Abstract
Carboxy-terminal binding protein (CtBP) is a well-known corepressor of several DNA binding transcription factors in Drosophila as well as in mammals. CtBP is implicated in Polycomb Group (PcG) complex-mediated transcriptional repression because it can bind to some PcG proteins, and mutation of the ctbp gene in flies results in lost PcG protein recruitment to Polycomb Response Elements (PREs) and lost PcG repression. However, the mechanism of reduced PcG DNA binding in CtBP mutant backgrounds is unknown. We show here that in a Drosophila CtBP mutant background, intergenic transcripts are induced across several PRE sequences and this corresponds to reduced DNA binding by PcG proteins Pleiohomeotic (PHO) and Polycomb (Pc), and reduced trimethylation of histone H3 on lysine 27, a hallmark of PcG repression. Restoration of CtBP levels by expression of a CtBP transgene results in repression of intergenic transcripts, restored PcG binding, and elevated trimethylation of H3 on lysine 27. Our results support a model in which CtBP regulates expression of intergenic transcripts that controls DNA binding by PcG proteins and subsequent histone modifications and transcriptional activity.
Keywords: polycomb, transcription, non-coding rna, repression
Hox gene expression in the Drosophila bithorax (BX-C) complex (Ubx, abd-A, and abd-B genes) is governed by both spatially and temporally regulated expression of the segmentation genes. After decay of the segmentation gene products, hox gene expression patterns are maintained by Polycomb Group (PcG) and Trithorax Group (TxG) proteins [Maeda and Karch, 2006]. PcG proteins comprise at least two complexes, PRC1 and PRC2, which are involved in the maintenance of the silent state, whereas TxG proteins maintain active gene expression [Simon and Tamkun, 2002]. PcG complexes bind to specific DNA regions termed Polycomb Response Elements (PREs) to mediate their effects on transcriptional repression. These elements are sometimes referred to as memory elements or maintenance elements (ME) because often they can switch between repressed and active modes depending upon the function of either PcG or TxG proteins, respectively [Rank et al., 2002]. The mechanism of specific DNA binding by PcG complexes was long considered an enigma because the individual PcG proteins do not possess DNA binding site specificity. This conundrum was partially solved with the molecular cloning of the cDNA encoding Drosophila PcG protein, Pleiohomeotic (PHO), which contains four zinc fingers and binds to specific DNA sequences within numerous PREs [Mihaly et al., 1998]. This suggested the role of specific DNA binding proteins in recruitment of PcG complexes to DNA. Association of PHO with E(z), Polyhomeotic (Ph), and Polycomb (Pc) [Mohd-Sarip et al., 2002; Wang et al., 2004], components of the PRC2 and PRC1 complexes, suggested a role for PHO in Polycomb recruitment to DNA [Schuettengruber et al., 2007; Schwartz and Pirrotta, 2007]. However, the molecular mechanism of PHO recruitment of PcG proteins to DNA is still not clear [Poux et al., 2001; Savla et al., 2008].
Yeast two-hybrid studies have shown direct interaction of Xenopus Pc with the corepressor protein, C-terminal binding protein (CtBP) [Sewalt et al., 1999]. Drosophila repressors such as Snail, Knirps, and Krüppel exert short-range transcriptional repressive activity by recruiting CtBP to DNA [Nibu and Levine, 2001; Nibu et al., 2003], but the association of CtBP with some PcG proteins suggests a role in PcG-mediated gene repression. We previously showed by transgenic studies that human YY1, the vertebrate counterpart of Drosophila PHO, is able to mediate transcriptional silencing in a PcG-dependent fashion and can phenotypically correct PHO mutant flies [Atchison et al., 2003]. We also found that, like PHO, YY1 is able to recruit PcG complexes to PRE sequences [Srinivasan and Atchison, 2004; Wilkinson et al., 2006]. Interestingly, we also showed that YY1 interacts with CtBP and is involved in PcG repression [Atchison et al., 2003]. Subsequently, we found that in a heterozygous CtBP mutant background (homozygous ctbp mutation is lethal), there is loss of YY1 DNA binding to PRE sequences, lost PcG recruitment, and decreased histone modification marks pertaining to Polycomb regulation [Srinivasan and Atchison, 2004]. This suggested a unique but still unknown role of CtBP in controlling PcG-mediated gene regulation.
Recently, it was found that gene regulation in the BX-C is accompanied by non-coding transcription through cis-regulatory sequences, and expression of these transcripts changes dynamically throughout development [Lempradl and Ringrose, 2008]. The first non-coding transcripts in the fly BX-C complex were described more than two decades ago [Lewis, 1985; Celniker and Lewis, 1987]. The pattern of non-coding transcription was similar to that of hox genes and was collinear with the regulatory domains of the hox gene clusters. The segmentation genes regulate these early transient transcripts as segmentation gene mutations alter the expression pattern of early non-coding RNAs (ncRNAs) [Sanchez-Herrero and Akam, 1989]. These transcripts may antagonize PcG function because there is a tight correlation between the reversal of PcG-mediated silencing and non-coding transcription through regulatory regions of the BX-C [Bae et al., 2002; Bender and Fitzgerald, 2002; Hogga and Karch, 2002; Rank et al., 2002; Schmitt et al., 2005]. Recruitment of Ash1 (a TxG protein) by transcripts located in the Ubx region mediates transcriptional activation [Sanchez-Elsner et al., 2006]. It has also been proposed that transcriptional elongation of bxd ncRNAs by TAC1 (a TxG protein) represses Ubx transcription [Petruk et al., 2006].
In this study, we demonstrate that short intergenic transcripts across PRED, sex combs reduced (scr) and engrailed (en) PRE regions are induced in ctbp+/− flies compared to wild-type flies. This is accompanied by reduced occupancy by PcG proteins at these PRE sequences, and reduced histone H3 trimethylation of lysine 27. We further show that these phenomena are reversed when transgenic CtBP expression is induced in the mutant background, suggesting that optimum levels of CtBP are essential for repression of intergenic transcripts and for PcG-mediated gene regulation.
MATERIALS AND METHODS
DROSOPHILA LINES, TRANSGENIC CtBP FLY LINE AND CROSSES
The BGUZ/CtBP;TM3SB and ctbp03463 fly lines were described previously [Srinivasan and Atchison, 2004; Wilkinson et al., 2006]. For generation of hspFLAG-dCtBP transgenic flies, CtBP full-length cDNA was cloned into hsp70-driven pry-derived vectors [Atchison et al., 2003] and coinjected with a transposase expressing plasmid (phsπ) into dechorionated ry506 embryos (Genetic Services, Inc.). Eclosed flies were backcrossed to ry506 and progeny were screened for transgene incorporation by appearance of ry+ eyes. For CtBP rescue experiments, hspFLAG-dCtBP flies were crossed to ctbp03463 flies and embryos were heat-shocked twice for 30 min at 37°C with an interval of 2 h.
ANALYSIS OF PRE-SEQUENCES FOR PUTATIVE PHO/YY1 BINDING SITES
Based on analysis and identification of various PRE regions in Drosophila by Ringrose et al. [2003], we chose PRED, scr, and en PRE regions for evaluating YY1/PHO binding sites. The details of the PRE sequences used for analysis are as follows. PRED: the sequence starts at 12,589,500 bp and ends at 12,590,299 bp. It is 23,000 bp upstream of the bxd gene that is known to be a Polycomb-regulated gene. The en PRE: the sequence starts at 6,592,700 bp and ends at 6,593,199 bp. Engrailed is the nearest gene regulated by this PRE element. The Scr PRE: the sequence spans 2,711,922–2,712,492 and is upstream of the protein coding region. Sequence analyses were done on version 3.1 of the Drosophila melanogaster genome. We used the MathInspector database (available with the Genomatix Software package), which predicts transcription factor binding sites (http://www.genomatix.de/), using a Core Similarity of 0.9–0.95 with the Matrix Similarity “Optimized.” The “core sequence” of a matrix is defined as the highest conserved positions of the matrix. The Matrix Similarity was determined by the highest conserved nucleotide of each position in the matrix of the binding sequence. Since YY1/PHO binding sites are quite degenerate, we chose to use “Optimized” Matrix Similarity, to minimize false positives. It should be noted that the binding sites analyzed for the PRED sequence corresponded to the sites validated by Fritsch et al. [1999].
RNA EXTRACTION AND RT-PCR ANALYSIS
Embryos were dechorionated with 50% bleach (Chlorox™) and then washed twice with PBS containing 0.01% Triton X-100. RNA was extracted from embryos using TRIZOL (Invitrogen) according to the manufacturer's protocol with minor modifications. Briefly, the washed embryo pellet was resuspended and homogenized in 200–300 μl of TRIZOL reagent using plastic pestles. The volume of TRIZOL was made up to 1 ml, samples were incubated at room temperature (RT) for 5 min, 200 μl of chloroform was added, and vigorously vortexed for 10 min. The remaining steps were performed according to the manufacturer's protocol. RNA pellets were dissolved in DEPC-treated water and 5 μg of total RNA was taken for first-strand cDNA synthesis (Superscript II Kit; Invitrogen) using both random hexamers and oligo(dT) primers according to the manufacturer's protocol. Conventional PCR was performed with Taq polymerase (ABI Biosystems) using primers (Table I) encompassing the PRE regions. Real-time PCR was performed with a Light Cycler System (Roche Molecular Biochemicals) at least twice in duplicate using SYBR Green (Sigma, St. Louis, MO). Normalization was performed with respect to β-actin and relative amounts were further calculated based on CT values of wild-type controls.
TABLE I.
Primers used for RT-PCR analyses to detect intergenic transcripts across PRE-D, scr and en PRE sequences.
| Primer name | Primer sequence |
|---|---|
| PRED sense 1F | CAGCCGTGCGGTATGGAGAG |
| PRED sense 1R | CTTGCAAAAGCGGCTATGAAAAG |
| PRED sense 2F | ATAAAACCCCAGTGCGAAATGC |
| PRED sense 2R | TGCGCGTAGTCTTATCTGTATCTCG |
| PRED sense 3F | TAAAGCGAGAGCGATCCGAGC |
| PRED sense 3R | AAACGGCCATTACGAACGACAG |
| PRED sense 6F | TTCGGGCTTGTATTCGTGTTTTG |
| PRED sense 6R | TTACGGCCCTTTTATAGATGTTGC |
| PRED sense 5F | TACGCACGTCAGACTTGGAATAGC |
| PRED sense 5R | CAAGCCCGAAAAAGAAGAAGAAGC |
| PRED sense 4F | AAAACGGCCATTACGAACGACAG |
| PRED sense 4R | GACGTGCGTAAGAGCGAGATACAG |
| PRED antisense 1F | CGGCCCTTTTATAGATGTTGCAAC |
| PRED antisense 1R | CCGCCGCTTCTTCTTCTTTTTC |
| PRED antisense 2F | ATAAAACCCCAGTGCGAAATGC |
| PRED antisense 2R | TGCGCGTAGTCTTATCTGTATCTCG |
| PRED antisense 3F | GCAAACATGGGCAAACACAACC |
| PRED antisense 3R | CAGCTCCGTCGCCATAACTGTC |
| PRED antisense 4F | CGTAATGGCCGTTTTAAGTGCG |
| PRED antisense 4R | TAAGCAAACATGGGCAAACACAAC |
| PRED antisense 5F | GCCCAGTGAAAATTTGGCAGC |
| PRED antisense 5R | ACAGCCGTGCGGTATGGAGAG |
| Scr 1F | AATCGGTCGAATTATTTAGCAAC |
| Scr 1R | ACTTCATCGGCAGTCTTGGAG |
| Scr 2F | GTAATTTTTATTTTTTGTTGC |
| Scr 2R | GCCCCTGCTTTCTACCATCTCC |
| Scr 3F | GAGAGGCCTTTGATTGTGTGG |
| Scr 3R | TCGACCGATTATGGAAAACTG |
| Scr4F | ATGCAGCTGGGAAATCGTTGG |
| Scr 4R | GGCCTCTCCTGGTTTATCTTTG |
| Scr 5F | AATGGCTGATTTGGGTTCTCTG |
| Scr 5R | ATAGGCCACCGGGTAACATTTTG |
| Engrailed 1F | ATCGTGTATTTAGCGTATTTTTG |
| Engrailed 1R | CAACTTTATCGACACCACCTTTAG |
| Engrailed 4F | CAGCATGCGCATAATAAAGTC |
| Engrailed 4R | TTTCGCCGGCTCACTCACTC |
| Engrailed 5F | GTTCGCATGGGGCAGGTGAC |
| Engrailed 5R | TAGCTGGCGAAGTGTGTGCG |
| Engrailed 6F | CTCGCTCGCTCGCACACAC |
| Engrailed 6R | TGACTACTTCGGAATCGCAGC |
CHROMATIN IMMUNOPRECIPITATION
Drosophila embryos from various egg-lays were dechorionated with 50% bleach, collected in PBS containing 0.04% Triton X-100, and subsequently fixed with 2% formaldehyde in 50 mM HEPES (pH 7.6), 1 mM EDTA, 0.5 mM EGTA, and 100 mM NaCl along with 3 volumes of n-heptane by vigorous rocking at RT for 15 min. Embryos were neutralized with PBS containing 0.125 M glycine and 0.01% Triton X-100 for 5 min at RT. Fixed embryos were washed with Wash Solution A (10 mM HEPES, pH 8.0, 10 mM EDTA, 0.5 mM EGTA, and 0.25% Triton X-100) for 10 min at RT and then with Wash Solution B (10 mM HEPES, pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, and 0.01% Triton X-100) for 10 min at RT. Embryos were resuspended in buffer containing 10 mM HEPES (pH 8.0), 1 mM EDTA, and 0.5 mM EGTA along with protease inhibitors (Sigma). Glass beads of 106-μm (G-4649; Sigma) were added and sonication was carried out at 6 W output power for 8 pulses of 20 s each (Ultrasonic Processor, Sonics & Materials, CT).
Sonicated chromatin (50–200 μg) was diluted in 16.7 mM Tris–HCl, pH 8.1, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton X-100 along with protease inhibitors (Sigma) and precleared for 1–2 h with protein A-ssDNA slurry (200 μg salmon sperm DNA/protein A agarose/0.5 mg/ml BSA) and 1% of the total chromatin was taken as the input sample. Immunoprecipitation was carried out at 4°C overnight with antibodies (anti-Polycomb, sc-25762; Santa Cruz Biotechnology; acetyl-K9 07-352, trimethyl K27 07-449, Upstate Biotechnology; or PHO antiserum, kindly provided by Dr. Judy Kassis). The DNA/protein/antibody complex was isolated using protein A agarose beads. Immune complexes were washed successively with low salt, high salt, and LiCl wash buffers followed by twice with TE. Two hundred microliters of elution buffer (0.1 M NaHCO3 and 1.0% SDS) was added to the beads along with RNaseA (50 μg/ml) and incubated at 37°C for 30 min followed by the addition of 4 μl of 0.5 M EDTA, 8 μl of 1 M Tris–HCl, pH 6.5, to a volume of 200 μl and Proteinase K to a final concentration of 0.5 mg/ml and incubation at 45°C for 2 h. Subsequently, 8 μl of 5 M NaCl was added and cross-linking was reversed at 65°C overnight.
DNA was extracted using the QIAquick Purification Kit (Qiagen). Real-time PCR was done using the SYBR Green system with primers encompassing different PRE regions to test for promoter occupancy. The primers used for chromatin immunoprecipitation (ChIP)-PCR are listed in Table II.
TABLE II.
Primers used for ChIP analyses across PRE-D, scr and en regions.
| Primer name | Primer sequence |
|---|---|
| ChIP_PRED F | TAAAGCGAGAGCGATCCGAGC |
| ChIP_PRED R | CAAGCCCGAAAAAGAAGAAGAAGC |
| ChIP_scr F | TCGACCGATTATGGAAAACTG |
| ChIP_scr R | ATGCAGCTGGGAAATCGTTGG |
| ChIP_eng F | ATCGTGTATTTAGCGTATTTTTG |
| ChIP_eng R | TTTCGCCGGCTCACTCACTC |
RESULTS
INDUCTION OF INTERGENIC TRANSCRIPTS IN CtBP+/− FLIES
Previously, using ChIP assays of chromatin isolated from fly embryos, and by immunofluorescence studies of salivary polytene chromosomes, we showed that in a Drosophila heterozygous CtBP mutant background, binding of PcG proteins to numerous PRE sequences is reduced compared to wild-type samples [Srinivasan and Atchison, 2004]. The mechanism of lost PcG DNA binding in ctbp+/− samples was perplexing because the wild-type ctbp allele should still provide roughly half the amount of CtBP protein. Several studies have shown that ncRNAs or intergenic transcripts are responsible for switching memory elements from a repressive mode to the active mode by interfering with PcG-mediated gene silencing [Hogga and Karch, 2002; Rank et al., 2002]. Therefore, we hypothesized that CtBP may serve to repress transcription arising from promoters that flank PRE sequences, and a 50% loss of CtBP may be sufficient to derepress these promoters allowing transcription to occur across PRE sequences thus interfering with PcG recruitment to PRE sequences.
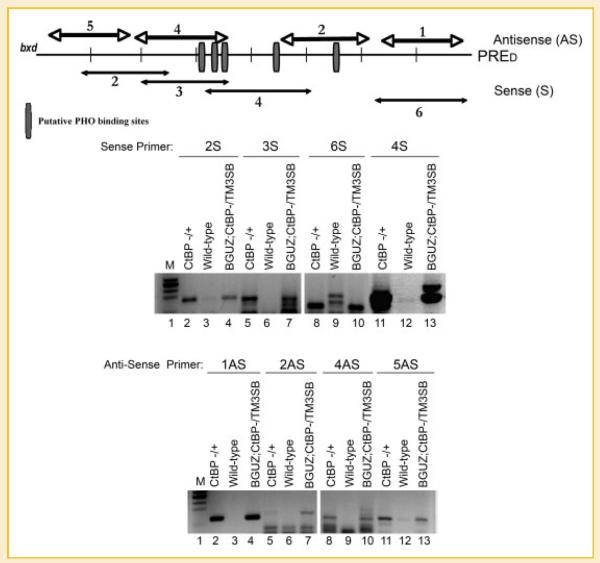
Based on computational analysis followed by experimental validation, many PRE sequences were identified by Ringrose et al. [2003]. Previous work from our lab showed that CtBP levels regulated YY1 DNA binding and PcG recruitment to different PREs in the fly genome [Srinivasan and Atchison, 2004]. Based on these data, we chose the PRED, scr, and en PRE sequences for our analyses. These sequences all contain PHO binding sites (see Figs. 1 and 2) and all exhibit reduced PcG DNA binding in a ctbp+/− background [Srinivasan and Atchison, 2004]. To search for potential transcripts across PREs that might be upregulated due to reduction in CtBP levels, we isolated RNA from fly embryos containing wild-type levels of CtBP (ry506) and from embryos containing heterozygous mutant levels of CtBP (ctbp+/− and BGUZ/CtBP;TM3SB). Using several primer pairs across the PRED sequence, we observed increased transcripts with most primer pairs in the ctbp+/− samples compared to wild type (Fig. 1). For instance, sense primer pair 2 (2S) showed an increase in transcript levels in ctbp+/− and BGUZ/CtBP;TM3SB lines compared to CtBP wild-type embryos (Fig. 1, middle panel, lanes 2–4). Likewise, primer pairs 3S, 4S, and 6S yielded transcripts in CtBP mutant backgrounds that were almost absent in wild-type embryos (Fig. 1, middle panel, lanes 5–13). In a similar manner, antisense primers 1AS, 2AS, 4AS, and 5AS detected transcripts in CtBP mutant backgrounds that were almost absent in CtBP wild-type embryos (Fig. 1, lower panel, lanes 2–13). Thus, in a CtBP mutant background, transcripts are induced across the PRED sequence.
Fig. 1.
CtBP controls intergenic transcripts at the PRED sequence. The upper panel shows the PRED region with predicted PHO binding sites (vertical rectangles). Double headed arrows show the location of primers used to detect RNA transcripts by RT-PCR along the PRED sequence. The middle and lower panels show the results of RT-PCR to detect RNA transcripts induced in CtBP mutant backgrounds compared to wild-type flies. RNA was isolated 14 h post-egg-lay. Genotypes of samples are shown above the lanes as well as primer pairs. Sizes of amplified products for sense primers 2S, 3S, 4S, and 6S are 166, 160, 175, and 121 bp, respectively. Antisense amplified products 1AS, 2AS, 4AS, and 5AS are 137, 156, 138, and 139 bp, respectively.
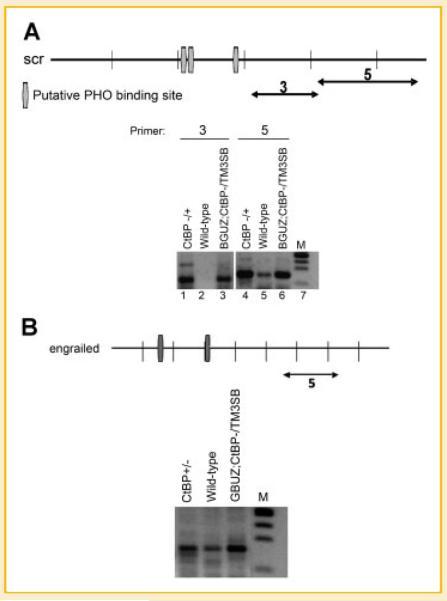
Fig. 2.
CtBP controls intergenic transcripts at the scr (A) and engrailed (B) PRE sequences. Maps of the scr and engrailed PRE regions are shown in the top panels with putative PHO binding sites represented as vertical rectangles. Primer pairs are represented by double-headed arrows. Lower panels show RT-PCR results with RNA isolated from 14 h embryos and the genotype of each sample shown above the lanes. Amplified products for scr primer pairs 3 and 5 are 122 and 150 bp, respectively, and the en amplified product is 152 bp.
These results were not unique to the PRED sequence. Similar results were obtained with two primer sets (3 and 5) at the scr PRE sequence (Fig. 2A, lanes 1–6), as well as the en PRE sequence (Fig. 2B, primer set 5). Time-course studies showed that transcripts across the PRED sequence were elevated in CtBP mutant backgrounds at 3, 10, and 14 h post-egg-lay but were near wild-type levels at 6 h (Fig. 1A, supplemental data). Time-course studies at the scr PRE showed highest transcript inductions at 3 and 10 h post-egg-lay compared to wild type (Fig. 1B, supplemental data) and at the en PRE at 14 h (Fig. 1C, supplemental data). Interestingly, all transcripts were detected with cDNA synthesized by random priming but not with oligo(dT) priming suggesting that these transcripts do not contain polyA tails (data not shown). In summary, we have detected intergenic transcripts across several PRE sequences that appear to be regulated by the levels of CtBP within the embryo.
RESTORATION OF CtBP LEVELS REVERSE TRANSCRIPT INDUCTION
To determine whether reduced levels of CtBP in the CtBP mutant backgrounds were indeed responsible for induction of transcripts, we crossed our ctbp+/− flies with a transgenic line containing a heat-shock inducible ctbp transgene (hspFLAG-dCTBP). Embryos were collected and either left untreated, or heat-shocked to induce CtBP expression. Strikingly, upon induction of CtBP, expression of the intergenic transcripts across the PRED sequence was reduced (Fig. 3, top panel, compare lanes 2 and 3, 6 and 7, 10 and 11, 14 and 15; bottom panel, lanes 2 and 3, 6 and 7, 10 and 11). As expected, very little change was observed in embryos where ctbp+/− flies were crossed with wild-type flies followed by heat-shock (Fig. 3, top panel, compare lanes 4 and 5, 8 and 9, 12 and 13, 16 and 17; bottom panel, lanes 4 and 5, 8 and 9, 12 and 13). Therefore, our data indicate that CtBP levels play a crucial role in regulation of expression of transcripts across PRE sequences.
Fig. 3.
Expression of transgenic CtBP restores repression of intergenic transcripts. ctbp+/− mutant flies were crossed to either FLAG-dCtBP transgenic flies or wild-type flies. Embryos were harvested at 3 h post-egg-lay, either left untreated or heat-shocked to induced CtBP expression, and RNA was isolated at 15 h and subjected to RT-PCR with the primers shown above the lanes. Genotypes and treatments are shown above each lane.
INTERGENIC TRANSCRIPTS CORRELATE WITH LEVELS OF PHO AND Pc DNA BINDING AND WITH HISTONE MODIFICATIONS
Previously we showed that YY1 is the functional mammalian homolog of Drosophila PHO and that YY1 can correct phenotypic defects in pho mutants flies [Atchison et al., 2003]. We also found that CtBP is required for YY1 or PHO DNA binding at PRE sequences and for subsequent recruitment of PcG proteins to DNA [Srinivasan and Atchison, 2004]. Our results above indicate that lowered levels of CtBP expression cause increased expression of intergenic transcripts across PRE regions. Therefore, we wanted to investigate whether induction of these transcripts resulted in the loss of PHO DNA binding at PRE sequences and subsequent loss of recruitment of PcG complexes. We performed ChIP experiments with wild-type and ctbp+/− embryos using various antibodies and DNA was quantitated by qPCR using primers that amplify the PRED, scr, and en PRE regions (Fig. 4). As anticipated, we found that in a CtBP mutant background, there was a reduction in occupancy by PHO at the PRED, scr, and en sequences compared to the wild type (Fig. 4A–C). This correlated with concomitant reduction in Polycomb (Pc) DNA binding and reduction in trimethylation of histone H3 on lysine 27 which is characteristic of PcG function (Fig. 4A–C). Interestingly, there was a corresponding increase in histone H3 acetylation on lysine 9 at the PRED sequence that correlates with active chromatin (Fig. 4A).
Fig. 4.

CtBP levels control PHO and Pc DNA binding and histone modifications. Wild-type and ctbp+/− embryos were harvested at 14 h and processed for ChIP assay with antibodies against PHO, Pc, H3 trimethyl-lysine 27, or H3 acetyl-lysine 9 followed by PCR with primers to the PRED (A), scr (B), or engrailed (C) PRE sequences.
CtBP MEDIATES PHO AND Pc BINDING TO PREs
We showed above that reduced levels of CtBP resulted in increased expression of intergenic transcripts across PRE sequences as well as loss of PHO binding. In addition, we showed that expression of intergenic transcripts could be reversed by overexpression of CtBP. Based on these results, we questioned whether increased CtBP levels in a ctbp+/− background would revert PHO and Pc binding to wild-type levels. To test this, ctbp+/− mutant flies were crossed with our transgenic hspFLAG-dCTBP line. Embryos were collected at 3 h, heat-shocked multiple times to induce CtBP expression, and chromatin was prepared 10 h post-egg-lay for ChIP assays with various antibodies. PCR was performed with primers spanning the PRED, scr, and en PRE sequences. Strikingly, after heat-shock we found significant increases in DNA binding by PHO and Pc to each PRE sequence, and a corresponding increase in levels of H3 trimethylation on lysine 27 compared to the non-heat-shock control samples (Fig. 5A–C). These results indicate that upon CtBP induction there is recovery of PcG complexes on PRE sequences that parallels repression of intergenic transcripts. Our results provide a direct correlation between levels of CtBP protein, upregulation of intergenic transcripts, and promoter occupancy by PRC complexes.
Fig. 5.
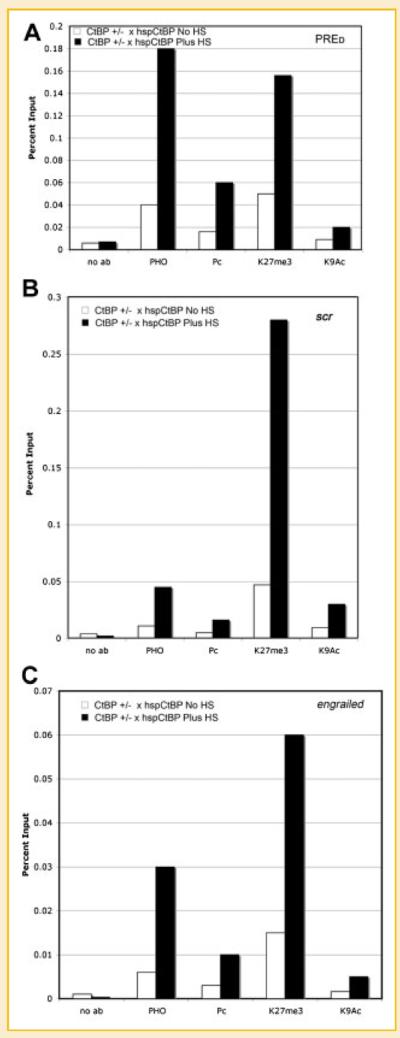
Expression of a CtBP transgene restores PHO and Pc DNA binding as well as methylation of H3 lysine 27. Wild-type and ctbp+/− embryos were harvested at 3 h, either left untreated or heat-shocked to induce CtBP expression, then processed for ChIP assay with antibodies against PHO, Pc, H3 methyl-lysine 27, or H3 acetyl-lysine 9 followed by PCR with primers to the PRED (A), scr (B), or engrailed (C) PRE sequences.
DISCUSSION
CtBP is a well-known corepressor of several DNA binding transcription factors in Drosophila (viz. Snail, Hairy, Krüppel, and Knirps) as well as in mammals [Nibu et al., 1998; Poortinga et al., 1998; Keller et al., 2000; Chinnadurai, 2007]. We show here that loss of CtBP results in elevated expression of intergenic transcripts, reduced PHO DNA binding, and concomitant loss of PcG recruitment. CtBP levels regulate intergenic transcripts across PRE sequences and these transcripts may control PHO DNA binding and subsequent PcG complex recruitment. We previously suggested that CtBP might act as a bridging molecule between YY1/PHO and PcG complexes to target PRC1/2 complexes to specific DNA sequences (PREs) [Srinivasan and Atchison, 2004]. While this remains possible, it does not readily explain the loss of PHO binding to PREs in a ctbp+/− background. Our results here are consistent with a new model in which CtBP represses transcription of intergenic transcripts that interfere with PHO DNA binding. In this model, wild-type levels of CtBP repress transcription of these intergenic transcripts thus allowing PHO DNA binding and subsequent PcG recruitment (Fig. 6). We suggest that reduced PHO binding is a secondary effect of increased intergenic transcripts. The use of intergenic transcripts to locally control PHO binding would allow a subset of PREs to be inactivated even though they have PHO binding sties.
Fig. 6.
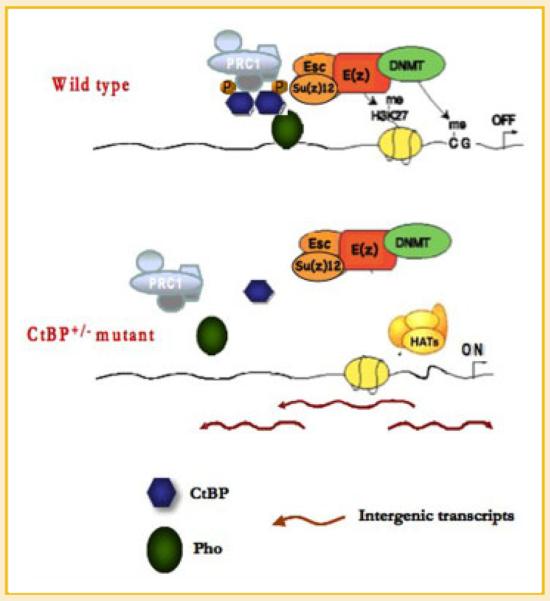
Model of CtBP regulation of intergenic transcripts and subsequent DNA binding by PHO and PcG function. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
It is well known that CtBP mediates both long-range and short-range transcriptional repression with Hairy, Knirps, Krüppel, and Snail. Hence, it might be well envisioned that a 50% loss of CtBP leads to sub-stoichiometric levels of CtBP resulting in transcriptional derepression with concomitant upregulation of intergenic transcripts across PREs. Our model (Fig. 6) predicts the existence of promoter/enhancer regions flanking PRE sequences that bind to sequence specific transcription factors that mediate their activity in a CtBP-dependent manner. Preliminary analyses of the sequences flanking PRED, scr, and en indicate putative binding sites for DNA-binding transcription factors such as Hairy, Knirps, and Snail, all of which are known to recruit CtBP for their transcriptional activity. It would be interesting to investigate whether these flanking regions do act as promoter regions for CtBP-mediated regulation, or whether more complex long-range enhancer activity is responsible for CtBP-mediated transcriptional repression of intergenic transcripts.
In Drosophila, earlier studies using in situ hybridization showed expression of non-coding transcripts across the regulatory domain of abd-A and abd-B in the same orientation as the hox genes [Zhou et al., 1999; Bae et al., 2002; Rank et al., 2002]. Interestingly, these non-coding BX-C transcripts are expressed before hox gene transcription leading to the proposition that their early expression may cause the BX-C region to adopt an open chromatin structure leading to the recruitment of activator proteins to regulatory regions [Bae et al., 2002; Hogga and Karch, 2002]. Transcription of ncRNA can regulate gene expression because forced transcription through regulatory sequences results in lost PcG repression [Hogga and Karch, 2002; Rank et al., 2002].
Our studies not only show the presence of short intergenic transcripts across various PRE regions during embryogenesis, but for the first time demonstrate that decreased expression of the corepressor CtBP (in ctbp+/− embryos) can cause induction of intergenic transcripts in vivo. The effects are striking, with reduced CtBP levels resulting in induction of intergenic transcripts, loss of DNA binding by PHO, loss of Polycomb recruitment to the cognate PRE sequences, and reduced trimethylation of histone H3 on lysine 27. When levels of CtBP protein are restored by heat-shock in transgenic flies, these transcripts are downregulated demonstrating that CtBP regulates these transcripts. With elevated CtBP levels and reduced intergenic transcription, PHO DNA binding and PcG recruitment are restored. The temporal expression pattern of intergenic transcripts in the ctbp+/− background yields transcripts peaking at the PRED and scr sequences at 3 and 10 h, while transcripts peak at 14 h at the en PRE. This correlates well with the protein expression profile. Ubx gene expression peaks in the embryonic stage, whereas engrailed expression peaks at later stages of development. Thus, these transcripts may naturally regulate PRE function during development. Transient changes in CtBP expression or post-translational mechanisms may regulate the function of CtBP to mediate transient changes in intergenic transcription levels leading to altered PHO DNA binding and PcG complex recruitment.
It is not yet clear whether the effect of intergenic transcripts on PHO DNA binding is direct or indirect. The act of transcription may directly dislodge PHO from DNA, or the mechanism of reduced PHO binding in response to lowered CtBP levels and elevated intergenic transcripts could be indirect. Histone modifications resulting from transcriptional activity could potentially provide a mechanism for controlling PHO DNA binding [Tie et al., 2009], or lowered CtBP levels might induce expression of genes that control PHO DNA binding in some other way. Additional details of the mechanism of CtBP regulating intergenic transcripts and PcG recruitment to DNA will require further studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Kassis for PHO antibody and F. Wilkinson for help with ChIP assays and for helpful discussions. We thank John Pehrson for critical reading of this manuscript. This work was supported by NIH grant GM071830.
Grant sponsor: NIH; Grant number: RO1 GM071830.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. The YY1 transcription factor functions as a PcG protein in vivo. EMBO J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae E, Calhoun VC, Levine M, Lewis EB, Drewell RA. Characterization of the intergenic RNA profile at abdominal-A and abdominal-B in the Drosophila bithorax complex. Proc Natl Acad Sci USA. 2002;99:16847–16852. doi: 10.1073/pnas.222671299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W, Fitzgerald DP. Transcription activates repressed domains in the Drosophila bithorax complex. Development. 2002;129:4923–4930. doi: 10.1242/dev.129.21.4923. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Lewis EB. Transabdominal, a dominant mutant of the Bithorax complex, produces a sexually dimorphic segmental transformation in Drosophila. Genes Dev. 1987;1:111–123. doi: 10.1101/gad.1.2.111. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Fritsch C, Brown JL, Kassis JA, Muller J. The DNA-binding Polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–3913. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- Hogga I, Karch F. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Poly-comb-mediated silencing. Development. 2002;129:4915–4922. doi: 10.1242/dev.129.21.4915. [DOI] [PubMed] [Google Scholar]
- Keller SA, Mao Y, Struffi P, Margulies C, Yurk CE, Anderson AR, Amey RL, Moore S, Ebels JM, Foley K, Corado M, Arnosti DN. dCtBP-dependent and -independent repression activities of the Drosophila Knirps protein. Mol Cell Biol. 2000;20:7247–7258. doi: 10.1128/mcb.20.19.7247-7258.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempradl A, Ringrose L. How does noncoding transcription regulate Hox genes? BioEssays. 2008;30:110–121. doi: 10.1002/bies.20704. [DOI] [PubMed] [Google Scholar]
- Lewis EB. Regulation of the genes of the bithorax complex in Drosophila. Cold Spring Harb Symp Quant Biol. 1985;50:155–164. doi: 10.1101/sqb.1985.050.01.021. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The ABC of the BX-C: The bithorax complex explained. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- Mihaly J, Mishra RK, Karch F. A conserved sequence motif in Polycomb-response elements. Mol Cell. 1998;1:1065–1066. doi: 10.1016/s1097-2765(00)80107-0. [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP. Pleiohomeotic can link Polycomb to DNA and mediate transcriptional repression. Mol Cell Biol. 2002;22:7473–7483. doi: 10.1128/MCB.22.21.7473-7483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y, Levine MS. CtBP-dependent activities of the short-range Giant repressor in the Drosophila embryo. Proc Natl Acad Sci USA. 2001;98:6204–6208. doi: 10.1073/pnas.111158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y, Zhang H, Bajor E, Barolo S, Small S, Levine M. dCtBP mediates transcriptional repression by Knirps, Kruppel, and Snail in the Drosophila embryo. EMBO J. 1998;17:7009–7020. doi: 10.1093/emboj/17.23.7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y, Senger K, Levine M. CtBP-independent repression in the Drosophila embryo. Mol Cell Biol. 2003;23:3990–3999. doi: 10.1128/MCB.23.11.3990-3999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: A Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S, McCabe D, Pirrotta V. Recruitment of components of Polycomb group chromatin complexes in Drosophila. Development. 2001;128:75–85. doi: 10.1242/dev.128.1.75. [DOI] [PubMed] [Google Scholar]
- Rank G, Prestel M, Paro R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol Cell Biol. 2002;22:8026–8034. doi: 10.1128/MCB.22.22.8026-8034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Rehmsmeier M, Dura J-M, Paro R. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev Cell. 2003;5:759–771. doi: 10.1016/s1534-5807(03)00337-x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E, Akam M. Spatially ordered transcription of regulatory DNA in the bithorax complex of Drosophila. Development. 1989;107:321–329. doi: 10.1242/dev.107.2.321. [DOI] [PubMed] [Google Scholar]
- Savla U, Benes J, Zhang J, Jones RS. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development. 2008;135:813–817. doi: 10.1242/dev.016006. [DOI] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R. Intergenic transcription through a Polycomb response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Sewalt RGAB, Gunster MJ, van der Vlag J, Satijn DPE, Otte AP. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol Cell Biol. 1999;19:777–787. doi: 10.1128/mcb.19.1.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Tamkun JW. Programming off and on states in chromatin: Mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev. 2002;12:210–218. doi: 10.1016/s0959-437x(02)00288-5. [DOI] [PubMed] [Google Scholar]
- Srinivasan L, Atchison ML. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 2004;18:2596–2601. doi: 10.1101/gad.1228204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. Hierarchical recruitment of Polycomb-group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci USA. 2006;103:19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Ashe H, Burks C, Levine M. Characterization of the transvection mediating region of the abdominal-B locus in Drosophila. Development. 1999;126:3057–3065. doi: 10.1242/dev.126.14.3057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.