transgenerational manner,36 thus providing potential adaptive benefits to the animals. Thus, COMPASS deficiency could also act by leading to the production of non-coding, double-stranded, RNAs, thus inducing a heritable silencing of pro-aging genes or even of members of the COMPASS complex themselves in the next generations.
Finally, at this stage, we cannot exclude that this instance of non-Mendelian inheritance of longevity could also be mediated by other types of molecules, for instance through yet unknown protein protagonists acting in a “prion-like” transmission paradigm.37
Concluding Remarks
Transgenerational inheritance of both simple and complex traits has been observed in multiple taxa, including plants, insects, fish and mammals.9,10,38-41 Interestingly, traits that have been found to have transgenerational inheritance include organismal responses to a number of environmental stimuli, such as tolerance to pesticides, resistance to toxins or heavy metals, metabolic gene expression, obesity and even behaviors.38,42-47
As epigenetic marks can respond to environmental cues,48,49 the function of the COMPASS complex might be sensitive to external stimuli (e.g., food availability or presence of toxins), and potentially relay this information to the next few generations, which might have evolutionary benefits. Another implication for the presence of transgenerational inheritance of acquired traits in many different species is that transgenerational inheritance of longevity might also be conserved in other species. Indeed, a transgenerational effect of nutrition of ancestors on the survival of subsequent generations through the male lineage seems to exist in humans.50 Thus, it is possible that a fraction of the heritable component of longevity in humans results from transgenerational epigenetic inheritance induced by environmental factors.
Acknowledgments
We thank Shuo Han, Jana Lim, and Aaron Daugherty for critical reading of the manuscript. This work was supported by NIH R01-AG31198 and ARRA-AG31198 grants to A.B.
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/19157
References
- 1.Cournil A, Kirkwood TB. If you would live long, choose your parents well. Trends Genet. 2001;17:233–5. doi: 10.1016/S0168-9525(01)02306-X. [DOI] [PubMed] [Google Scholar]
- 2.Gudmundsson H, Gudbjartsson DF, Frigge M, Gulcher JR, Stefánsson K. Inheritance of human longevity in Iceland. Eur J Hum Genet. 2000;8:743–9. doi: 10.1038/sj.ejhg.5200527. [DOI] [PubMed] [Google Scholar]
- 3.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–4. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 4.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–12. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 5.Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–71. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–3. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–6. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 8.Furuhashi H, Kelly WG. The epigenetics of germ-line immortality: lessons from an elegant model system. Dev Growth Differ. 2010;52:527–32. doi: 10.1111/j.1440-169X.2010.01179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho DH, Burggren WW. Epigenetics and transgenerational transfer: a physiological perspective. J Exp Biol. 2010;213:3–16. doi: 10.1242/jeb.019752. [DOI] [PubMed] [Google Scholar]
- 10.Rando OJ, Verstrepen KJ. Timescales of genetic and epigenetic inheritance. Cell. 2007;128:655–68. doi: 10.1016/j.cell.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 11.Daxinger L, Whitelaw E. Transgenerational epigenetic inheritance: more questions than answers. Genome Res. 2010;20:1623–8. doi: 10.1101/gr.106138.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuzin F, Grandjean V, Rassoulzadegan M. Inherited variation at the epigenetic level: paramutation from the plant to the mouse. Curr Opin Genet Dev. 2008;18:193–6. doi: 10.1016/j.gde.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–8. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barth TK, Imhof A. Fast signals and slow marks: the dynamics of histone modifications. Trends Biochem Sci. 2010;35:618–26. doi: 10.1016/j.tibs.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Greer EL, Maures TJ, Hauswirth AG, Green EM, Leeman DS, Maro GS, et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature. 2010;466:383–7. doi: 10.1038/nature09195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin C, Li J, Green CD, Yu X, Tang X, Han D, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14:161–72. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Maures TJ, Greer EL, Hauswirth AG, Brunet A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell. 2011;10:980–90. doi: 10.1111/j.1474-9726.2011.00738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolff S, Dillin A. The trifecta of aging in Caenorhabditis elegans. Exp Gerontol. 2006;41:894–903. doi: 10.1016/j.exger.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 19.Greer EL, Brunet A. Signaling networks in aging. J Cell Sci. 2008;121:407–12. doi: 10.1242/jcs.021519. [DOI] [PubMed] [Google Scholar]
- 20.Katz DJ, Edwards TM, Reinke V, Kelly WG. A C. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell. 2009;137:308–20. doi: 10.1016/j.cell.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao Y, Bedet C, Robert VJ, Simonet T, Dunkelbarger S, Rakotomalala C, et al. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc Natl Acad Sci U S A. 2011;108:8305–10. doi: 10.1073/pnas.1019290108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–72. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 23.Muramoto T, Müller I, Thomas G, Melvin A, Chubb JR. Methylation of H3K4 Is required for inheritance of active transcriptional states. Curr Biol. 2010;20:397–406. doi: 10.1016/j.cub.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 24.Wang X, Zhao Y, Wong K, Ehlers P, Kohara Y, Jones SJ, et al. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics. 2009;10:213. doi: 10.1186/1471-2164-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuhashi H, Takasaki T, Rechtsteiner A, Li T, Kimura H, Checchi PM, et al. Trans-generational epigenetic regulation of C. elegans primordial germ cells. Epigenetics Chromatin. 2010;3:15. doi: 10.1186/1756-8935-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rechtsteiner A, Ercan S, Takasaki T, Phippen TM, Egelhofer TA, Wang W, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6:6. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puschendorf M, Terranova R, Boutsma E, Mao X, Isono K, Brykczynska U, et al. PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos. Nat Genet. 2008;40:411–20. doi: 10.1038/ng.99. [DOI] [PubMed] [Google Scholar]
- 28.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suter CM, Martin DI. Paramutation: the tip of an epigenetic iceberg? Trends Genet. 2010;26:9–14. doi: 10.1016/j.tig.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–74. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 31.Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, et al. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–9. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 32.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 33.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 34.Kaufman EJ, Miska EA. The microRNAs of Caenorhabditis elegans. Semin Cell Dev Biol. 2010;21:728–37. doi: 10.1016/j.semcdb.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Alcazar RM, Lin R, Fire AZ. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–88. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rechavi O, Minevich G, Hobert O. Transgenerational Inheritance of an Acquired Small RNA-Based Antiviral Response in C. elegans. Cell. 2011;147:1248–56. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halfmann R, Lindquist S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science. 2010;330:629–32. doi: 10.1126/science.1191081. [DOI] [PubMed] [Google Scholar]
- 38.Ashe A, Whitelaw E. Another role for RNA: a messenger across generations. Trends Genet. 2007;23:8–10. doi: 10.1016/j.tig.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Erhard KF, Jr., Hollick JB. Paramutation: a process for acquiring trans-generational regulatory states. Curr Opin Plant Biol. 2011;14:210–6. doi: 10.1016/j.pbi.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Seong KH, Li D, Shimizu H, Nakamura R, Ishii S. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011;145:1049–61. doi: 10.1016/j.cell.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 41.Nelson VR, Nadeau JH. Transgenerational genetic effects. Epigenomics. 2010;2:797–806. doi: 10.2217/epi.10.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sánchez M, Andreu-Moliner E, Ferrando MD. Laboratory investigation into the development of resistance of Daphnia magna to the herbicide molinate. Ecotoxicol Environ Saf. 2004;59:316–23. doi: 10.1016/j.ecoenv.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 43.Gluckman PD, Hanson MA, Beedle AS. Non-genomic transgenerational inheritance of disease risk. Bioessays. 2007;29:145–54. doi: 10.1002/bies.20522. [DOI] [PubMed] [Google Scholar]
- 44.Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143:1084–96. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–6. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 46.Franklin TB, Linder N, Russig H, Thöny B, Mansuy IM. Influence of early stress on social abilities and serotonergic functions across generations in mice. PLoS One. 2011;6:e21842. doi: 10.1371/journal.pone.0021842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franklin TB, Russig H, Weiss IC, Gräff J, Linder N, Michalon A, et al. Epigenetic transmission of the impact of early stress across generations. Biol Psychiatry. 2010;68:408–15. doi: 10.1016/j.biopsych.2010.05.036. [DOI] [PubMed] [Google Scholar]
- 48.Mathers JC, Strathdee G, Relton CL. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet. 2010;71:3–39. doi: 10.1016/B978-0-12-380864-6.00001-8. [DOI] [PubMed] [Google Scholar]
- 49.Franklin TB, Mansuy IM. Epigenetic inheritance in mammals: evidence for the impact of adverse environmental effects. Neurobiol Dis. 2010;39:61–5. doi: 10.1016/j.nbd.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 50.Kaati G, Bygren LO, Pembrey M, Sjöström M. Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet. 2007;15:784–90. doi: 10.1038/sj.ejhg.5201832. [DOI] [PubMed] [Google Scholar]