Abstract
In this commentary, we discuss how our recent paper by Yang et al. contributes a new wrinkle to the already somewhat curious Wnt signaling pathway in C. elegans. We begin with a historical perspective on the Wnt pathway in the worm, followed by a summary of the key salient point from Yang et al., 2011, namely demonstration of mutually inhibitory binding of a β-catenin SYS-1 to the N-terminus and another β-catenin WRM-1 to the C-terminus of the TCF protein POP-1, and a plausible structural explanation for these differential binding specificities. The mutually inhibitory binding creates one population of POP-1 that is bound by WRM-1, phosphorylated by the NLK kinase and exported from the nucleus, and another bound by coactivator SYS-1 that remains in the nucleus. We speculate on the evolutionary history of the four β-catenins in C. elegans and suggest a possible link between multiple β-catenin gene duplications and the requirement to reduce nuclear POP-1 levels to activate Wnt target genes.
Keywords: asymmetry, embryo, endoderm, POP-1, SYS-1, Wnt signaling, WRM-1
Some Background
In the late 1990s it was determined by standard genetic analysis and cloning that C. elegans had three β-catenin genes.1-3 This number was confirmed when the C. elegans genome sequence was published in 1998 and completed in 2002,4 as no additional β-catenin related genes were revealed by sequence homology. C. elegans was therefore something of a Wnt signaling oddity—most organisms, including Hydra, Drosophila, mouse and man, have a single β-catenin protein. This bifunctional protein participates in at least two quite different processes: First, cell-cell adhesion through interaction with cadherin and α-catenin and localization at adherens junctions, and second, canonical Wnt pathway signaling, in which cytoplasmic β-catenin is stabilized as a result of the Wnt signal, translocates to the nucleus, and functions as a coactivator of the TCF protein in activating transcription of Wnt target genes (reviewed in refs. 5 and 6). The genetic analysis in C. elegans was consistent with these two functions being split between the three β-catenins, HMP-2, BAR-1 and WRM-1.7,8 HMP-2 seemed fairly straightforward, as it appeared to function in adhesion only.1 HMP-2 bound to cadherin (HMR-1) and α-catenin (HMP-1), localized to adherens junctions, and the hmp-2 mutant phenotype was consistent with a single function in cell-cell adhesion.1 BAR-1 also appeared relatively straightforward: it functioned as a coactivator with the sole C. elegans TCF protein, POP-1, in a canonical Wnt pathway during post-embryonic development.2 BAR-1 bound to POP-1 via a conserved N-terminal β-catenin binding domain, did not bind to α-catenin, did not localize at adherens junctions, and its mutant phenotype was completely consistent with a function only in Wnt signaling.
It was the third β-catenin, WRM-1, that was puzzling for some time. WRM-1 was shown by RNAi to be required for the specification of the endoderm precursor in the early embryo.3 This process was shown to require Wnt and MAPK signaling from P2 to the EMS blastomere at the 4-cell stage, resulting in E, the posterior daughter of EMS, being specified as the sole endoderm (gut) precursor for the worm.3,9-12 The observation that embryos depleted of wrm-1 by RNAi resembled embryos lacking Wnt signaling3 led to an early assumption that WRM-1 was probably the β-catenin transcriptional coactivator for POP-1 and activated Wnt target genes in the E nucleus. However, WRM-1 showed almost no signs of functioning as such. WRM-1 did not function as a coactivator of POP-1 in the standard reporter assays.7 While two groups failed to detect a physical interaction between WRM-1 and POP-1 by co-immunoprecipitation,7,13 one of these two groups and another group reported a very weak interaction in yeast two hybrid assays.8,13 Natarajan et al. noted that the weak interaction did not require the POP-1 N-terminal domain, an evolutionarily conserved domain through which β-catenins bind to TCFs.8 The Mello lab subsequently identified a biochemical function for WRM-1 which explained the requirement for WRM-1 in E specification. They showed that WRM-1 binds to the MAP kinase LIT-1, and that this binding was required for LIT-1 kinase activity.13 The LIT-1 kinase activity is required for the nuclear levels of POP-1 to be lowered in the E nucleus, which is a requirement for activation of Wnt target genes.3,14,15
While the role of WRM-1 in the specification of the endoderm precursor was becoming more clear, the nature of the Wnt pathway and its role in endoderm/gut specification remained murky. Depletion of the TCF protein POP-1 resulted in a phenotype opposite to that from depletion of wrm-1 or the gene encoding the Wnt ligand, mom-2.3,12,16 These results initially suggested that POP-1 functioned to repress Wnt target genes in the E blastomere. TCF proteins, by virtue of being able to bind to corepressors such as Groucho, can also function as potent repressors of Wnt target genes.17-19 Therefore, it was initially concluded that high levels of POP-1 repressed Wnt targets, and the Wnt-dependent lowering of POP-1 levels in the E nucleus resulted in transcriptional derepression of these genes. The POP-1/WRM-1 Wnt pathway was initially referred to as being non-canonical (not in the PCP or Ca2+ sense) because it appeared to deviate significantly from the canonical pathway.20-23
“If there are four equations and only three variables, and no one of the equations is derivable from the others by algebraic manipulation then there is another variable missing.”—Talcott Parsons
“I need more.” —Iggy Pop
However, analyses of Wnt target gene expression in E showed that the lowered POP-1 levels in the E nucleus did not result in simple derepression of Wnt target genes. Instead, lowered POP-1 level was shown to be a requirement for activated, higher level expression of Wnt target genes.15,24 These results called for a coactivator of POP-1 whose level is limiting relative to POP-1.15 It was proposed that when nuclear POP-1 levels are too high, excess POP-1 that is not bound by the limiting coactivator would be available to bind to corepressors, resulting in transcriptional repression of Wnt targets.20,25 None of the three previously identified C. elegans β-catenins fit the bill for this candidate coactivator for POP-1. Enter the fourth worm β-catenin, SYS-1, which was identified not through sequence homology, but solely through its function as a POP-1 coactivator via a genetic screen that identified regulators of the asymmetric cell division of the somatic gonadal precursor (SGPs) cells Z1 and Z4.26,27 Structural analysis confirmed the β-catenin nature of SYS-1, which only exhibits approximately 10% sequence identity with human β-catenin.28 We and others subsequently showed SYS-1 to be the limiting coactivating β-catenin for POP-1 in the specification of endoderm in embryos, while others showed the same to be true for the SGPs.26,29,30 SYS-1 levels (both cytoplasmic and nuclear) are higher in E compared with the sister cell of E, MS, and the high level of SYS-1 is dependent on mom-2 and other genes in the Wnt signaling pathway.29,30 We also showed that forced expression of SYS-1 in MS could lead to MS developing into endoderm,29 supporting the proposed model that expression of Wnt target genes depends on the abundance of SYS-1 relative to POP-1.
POP-1 can be Bound by SYS-1 and WRM-1, but not at the Same Time
The level of SYS-1 is only regulated by the Wnt pathway, and not by LIT-1, WRM-1 or MOM-4, a MAPKKK that functions upstream of LIT-1.29,30 The signal from P2 to EMS results in a simultaneous increase in the level of SYS-1 (Wnt signal) and decrease in the level of nuclear POP-1 (MAPK signal) in E. This immediately presents a problem: how can cells maintain a high level of SYS-1 in the nucleus while simultaneously exporting from the same nucleus the SYS-1 binding partner, POP-1? That this in fact happens argues that there must be two separate populations of POP-1, one which is not bound by SYS-1 that is exported from the nucleus, and another, bound by SYS-1, that remains in the nucleus and transcriptionally activates Wnt target genes. Our recent paper31 directly addresses this question, and presents a mechanism whereby this partitioning of POP-1 can be achieved. We showed that WRM-1 interacts robustly and specifically with the C-terminus of POP-1, and not via the conserved N-terminal β-catenin binding domain. In addition, we showed that WRM-1 has two functions: first, as the substrate-binding unit of the LIT-1/WRM-1 kinase complex, and second, as an activator of LIT-1 kinase activity that is independent of its ability to bind POP-1. Most importantly, however, we showed that SYS-1 binding to the N-terminus and WRM-1 binding to the C-terminus of POP-1 are mutually inhibitory. This mutual inhibition generates the two distinct populations of POP-1 in the Wnt responsive cell. POP-1 bound by WRM-1 is phosphorylated and exported from the nucleus, whereas POP-1 bound by SYS-1 is not LIT-1 phosphorylated, remains in the nucleus, and activates transcription of Wnt target genes. The final nuclear level of POP-1 will be determined primarily by competition between SYS-1 and WRM-1 binding to POP-1.
But how does this actually work? An unexpected finding that the C-terminal sequence of POP-1 bore limited but significant similarity to its N-terminal β-catenin binding domain gave a hint. This suggested to us that the two domains might exhibit structural similarity, and, furthermore, that structural variations between these domains could possibly explain the differential binding specificities of the two β-catenins. Taking advantage of several solved vertebrate TCF/β-catenin structures,32-34 including the POP-1/SYS-1 structure,28 we performed computer modeling of a POP-1/WRM-1 interaction, and presented a plausible structural explanation for how SYS-1 and WRM-1 bind specifically to the opposite termini of POP-1.31
β-catenins are comprised of a central core of 12 armadillo repeats, along with N-terminal and C-terminal tails.35 Each armadillo repeat is comprised of typically three helices, with each repeat packing together with neighboring repeats to form a superhelix with a continuous hydrophobic (positively charged) groove on the concave surface of the superhelix.36 It is via this hydrophobic groove that β-catenin contacts most of its binding partners. The β-catenin binding domain of vertebrate TCFs contains two separate structural motifs that contact the β-catenin positive groove: first, an extended strand which interacts with ARM repeats 5–9 primarily via charged amino acids, followed by an α helix, which interacts with ARM repeats 3–4, mostly through hydrophobic interactions.32,37-39 Interaction via the extended strand motif in human TCF4 is stabilized when a conserved aspartate (D16) forms a crucial salt bridge with K435, termed a “charged button,” in ARM repeat 8 of β-catenin. The structure determined for the complex of SYS-1 interacting with the N-terminus of POP-1 shows interaction of an extended strand of POP-1 with SYS-1 ARM repeats 5–9, including a conserved “charged button.”28 However, the POP-1 N-terminus did not appear to form an α helix and was unstructured in the crystals, suggesting that it played little direct role in the interaction with SYS-1. Computer modeling suggested that WRM-1 had the conserved lysine in the right position to form the conserved salt bridge. However, the large sidechain of a residue near the “charged button” would preclude the formation of the salt bridge.28 Expanding upon these studies, we undertook a computer-based modeling of an interaction between WRM-1 and the C-terminal domain of POP-1.31 The very C-terminus of POP-1 was predicted to have a high propensity to adopt an α-helical conformation and to interact with WRM-1 ARM repeats 3–5 through hydrophobic interactions.
“Pop changes week to week, month to month.” —Ravi Shankar
It appears, therefore, that at some point during an expansion of the β-catenin gene family during the evolutionary history of C. elegans, duplicated β-catenin proteins (see further below) diverged to the point that they bound to separate domains of POP-1. In doing so, each of the β-catenins adapted to a different binding motif in POP-1 from the two available (extended strand vs. α helix) to predominate each interaction. It is intriguing that the small first exon at the pop-1 locus encodes the β-catenin binding domain and little else. The fact that the C-terminal domain of POP-1 shares sequence and structure similarity with the N-terminal domain suggested to us the possibility that a shuffling of the first pop-1 exon to the 3′ end of the genomic locus might account for the two termini bearing some resemblance. Although highly speculative, this scenario does beg the question whether the exon duplication and shuffle happened first, with duplicated β-catenins adapting to the presence of two potential binding sites, or whether duplicated and diverging β-catenins competing for a single N-terminal binding site on POP-1 drove selection for an exon shuffle event.
“If evolution really works, how come mothers only have two hands?” —Milton Berle
Most organisms, including humans, make do with a single, multifunctional β-catenin. It is not clear at all why the ~1,000-cell worm maintains four functional β-catenin genes, especially in light of the very compact nature of its genome. Here we would like to speculate on the evolutionary history of the four C. elegans β-catenins, and, specifically, how those gene duplications might have led to the requirement to lower POP-1 nuclear levels in order to activate Wnt target genes.
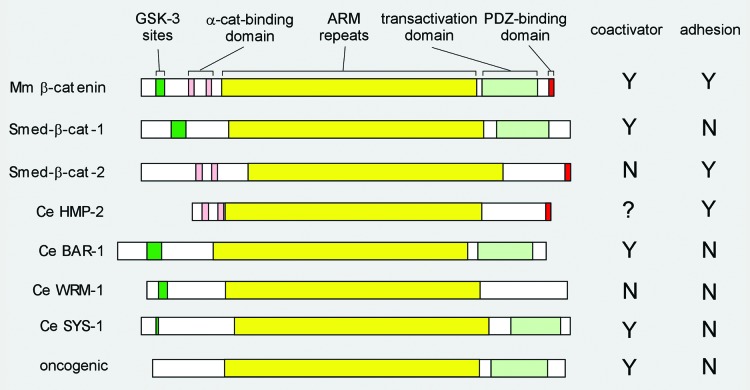
A typical bifunctional β-catenin contains GSK-3 phosphorylation sites required for ubiquitination and degradation located near the N-terminus,40,41 followed by two small domains involved in α-catenin binding, the central 12 ARM repeats, followed by a C-terminal transactivation domain and a short PDZ binding motif at the very C-terminus (Fig. 1).42 The α-catenin binding domains and the PDZ binding domain are absolutely required for cell-cell adhesion but are dispensable for Wnt signaling, whereas the GSK-3 motif and the C-terminal transactivation domains are required for Wnt signaling but are dispensable for the adhesion function. In addition, there are specific residues within the ARM repeat domain that are critical for the interaction of β-catenin with APC, axin and TCF but are not required for cadherin binding, and vice versa.
Figure 1. β-catenin domain structure. Schematic representation of the indicated β-catenin proteins with functional domains referred to in the text highlighted in color: central ARM repeat domain in yellow; coactivator-associated domains in shades of green: GSK-3 phosphorylation sites (dark green) and C-terminal transactivation domain (light green); cell-cell adhesion-associated domains in shades of red: N-terminal α-catenin-binding domain (pink) and C-terminal PDZ-binding domain (red). The bifunctional mouse (Mm, top) β-catenin is used here to represent the canonical single β-catenins in human, Xenopus, and flies. The two monofunctional planarian (Smed) proteins are above the four C. elegans (Ce) proteins, followed by the oncogenic form of β-catenin (bottom). GSK-3 phosphorylation motif: typically DpSGΦXpS/T, where Φ is a hydrophobic amino acid (V, I, L, M, F, W, or C). This motif is often coupled with additional S/TxxxS/T motifs. Mouse β-catenin, planaria β-catenin-1, and Ce BAR-1 contain this motif near their N-termini. The three other Ce proteins lack this motif, but do have 2–3 possible GSK-3 sites (simple (S)/(T)xxx(S)/(T) near their N-termini. α-catenin-binding and the C-terminal PDZ-binding domains: Both planaria β-catenin-2 and C. elegans HMP-2 bind α-catenin (experimentally tested) through a domain similar to the α-catenin-binding domain in mouse (TRAQRVRAAMFPE ..21.. VQRLAEPSQMLK). All three proteins also have a PDZ-binding domain at their very C-terminus (mouse: WFDTDL[stop]). The other three C. elegans β-catenins do not exhibit sequence related to either motif and have been tested negative in α-catenin binding assays. C-terminal transactivating domain: Transcription activation domain was normally tested in a TOPFLASH reporter assay. Only BAR-1 and SYS-1 activate a TOPFLASH reporter in tissue culture cells. However, this assay does not accurately measure the transactivation domain of β-catenin as this assay requires binding of β-catenin to the N-terminal domain of the TCF/POP-1 protein. Natarajan (2001) reported that both BAR-1 and HMP-2 can activate transcription in a yeast 1-hybrid assay, but BAR-1 did so significantly better than HMP-2. WRM-1 was shown to have a very weak, slightly above background activation capability in such an assay.
β-catenin is a very ancient protein,43 and both the primary sequence and the functional domain structure of β-catenin tend to be highly conserved during evolution. The single β-catenin detected in Hydra magnipapillata bears a striking similarity to human β-catenin, given the evolutionary distance between hydra and humans.44,45 This protein is 62% identical to human β-catenin, and includes all the diagnostic motifs present in well-characterized bifunctional β-catenins, and functions in both cell-cell adhesion and Wnt signaling. Similarly, a single bifunctional GSK-3β-sensitive β-catenin in the annelid Platynereis dumerilii contains all of the same domains and exhibits 84% amino acid identity to human β-catenin in the ARM repeats.46 The unicellular amoeba Dictyostelium discoideum has a single β-catenin-related protein, Aardvark, which functions both in adherens junctions in the stalk cells of the fruiting body as well as a GSK-3 dependent transcriptional regulator during spore cell differentiation, and exhibits approximately 50% similarity in the ARM repeats with human β-catenin.47
The Four β-Catenins in C. elegans
One major surprise from the sequence of the four C. elegans β-catenins was the degree to which they have diverged in sequence from human β-catenin. Indeed, the four C. elegans β-catenins are more highly diverged from vertebrate β-catenin than any other characterized β-catenins.1-3,7,8,26 The C. elegans β-catenins exhibit identity values ranging from only ~10% for SYS-1 up to ~28% for HMP-2 when compared with human β-catenin. Furthermore, these four worm proteins are almost as diverged from one another as they each are from human β-catenin. This is a remarkable degree of sequence divergence, to the point that one of the C. elegans β-catenins, SYS-1, was not identified as such even following completion of the genome sequence. Although there are indications that protein sequence divergence rates might be higher in C. elegans than other animals,48 this level of divergence was nonetheless surprising. Because the C. elegans genome encodes at least four bone fide β-catenin proteins, with each protein appearing to retain only one of β-catenins several functions, the argument was made that this then freed the individual β-catenin genes to undergo more extensive sequence divergence. However, this argument is undermined by the β-catenin gene duplication and functional divergence observed in the planaria Schmidtea mediterranea.49,50 S. mediterranea has two β-catenins, Smed-β-catenin-1 and Smed-β-catenin-2, which have diverged such that each appears to have preserved only one of the two main functions of human β-catenin: Smed-β-catenin-2 functions solely in adhesion, whereas Smed-β-catenin-1 functions solely in Wnt signal transduction. Despite divergent functional domains, these two proteins still maintain significant similarity (~44% each) to human β-catenin.
The duplicated β-catenins in planaria clearly illustrate how the multiple functional domains present in the bifunctional protein can be partitioned between two monofunctional proteins (Fig. 1).49 Smed-β-catenin-1, which functions solely in Wnt signaling, has the GSK-3 motif, has a functional C-terminal transactivation domain, and binds TCF but not α-catenin. It lacks the α-catenin binding domain and the very C-terminal PDZ binding domain required for the cell-cell adhesion function. Smed-β-catenin-1 can induce axis formation when assayed in Xenopus embryos.50 Conversely, Smed-β-catenin-2, which functions solely in cell adhesion, has the α-catenin binding domain and the C-terminal PDZ binding domain, and binds cadherin/α-catenin but not TCF or other Wnt pathway factors. The N-terminal GSK-3 motif of Smed-β-catenin-2 is missing and it lacks a functional C-terminal transactivation domain. Smed-β-catenin-2 does not induce axis formation when assayed in Xenopus embryos.50
Despite the very low level of identity to each other and to human β-catenin, the C. elegans β-catenins do not appear to be as fully diverged in function as the two planaria proteins (Fig. 1).49 However, it has been shown that overexpression of HMP-2 or WRM-1 from the bar-1 promoter can rescue the developmental defects in bar-1(-) mutants.8 Two recent papers have also suggested that HMP-2, when released from the adherens junction, can enter the nucleus and partially suppress the phenotype due to a lack of Wnt signaling.51,52 However, the molecular mechanisms by which HMP-2 or WRM-1 should exert these effects remains unknown as no direct molecular or biochemical analyses were performed. Whether HMP-2 can function in POP-1 nuclear export, like WRM-1, or in transcription activation, like BAR-1 and SYS-1, is unknown. Similarly, it is not clear whether WRM-1 rescues bar-1(-) by functioning as a coactivator or through an indirect effect. Unlike BAR-1, which exhibits a classic N-terminal GSK-3 phosphorylation motif,8,40,41 the HMP-2 protein has an N-terminal truncation which deletes the region which would contain the GSK-3 phosphorylation motif. Therefore, were HMP-2 to function as a coactivator for POP-1, it would not be subject to the same APC-Axin-GSK-3 cytoplasmic degradation that establishes the unstimulated ground state for canonical Wnt signaling.
It appears that the four β-catenins in C. elegans have each acquired mutations in different domains that restrict their function to one or a subset of those for the ancestral β-catenin. However, despite accumulating mutations, some functional domains remain at least partially operational and can be detected under certain experimental conditions. Why the C. elegans β-catenin proteins are so divergent at the amino acid sequences is unclear, although a high sequence divergence has also been noted for other worm proteins functioning in the Wnt pathway. For example, the two axin-related proteins PRY-1 and AXL-1, are both functional Axin orthologs and yet display only 20% amino acid identity when compared with each other, and are only 14–16% identical to Drosophila and vertebrate Axins.53
“In trying to make something new, half the undertaking lies in discovering whether it can be done. Once it has been established that it can, duplication is inevitable.” —Helen Gahagan
Did the β-Catenin Gene Duplications Drive the Requirement for POP-1 Reduction?
We suggest that immediately following a β-catenin gene duplication, the overall level of β-catenin is likely to be elevated, leading to an undesirable hyperstimulation of the Wnt pathway. There would therefore be considerable selective pressure to curb the over activation of Wnt target genes. This could be achieved by one or more of the following mechanisms. First, expression at one, or the other, or both of the duplicated loci could be reduced, or even shut down completely as in the case of pseudogenes. There is accumulating evidence from both yeast and mammals that a substantial decrease in the level of gene expression occurs following gene duplication, and in certain cases this can be shown to be beneficial by rebalancing gene dosage after duplication.54 Second, β-catenin could be sequestered to other subcellular locations outside of the nucleus. Third, the expression of different β-catenins could be temporally and spatially separated. Fourth, the level of nuclear TCF protein could be lowered. We propose that a combination of all these possible adaptations took place following multiple β-catenin gene duplication events during C. elegans evolution.
At an early point in C. elegans evolutionary history, an ancestral multifunctional β-catenin gene duplicated. We suggest that worms responded to the elevated level of β-catenin initially by reducing the overall expression of β-catenin. In addition, sequence divergence in one of the β-catenin genes (“adhesion” β-catenin) led to a progressive reduction in binding to Wnt components. As the function of this protein became more adhesion-specific, it was sequestered away from the nucleus. This “adhesion” β-catenin subsequently lost the GSK-3 phosphorylation sites as a result of N-terminal truncation. We suggest that this is how the hmp-2 gene came to be.
The other β-catenin gene that resulted from the duplication event maintained primarily a Wnt pathway coactivator function (“Wnt” β-catenin). It was relieved, therefore, from selective pressure to maintain functional adhesion domains, and began to accumulate inactivating mutations in those domains required solely for the cell-cell adhesion function—i.e., the α-catenin binding and the C-terminal PDZ binding domains. Following the initial gene duplication event, we propose that a subsequent gene duplication event occurred involving specifically this “Wnt” β-catenin gene. In addition to the worms responding with a further reduction in the level of the two β-catenins, both capable of coactivating Wnt targets, we propose that changes in the cis regulatory sequences resulted in their non-overlapping temporal and spatial expression patterns during development. The bar-1 precursor was expressed only post-embryonically. The sys-1 precursor was expressed in embryos and, when expressed post-embryonically, was expressed in distinct tissues from those expressing bar-1.
Reducing the level of nuclear TCF protein could also reduce Wnt signal strength in cells expressing abnormally high levels of β-catenin. We propose that the need to reduce nuclear levels of POP-1 in order to generate the correct Wnt signal coevolved with the reductions in β-catenin expression that followed each β-catenin gene duplication. As β-catenin expression levels dropped over evolutionary time, the level of coactivating β-catenin in any given cell became limiting as the functions of duplicated β-catenin started to diverge. If POP-1 levels had remained unchanged, it would have led to an abundance of POP-1/corepressor interactions and permanent repression of Wnt target genes. Therefore, the nuclear level of POP-1 was reduced to accommodate the limiting level of coactivating β-catenin.
POP-1 levels must be reduced for Wnt target gene activation, and the worm achieves this is by exporting POP-1 from the nucleus of the Wnt-responsive cell. The fourth C. elegans variant β-catenin, WRM-1, functions, along with the NLK homolog LIT-1, to promote POP-1 nuclear export. Is the WRM-1 function the result of a worm-specific adaptation for the need to reduce POP-1 levels, or might this instead indicate another general function of β-catenins? A recent report shows that in Xenopus, phosphorylation of certain TCF proteins by the HIPK2 kinase requires β-catenin, which appears to function as a scaffold bringing the TCF and kinase together.55,56 The end result is the removal of a repressive TCF/LEF from target promoters and the replacement by an activating TCF. These findings and our results on WRM-1 are consistent with an ancestral form of β-catenin having a function promoting TCF phosphorylation and its subsequent removal from target genes or nucleus. In vertebrates, NLK phosphorylation of certain TCFs has also been shown to inhibit their DNA-binding ability.57,58 Therefore, although the mechanistic details differ between the worm and vertebrates with respect to how NLK regulates TCF proteins, the end result appears to be the same—NLK phosphorylation of a TCF results in removal of that TCF from the Wnt target genes and subsequent activation of the same genes. We suggest that WRM-1 maintains the scaffold function of the ancestral β-catenin, becoming a subunit for the LIT-1 kinase complex, which has only become apparent due to the unique set of experimental conditions afforded by C. elegans: (1) Subfunctionalization of the β-catenins has occurred to a point where WRM-1, during wildtype development, may retain only this function; (2) the invariant C. elegans cell lineage which permits live analysis of a precisely defined Wnt signal event at single-cell resolution (i.e., one can distinguish responding vs. non-responding sister cells) over multiple cell divisions; and (3) the developmental readout of the reception of that Wnt signal by a single cell is dramatic (normal gut or no gut), rapid, and genetically tractable.
“You have made your way from worm to man, and much in you is still worm.” —Friedrich Nietzsche
And So… What Might this all Mean?
The β-catenin evolutionary history presented here is, of course, highly speculative. However, the addition of complete genome sequences from organisms that are informative regarding the evolutionary relationship between protostomes (e.g., worms, arthropods, mollusks) and deuterostomes (echinoderms and chordates) should lead to a clearer understanding of the evolutionary history of the β-catenin genes.
Wnt signal strength not only plays a critical role during development, but, if deregulated, can contribute to embryonic defects and a number of human cancers. The key pathway changes associated with cancer lead to aberrant stabilization of β-catenin and hyperstimulation of Wnt pathway target genes. We argue here that studies in C. elegans suggest that in response to reductions in β-catenin levels, the worm may compensate by lowering nuclear TCF levels in order to maintain a balanced Wnt signal in the presence of a limiting amount of coactivator and a relative preponderance of corepressor (a form of dosage compensation). Can vertebrates/humans also modify Wnt signal strength through modification of nuclear TCF levels? There is evidence that certain human tumor cell lines exhibit selective export from the nucleus of particular TCF/LEFs, perhaps in an attempt to rebalance the Wnt signal strength by varying the β-catenin to TCF ratio within the nucleus.59,60 In the face of aberrantly high β-catenin levels as a result of pro-oncogenic mutations in APC or β-catenin itself, dialing down the level of nuclear TCF available would lead to a reduction in Wnt signal strength. In addition, there is a clear relationship in mammals between loss of cell-cell adhesion and the transformed phenotype (which includes uncontrolled growth and increased cell migration/metastasis).61 Increased expression of cadherins can repress the transformed phenotype, presumably by binding β-catenin and sequestering it from the nucleus.62,63 These studies suggest that there is a very dynamic equilibrium between the different sub-cellular pools of β-catenin. The study of this homeostatic regulation of Wnt signal strength has been predominated to date by analysis of β-catenin levels. We believe that our understanding of Wnt signaling in mammalian development and tumorigenesis can be enhanced by a more careful analysis of the possible role of TCF level modulation.
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/19156
References
- 1.Costa M, Raich W, Agbunag C, Leung B, Hardin J, Priess JR. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J Cell Biol. 1998;141:297–308. doi: 10.1083/jcb.141.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisenmann DM, Maloof JN, Simske JS, Kenyon C, Kim SK. The beta-catenin homolog BAR-1 and LET-60 Ras coordinately regulate the Hox gene lin-39 during Caenorhabditis elegans vulval development. Development. 1998;125:3667–80. doi: 10.1242/dev.125.18.3667. [DOI] [PubMed] [Google Scholar]
- 3.Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, et al. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell. 1997;90:707–16. doi: 10.1016/S0092-8674(00)80531-0. [DOI] [PubMed] [Google Scholar]
- 4.C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–8. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 5.Brembeck FH, Ros´rio M, Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr Opin Genet Dev. 2006;16:51–9. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Stepniak E, Radice GL, Vasioukhin V. Adhesive and signaling functions of cadherins and catenins in vertebrate development. Cold Spring Harb Perspect Biol. 2009;1:a002949. doi: 10.1101/cshperspect.a002949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korswagen HC, Herman MA, Clevers HC. Distinct beta-catenins mediate adhesion and signalling functions in C. elegans. Nature. 2000;406:527–32. doi: 10.1038/35020099. [DOI] [PubMed] [Google Scholar]
- 8.Natarajan L, Witwer NE, Eisenmann DM. The divergent Caenorhabditis elegans beta-catenin proteins BAR-1, WRM-1 and HMP-2 make distinct protein interactions but retain functional redundancy in vivo. Genetics. 2001;159:159–72. doi: 10.1093/genetics/159.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein B. Induction of gut in Caenorhabditis elegans embryos. Nature. 1992;357:255–7. doi: 10.1038/357255a0. [DOI] [PubMed] [Google Scholar]
- 10.Meneghini MD, Ishitani T, Carter JC, Hisamoto N, Ninomiya-Tsuji J, Thorpe CJ, et al. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399:793–7. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- 11.Shin TH, Yasuda J, Rocheleau CE, Lin R, Soto M, Bei Y, et al. MOM-4, a MAP kinase kinase kinase-related protein, activates WRM-1/LIT-1 kinase to transduce anterior/posterior polarity signals in C. elegans. Mol Cell. 1999;4:275–80. doi: 10.1016/S1097-2765(00)80375-5. [DOI] [PubMed] [Google Scholar]
- 12.Thorpe CJ, Schlesinger A, Carter JC, Bowerman B. Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell. 1997;90:695–705. doi: 10.1016/S0092-8674(00)80530-9. [DOI] [PubMed] [Google Scholar]
- 13.Rocheleau CE, Yasuda J, Shin TH, Lin R, Sawa H, Okano H, et al. WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell. 1999;97:717–26. doi: 10.1016/S0092-8674(00)80784-9. [DOI] [PubMed] [Google Scholar]
- 14.Lo MC, Gay F, Odom R, Shi Y, Lin R. Phosphorylation by the beta-catenin/MAPK complex promotes 14-3-3-mediated nuclear export of TCF/POP-1 in signal-responsive cells in C. elegans. Cell. 2004;117:95–106. doi: 10.1016/S0092-8674(04)00203-X. [DOI] [PubMed] [Google Scholar]
- 15.Shetty P, Lo MC, Robertson SM, Lin R. C. elegans TCF protein, POP-1, converts from repressor to activator as a result of Wnt-induced lowering of nuclear levels. Dev Biol. 2005;285:584–92. doi: 10.1016/j.ydbio.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 16.Lin R, Thompson S, Priess JR. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell. 1995;83:599–609. doi: 10.1016/0092-8674(95)90100-0. [DOI] [PubMed] [Google Scholar]
- 17.Calvo D, Victor M, Gay F, Sui G, Luke MP, Dufourcq P, et al. A POP-1 repressor complex restricts inappropriate cell type-specific gene transcription during Caenorhabditis elegans embryogenesis. EMBO J. 2001;20:7197–208. doi: 10.1093/emboj/20.24.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavallo RA, Cox RT, Moline MM, Roose J, Polevoy GA, Clevers H, et al. Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature. 1998;395:604–8. doi: 10.1038/26982. [DOI] [PubMed] [Google Scholar]
- 19.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–12. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 20.Herman MC. C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity. Development. 2001;128:581–90. doi: 10.1242/dev.128.4.581. [DOI] [PubMed] [Google Scholar]
- 21.Herman MA. Control of cell polarity by noncanonical Wnt signaling in C. elegans. Semin Cell Dev Biol. 2002;13:233–41. doi: 10.1016/S1084-9521(02)00051-4. [DOI] [PubMed] [Google Scholar]
- 22.Korswagen HC. Canonical and non-canonical Wnt signaling pathways in Caenorhabditis elegans: variations on a common signaling theme. Bioessays. 2002;24:801–10. doi: 10.1002/bies.10145. [DOI] [PubMed] [Google Scholar]
- 23.Thorpe CJ, Schlesinger A, Bowerman B. Wnt signalling in Caenorhabditis elegans: regulating repressors and polarizing the cytoskeleton. Trends Cell Biol. 2000;10:10–7. doi: 10.1016/S0962-8924(99)01672-4. [DOI] [PubMed] [Google Scholar]
- 24.Maduro MF, Kasmir JJ, Zhu J, Rothman JH. The Wnt effector POP-1 and the PAL-1/Caudal homeoprotein collaborate with SKN-1 to activate C. elegans endoderm development. Dev Biol. 2005;285:510–23. doi: 10.1016/j.ydbio.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 25.Siegfried KR, Kidd AR, 3rd, Chesney MA, Kimble J. The sys-1 and sys-3 genes cooperate with Wnt signaling to establish the proximal-distal axis of the Caenorhabditis elegans gonad. Genetics. 2004;166:171–86. doi: 10.1534/genetics.166.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kidd AR, 3rd, Miskowski JA, Siegfried KR, Sawa H, Kimble J. A beta-catenin identified by functional rather than sequence criteria and its role in Wnt/MAPK signaling. Cell. 2005;121:761–72. doi: 10.1016/j.cell.2005.03.029. [DOI] [PubMed] [Google Scholar]
- 27.Miskowski J, Li Y, Kimble J. The sys-1 gene and sexual dimorphism during gonadogenesis in Caenorhabditis elegans. Dev Biol. 2001;230:61–73. doi: 10.1006/dbio.2000.9998. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Phillips BT, Amaya MF, Kimble J, Xu W. The C. elegans SYS-1 protein is a bona fide beta-catenin. Dev Cell. 2008;14:751–61. doi: 10.1016/j.devcel.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang S, Shetty P, Robertson SM, Lin R. Binary cell fate specification during C. elegans embryogenesis driven by reiterated reciprocal asymmetry of TCF POP-1 and its coactivator beta-catenin SYS-1. Development. 2007;134:2685–95. doi: 10.1242/dev.008268. [DOI] [PubMed] [Google Scholar]
- 30.Phillips BT, Kidd AR, 3rd, King R, Hardin J, Kimble J. Reciprocal asymmetry of SYS-1/beta-catenin and POP-1/TCF controls asymmetric divisions in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:3231–6. doi: 10.1073/pnas.0611507104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang XD, Huang S, Lo MC, Mizumoto K, Sawa H, Xu W, et al. Distinct and mutually inhibitory binding by two divergent β-catenins coordinates TCF levels and activity in C. elegans. Development. 2011;138:4255–65. doi: 10.1242/dev.069054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graham TA, Ferkey DM, Mao F, Kimelman D, Xu W. Tcf4 can specifically recognize beta-catenin using alternative conformations. Nat Struct Biol. 2001;8:1048–52. doi: 10.1038/nsb718. [DOI] [PubMed] [Google Scholar]
- 33.Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Crystal structure of a beta-catenin/Tcf complex. Cell. 2000;103:885–96. doi: 10.1016/S0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- 34.Poy F, Lepourcelet M, Shivdasani RA, Eck MJ. Structure of a human Tcf4-beta-catenin complex. Nat Struct Biol. 2001;8:1053–7. doi: 10.1038/nsb720. [DOI] [PubMed] [Google Scholar]
- 35.Peifer M, Berg S, Reynolds AB. A repeating amino acid motif shared by proteins with diverse cellular roles. Cell. 1994;76:789–91. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 36.Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–82. doi: 10.1016/S0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- 37.Eastmond PJ, Graham IA. The multifunctional protein AtMFP2 is co-ordinately expressed with other genes of fatty acid beta-oxidation during seed germination in Arabidopsis thaliana (L.) Heynh. Biochem Soc Trans. 2000;28:95–9. doi: 10.1042/bst0280095. [DOI] [PubMed] [Google Scholar]
- 38.Kimelman D, Xu W. beta-catenin destruction complex: insights and questions from a structural perspective. Oncogene. 2006;25:7482–91. doi: 10.1038/sj.onc.1210055. [DOI] [PubMed] [Google Scholar]
- 39.Xu W, Kimelman D. Mechanistic insights from structural studies of beta-catenin and its binding partners. J Cell Sci. 2007;120:3337–44. doi: 10.1242/jcs.013771. [DOI] [PubMed] [Google Scholar]
- 40.Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 1997;16:3797–804. doi: 10.1093/emboj/16.13.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–54. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 42.Schneider SQ, Finnerty JR, Martindale MQ. Protein evolution: structure-function relationships of the oncogene beta-catenin in the evolution of multicellular animals. J Exp Zool B Mol Dev Evol. 2003;295:25–44. doi: 10.1002/jez.b.6. [DOI] [PubMed] [Google Scholar]
- 43.Wikramanayake AH, Hong M, Lee PN, Pang K, Byrum CA, Bince JM, et al. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–50. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- 44.Hobmayer B, Rentzsch F, Kuhn K, Happel CM, von Laue CC, Snyder P, et al. WNT signalling molecules act in axis formation in the diploblastic metazoan Hydra. Nature. 2000;407:186–9. doi: 10.1038/35025063. [DOI] [PubMed] [Google Scholar]
- 45.Hobmayer E, Hatta M, Fischer R, Fujisawa T, Holstein TW, Sugiyama T. Identification of a Hydra homologue of the beta-catenin/plakoglobin/armadillo gene family. Gene. 1996;172:155–9. doi: 10.1016/0378-1119(96)00162-X. [DOI] [PubMed] [Google Scholar]
- 46.Schneider SQ, Bowerman B. beta-Catenin asymmetries after all animal/vegetal- oriented cell divisions in Platynereis dumerilii embryos mediate binary cell-fate specification. Dev Cell. 2007;13:73–86. doi: 10.1016/j.devcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 47.Grimson MJ, Coates JC, Reynolds JP, Shipman M, Blanton RL, Harwood AJ. Adherens junctions and beta-catenin-mediated cell signalling in a non-metazoan organism. Nature. 2000;408:727–31. doi: 10.1038/35047099. [DOI] [PubMed] [Google Scholar]
- 48.Gamulin V, Muller IM, Muller WEG. Sponge proteins are more similar to those of Homo sapiens than to Caenorhabditis elegans. Biol J Linn Soc Lond. 2000;71:821–8. doi: 10.1111/j.1095-8312.2000.tb01293.x. [DOI] [Google Scholar]
- 49.Chai G, Ma C, Bao K, Zheng L, Wang X, Sun Z, et al. Complete functional segregation of planarian beta-catenin-1 and -2 in mediating Wnt signaling and cell adhesion. J Biol Chem. 2010;285:24120–30. doi: 10.1074/jbc.M110.113662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iglesias M, Gomez-Skarmeta JL, Saló E, Adell T. Silencing of Smed-betacatenin1 generates radial-like hypercephalized planarians. Development. 2008;135:1215–21. doi: 10.1242/dev.020289. [DOI] [PubMed] [Google Scholar]
- 51.Putzke AP, Rothman JH. Repression of Wnt signaling by a Fer-type nonreceptor tyrosine kinase. Proc Natl Acad Sci U S A. 2010;107:16154–9. doi: 10.1073/pnas.1006600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sumiyoshi E, Takahashi S, Obata H, Sugimoto A, Kohara Y. The β-catenin HMP-2 functions downstream of Src in parallel with the Wnt pathway in early embryogenesis of C. elegans. Dev Biol. 2011;355:302–12. doi: 10.1016/j.ydbio.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 53.Oosterveen T, Coudreuse DY, Yang PT, Fraser E, Bergsma J, Dale TC, et al. Two functionally distinct Axin-like proteins regulate canonical Wnt signaling in C. elegans. Dev Biol. 2007;308:438–48. doi: 10.1016/j.ydbio.2007.05.043. [DOI] [PubMed] [Google Scholar]
- 54.Qian W, Liao BY, Chang AY, Zhang J. Maintenance of duplicate genes and their functional redundancy by reduced expression. Trends Genet. 2010;26:425–30. doi: 10.1016/j.tig.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hikasa H, Ezan J, Itoh K, Li X, Klymkowsky MW, Sokol SY. Regulation of TCF3 by Wnt-dependent phosphorylation during vertebrate axis specification. Dev Cell. 2010;19:521–32. doi: 10.1016/j.devcel.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hikasa H, Sokol SY. Phosphorylation of TCF proteins by homeodomain-interacting protein kinase 2. J Biol Chem. 2011;286:12093–100. doi: 10.1074/jbc.M110.185280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ishitani T, Ninomiya-Tsuji J, Matsumoto K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:1379–89. doi: 10.1128/MCB.23.4.1379-1389.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishitani T, Ninomiya-Tsuji J, Nagai S, Nishita M, Meneghini M, Barker N, et al. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature. 1999;399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- 59.Najdi R, Holcombe RF, Waterman ML. Wnt signaling and colon carcinogenesis: beyond APC. J Carcinog. 2011;10:5. doi: 10.4103/1477-3163.78111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Najdi R, Syed A, Arce L, Theisen H, Ting JH, Atcha F, et al. A Wnt kinase network alters nuclear localization of TCF-1 in colon cancer. Oncogene. 2009;28:4133–46. doi: 10.1038/onc.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gottardi CJ, Wong E, Gumbiner BM. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J Cell Biol. 2001;153:1049–60. doi: 10.1083/jcb.153.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong AS, Gumbiner BM. Adhesion-independent mechanism for suppression of tumor cell invasion by E-cadherin. J Cell Biol. 2003;161:1191–203. doi: 10.1083/jcb.200212033. [DOI] [PMC free article] [PubMed] [Google Scholar]