Introduction
A fundamental feature of eukaryotic cells is the use of membrane-bound organelles to compartmentalize activities and serve as scaffolds for signal transduction. The best-characterized signaling pathways involve membrane-bound receptors that respond to extracellular or lumenal stimuli. In these instances, the spatial separation of an extracellular stimulus from the cytosol mandates the use of organelles as signaling platforms, as transmembrane receptors must transmit information across a lipid bilayer. However, an important gap exists in our knowledge of how stimuli from the cytosol are able to initiate specific signaling events.
How common is the use of organelles in signal transduction from cytosolic receptors? An example of this situation can be found in the study of virus-host interactions. The ability to detect cytosolic viruses depends on the RIG-I-like Receptor (RLR) family of proteins, which are soluble RNA helicases that detect viruses containing RNA (and in some cases DNA) genomes (Ablasser et al., 2009; Chiu et al., 2009; Yoneyama et al., 2004). The best characterized RLRs, RIG-I and MDA-5, detect 5’-triphosphate-containing short dsRNA and long dsRNA, respectively (Kato et al., 2008; Pichlmair et al., 2006). RLRs can either detect viral RNA directly or after RNA polymerase III-mediated transcription of microbial DNA (Ablasser et al., 2009; Chiu et al., 2009; Kato et al., 2008). Mice deficient in either of these RLRs are sensitive to different classes of viruses, underscoring both their specificity of action and their importance in immune defense (Gitlin et al., 2006; Kato et al., 2006).
Although RIG-I and MDA-5 have specificities for different ligands, both induce a common signaling pathway that triggers the expression of Type I Interferons (IFNs) and IFN-stimulated genes (ISGs). Many ISGs function as direct antiviral effectors, acting to prevent viral genome replication, viral particle assembly, or virion release from infected cells. Generally, it is thought that RLRs induce the expression of IFNs that act in both autocrine and paracrine manners to amplify ISG expression. However, ISGs can also be induced directly upon viral infection, without the need for IFN signaling (Collins et al., 2004; Mossman et al., 2001). At the receptor-proximal level, RLR-dependent responses are regulated by the adaptor protein MAVS (also called IPS-1, Cardif or VISA) (Nakhaei et al., 2009). Upon viral detection, MAVS binds to RLRs and promotes the activation of NF-κB, AP-1 and various Interferon Regulatory Factors (IRFs), which act to induce ISGs and create an antiviral state in the cell. Although much has been learned about the genetics of RLR signaling, less is known about where within the cell signal transduction occurs. Identifying the sites of RLR signal transduction is critical to understanding how antiviral networks are integrated into the general cellular infrastructure within which they operate.
The first clue that cytosolic RLR signaling may occur from organelles came from studies of the MAVS adaptor. MAVS contains a C-terminal transmembrane domain that anchors it to the mitochondrial outer membrane (Seth et al., 2005). It is from this location that MAVS is thought to engage active RLRs and induce signal transduction. Whether mitochondria are the only organelles that promote RLR-mediated signaling has not been addressed.
Mitochondria have long been appreciated to have an intimate functional relationship with peroxisomes (Hettema and Motley, 2009). Both are membrane-bound organelles found in mammalian cells and are involved in the metabolism of lipids and reactive oxygen species. However, while mitochondria are well-established sites of both antiviral signaling and antiviral apoptosis, peroxisomes are thought to function solely as metabolic organelles.
Recently, several mitochondrial proteins have been found to reside also on peroxisomes. Included in this group are the outer membrane proteins Fis1 and Mff, which regulate the morphology of both organelles (Gandre-Babbe and van der Bliek, 2008; Koch et al., 2005). Interestingly, Fis1, Mff and MAVS all have similar domain structures: each contains an N-terminal effector domain and a C-terminal localization motif, which consists of a transmembrane domain and a short lumenal tail containing basic amino acids. That other so-called “tail-anchored” mitochondrial outer membrane proteins operate from peroxisomes raised the possibility that MAVS also functions from these organelles.
We have discovered that MAVS does indeed reside on peroxisomes and can induce antiviral signaling from this organelle. Our work supports a model whereby peroxisomal MAVS induces the immediate expression of antiviral factors that function to contain a nascent infection. Long-term containment of the infection, however, requires the function of mitochondrial MAVS as well. These data demonstrate that peroxisomes are not simply metabolic organelles, but rather serve as critical subcellular hubs that promote MAVS-dependent antiviral immunity.
Results
MAVS is located on both mitochondria and peroxisomes
MAVS has a similar domain organization to other tail-anchored membrane proteins that function from mitochondria and peroxisomes (Gandre-Babbe and van der Bliek, 2008; Koch et al., 2005). We therefore sought to determine whether MAVS also resides on peroxisomes. The subcellular localization of MAVS was examined in mouse embryonic fibroblasts (MEFs) whose peroxisomes were marked by a DsRed allele containing a type I peroxisomal targeting sequence (PTS1). In addition to staining structures that appeared to be mitochondria, MAVS was detected on PTS1-positive peroxisomes scattered throughout the cell (Figure 1A). A similar staining pattern was seen for Mff (Figure 1A), which functions from both peroxisomes and mitochondria (Gandre-Babbe and van der Bliek, 2008). In contrast, the Toll-Like Receptor (TLR) adaptor protein TIRAP (Fitzgerald et al., 2001; Horng et al., 2001) was not detected on peroxisomes (Figure 1A). To confirm that the peroxisomal staining was distinct from mitochondria, cells were additionally stained with MitoTracker. While no co-staining was detected between PTS1 and MitoTracker, MAVS was detected on both PTS1-positive peroxisomes and MitoTracker-positive mitochondria (Figure 1B). Similar results were obtained when examining epitope-tagged MAVS in murine macrophages (Figure S1) or endogenous MAVS in human hepatocytes (Figure 1C). As an independent means of assessing MAVS localization, hepatocytes were biochemically fractionated to separate peroxisomes and mitochondria, which were respectively distinguished by Pex14 and mtHSP70 (Figure 1D). Both MAVS and Fis1 (a protein that occupies both organelles (Koch et al., 2005)) were detected in fractions containing either peroxisomes or mitochondria. Collectively, based on studies in both human and mouse cells, these data establish that peroxisomes are a bona fide reservoir of the RLR adaptor protein MAVS.
Figure 1. MAVS resides on mitochondria and peroxisomes.
(A) MEFs were transfected with the peroxisomal marker DsRed-PTS1 and either Flag-MAVS, myc-MFF or Flag-TIRAP. Cells were stained using anti-MAVS, anti-myc or anti-Flag antibodies, respectively. All images for all panels are representative of at least three independent experiments where over 500 cells were examined per condition and >95% of the cells displayed similar staining.
(B) MEFs expressing Flag-MAVS as well as EGFP-PTS1 and Pex19 from a bicistronic construct were stained with anti-MAVS antibody and MitoTracker to visualize mitochondria.
(C) Huh-7 hepatocytes were transfected with DsRed-PTS1 and endogenous MAVS was detected using anti-MAVS antisera.
(D) Peroxisomes were separated from mitochondria on a Nycodenz gradient using HepG2 hepatocyte lysates. Selected fractions of the gradient were analyzed by immunoblotting with Pex14, mtHSP70, Fis1 or MAVS antisera.
(E) Pex19-deficient human fibroblasts were stained for endogenous MAVS before and after introduction of a functional Pex19 allele as indicated. Mitochondria were stained with anti-mtHSP70 antibody. Peroxisomes were visualized by transfection with a bicistronic construct encoding EGFP-PTS1 and Pex19. See also Figure S1.
One possible reason why MAVS is present on peroxisomes is that newly synthesized MAVS might first pass through peroxisomes en route to mitochondria. To address this possibility, we used human fibroblasts from a patient lacking a functional Pex19 protein. Pex19 controls peroxisome biogenesis, and thus Pex19-deficient cells contain no peroxisomes or peroxisomal remnant structures (Matsuzono et al., 1999; Sacksteder et al., 2000). Notably, MAVS was delivered to mitochondria in Pex19-deficient cells (Figure 1E), indicating that the pathway to mitochondria does not require a peroxisomal intermediate. Moreover, MAVS localized to both peroxisomes and mitochondria in Pex19-deficient cells that expressed Pex19 after transient transfection or retroviral gene transfer (Figure 1E). It is therefore unlikely that localization of MAVS to peroxisomes is the result of a biosynthetic pathway for delivering outer membrane proteins to mitochondria.
A systematic strategy to separate functions of peroxisomal and mitochondrial MAVS
Our finding that MAVS is located on peroxisomes raised the possibility that these organelles serve as a site of antiviral signal transduction. We first considered using the Pex19-deficient cells to address sufficiency of mitochondrial MAVS in antiviral signaling, but since peroxisomes are required for biochemical processes that occur in mitochondria, Pex19-deficient cells have profound defects in mitochondrial function (Wanders, 2004). We therefore used the alternative approach of genetically separating the putative mitochondrial and peroxisomal functions of MAVS. This was accomplished by replacing the previously defined MAVS localization motif (Seth et al., 2005) with a set of domains that instead direct the protein to a single compartment (Figure 2A). Using the localization motif of the peroxin Pex13 (Fransen et al., 2001), we created a protein called MAVS-Pex. By deleting the MAVS localization motif, we also created a cytosolic allele (MAVS-Cyto) (Seth et al., 2005). Because the fidelity of mitochondrial sorting signals is not always transferrable to other proteins (Ingelmo-Torres et al., 2009), we lastly created two different alleles of MAVS containing a sorting signal derived from two proteins residing on the mitochondrial outer membrane protein, either OMP25 or Fis1 (Koch et al., 2005; Nemoto and De Camilli, 1999).
Figure 2. Targeting of MAVS to distinct subcellular compartments by replacement of its transmembrane domain.
(A) Schematic of WT and mutant MAVS alleles to be tested for signaling from peroxisomes and mitochondria.
(B) Stable cell lines expressing the MAVS alleles listed in (A) were generated by retroviral transduction of MAVS-KO cells. Resulting transgenic cells expressed a MAVS allele and GFP, whose translation is directed by an IRES. Shown are overlaid histograms of stable populations of each cell expressing equivalent levels of the bicistronic mRNAs encoding MAVS and GFP.
(C) Lysates from stable cell lines described in (B) and parental MAVS KO MEFs were analyzed by immunoblotting with anti-MAVS antibody.
(D) Micrographs of MAVS chimeric cell lines indicated were stained with anti-MAVS antibody. Mitochondria were stained with anti-mtHSP70 antibody. Peroxisomes were visualized by transfection with DsRed-PTS1. Note that MAVS-Pex resides on peroxisomes, MAVS-Mito on mitochondria, MAVS-Mimic on both organelles and MAVS-Cyto localizes on neither of the two organelles. All images for all panels are representative of at least three independent experiments where over 500 cells were examined per condition and >95% of the cells displayed similar staining. See also Figure S2.
Using retroviral gene transfer of MAVS-KO MEFs, we created cell lines expressing comparable levels of each MAVS allele (Figure 2B, C) and determined their localizations by confocal microscopy. Full-length MAVS (MAVS-WT) was located on both mitochondria and peroxisomes (data not shown), and MAVS-Cyto was found on neither organelle (Figure 2D, S2). As expected, MAVS-Pex was found exclusively on peroxisomes (Figure 2D, S2). Of the alleles containing the putative mitochondrial targeting sequences, the allele harboring the Fis1 transmembrane domain was found primarily on mitochondria, whereas the one containing the OMP25 transmembrane domain was located on both mitochondria and peroxisomes (Figure 2D, S2). We therefore refer to the mitochondria-specific allele as MAVS-Mito to indicate its exclusive localization to mitochondria and the allele found on both organelles as MAVS-Mimic to indicate its ability to copy the localization pattern of MAVS-WT. Collectively, this set of MAVS-expressing MEF lines differs only in the subcellular positioning of the signaling domain of MAVS and thereby provides an ideal system to determine the relative roles of mitochondrial and peroxisomal localization in MAVS-dependent signal transduction.
MAVS-dependent signaling occurs from both peroxisomes and mitochondria
To address the function of peroxisomal MAVS, we monitored the expression of antiviral factors in response to infection with reovirus. We chose reovirus because it is a known inducer of both RIG-I and MDA-5 signaling pathways (Loo et al., 2008), allowing direct examination of both RLRs in a single experiment.
Cells were infected with reovirus, and extracts were examined at various times for expression of viperin, a well-characterized ISG (Chin and Cresswell, 2001; Severa et al., 2006). MAVS-WT, -Mimic or -Mito expressing cells induced viperin expression in response to infection (Figure 3A). This response was MAVS dependent, as MAVS-KO cells showed no change in viperin expression. MAVS-Cyto cells were unable to induce viperin expression, confirming that membrane localization is necessary for MAVS function (Seth et al., 2005). Interestingly, despite the fact that MAVS-Pex is found only on peroxisomes, MAVS-Pex cells induced viperin expression after infection (Figure 3A).
Figure 3. Peroxisomal MAVS mediates ISG expression, but does not induce Type I IFN secretion.
(A) MAVS-expressing MEFs and MAVS-KO cells were infected with reovirus. At indicated times, cell-associated ISG expression was determined by immunoblotting with an anti-viperin antibody. (B) Similar to (A) except for infection with influenza virus strain ΔNS1 in lieu of reovirus. (C and D) Cell culture media from (A) and (B) were tested for Type I IFN activity using a bioassay. Error bars show standard deviation of triplicate infections (E) MAVS-expressing MEFs and parental MAVS-KO cells were treated with 100 IU/ml IFNβ. At indicated times, cell associated ISG expression was determined by immunoblotting with anti-viperin antibody. Note that all cell lines respond similarly to IFNβ, indicating intact Type I IFN signaling. All data are the result of at least 2 independent experiments. See also Figure S3.
An examination of the kinetics of ISG induction indicated that cells containing MAVS on peroxisomes (MAVS-WT, -Mimic and -Pex) induced viperin expression within 4 h of infection. In contrast, exclusive localization to mitochondria (MAVS-Mito) resulted in viperin expression with delayed kinetics (Figure 3A). These results suggest that localization of MAVS to either peroxisomes or mitochondria is sufficient to induce antiviral signaling but that peroxisomal residence allows for more rapid expression of ISGs. Interestingly, rapid expression of ISGs by MAVS-Pex appeared transient, as viperin expression decreased at later times of infection (Figure 3A).
To determine if peroxisomal signaling by MAVS requires signaling by both RIG-I and MDA-5, we performed similar experiments using influenza virus, which activates the RIG-I pathway exclusively (Gitlin et al., 2006; Kato et al., 2006). A similar pattern of viperin expression was observed with influenza as with reovirus, although the kinetic differences between MAVS-Pex and -Mito were even more pronounced with influenza (Figure 3B). Thus, RIG-I signaling alone is sufficient to induce MAVS-dependent signaling from peroxisomes. In sum, these data indicate that peroxisomal MAVS induces rapid but transient viperin expression, whereas mitochondrial MAVS induces delayed but stable viperin expression. Signaling from both organelles thus contributes to the rapid and stable expression of viperin that is observed in MAVS-WT cells.
While the above studies provide strong genetic evidence for MAVS signaling from peroxisomes in a population of cells, they do not allow us to examine individual cells for compartment-specific signaling events. To address this, we took advantage of the fact that various signaling pathways induce morphological changes in the organelles where signaling occurs, including RLR-dependent activities on mitochondria (Castanier et al., ; Yasukawa et al., 2009). We reasoned that if signaling was actually occurring on these organelles, then their morphology may change during viral infection. In support of this prediction, reovirus infection induced peroxisomal aggregation and the formation of peroxisomal tubules (Figure S3A, B). The tubes formed ranged from approximately 2 microns in length to over 5 microns, and depended on MAVS localization to peroxisomes. Cells expressing MAVS-WT exhibited this activity, and cells expressing MAVS-Pex have greatly exaggerated behavior in these assays, with nearly all cells displaying peroxisomes over 5 microns in length (Figure S3A, B). Cells expressing MAVS-Mito or MAVS-Cyto exhibited little or no changes in peroxisome morphology. These data suggest that RLRs engage peroxisomal MAVS to induce peroxisomal tubules, and that the extent of tubulation is determined by the concentration of MAVS on these organelles. These independent assays demonstrate that MAVS-dependent signaling occurs locally (on the peroxisome).
Peroxisomal MAVS triggers an IFN-independent signaling pathway that promotes ISG expression
The different kinetics of viperin induction by peroxisomal and mitochondrial MAVS suggest that more than one mechanism of RLR-induced ISG expression may operate in virus-infected cells. ISG expression can be induced directly, or indirectly through the action of secreted Type I IFNs (Collins et al., 2004; Mossman et al., 2001). To determine if IFNs contribute to expression of ISGs induced by mitochondrial or peroxisomal MAVS, we monitored the rate of IFN production by reovirus infected cells. As expected, IFN production was dependent on membrane-localized MAVS, and all mitochondrial MAVS proteins (MAVS-WT, -Mimic and -Mito) triggered IFN production, though with delayed kinetics in the case of MAVS-Mito (Figure 3C). These data indicate that in the case of the mitochondria-localized MAVS proteins, IFN expression coincides with ISG induction. Surprisingly, no detectable IFNs were produced by MAVS-Pex cells. Similar results were obtained with cells infected with influenza (Figure 3D), though the relative amounts of IFNs produced with these two viruses differed dramatically, reflecting unique aspects of each virus lifecycle. However, our inability to detect a role for IFNs in promoting viperin expression in MAVS-Pex cells was not due to an inability of the cells to respond to IFNs, because addition of recombinant IFNβ was sufficient to induce viperin expression in all cells examined (Figure 3E).
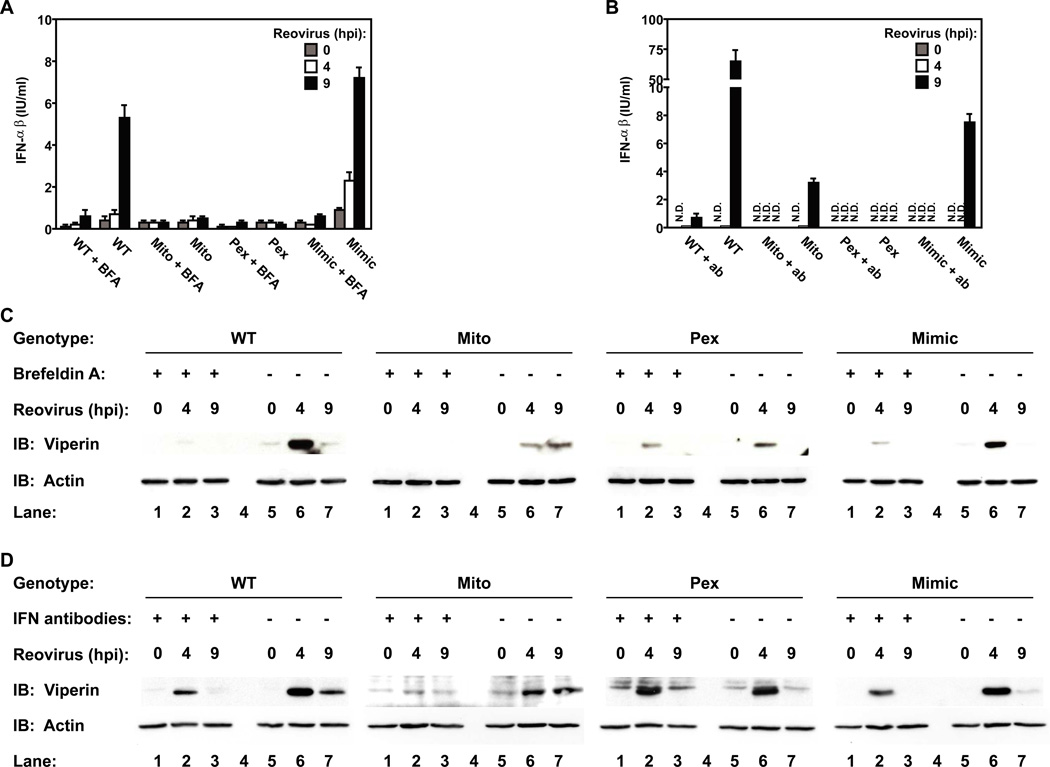
The observation that viperin can be expressed in the absence of Type I IFN induction suggests that IFNs may not contribute to ISG expression induced by peroxisomal MAVS. This possibility was tested by infecting cells under conditions in which the functions of IFNs are prevented, by either disrupting protein secretion with brefeldin A (BFA) or by utilizing neutralizing antibodies to secreted IFNs. Both treatments disrupted the activity of Type I IFNs produced during reovirus infection (Figure 4A, B) and inhibited the expression of viperin by cells expressing mitochondrial MAVS (MAVS-WT, -Mimic, -Mito) (Figure 4C, D). These data indicate that signaling by IFNs promotes viperin expression. However, because these treatments did not completely abolish viperin expression, an IFN-independent pathway of viperin induction must also exist. Interestingly, MAVS signaling from peroxisomes primarily utilized the IFN-independent pathway, as viperin expression within MAVS-Pex cells was largely resistant to these treatments (Figure 4C, 4D). These data therefore indicate that the subcellular positioning of MAVS determines the type of signaling pathway activated during viral infection. Peroxisomal MAVS induces the rapid and direct expression of viperin, which is followed by mitochondrial MAVS triggering viperin expression directly, as well as indirectly through the IFN-mediated feed-forward loop.
Figure 4. Peroxisomal MAVS directly induces viperin expression.
(A) MAVS-expressing MEFs and MAVS-KO cells were pretreated with 20µg/ml BFA before infection with reovirus in presence of the drug. At indicated times, cell supernatants were tested for Type I IFN activity using a bioassay. (B) Similar to (A) except Type I IFN activity was blocked by addition of 250 NU/ml anti-IFNβ and 500 NU/ml anti-IFNα antibodies after infection with reovirus. (C and D) Cell lysates from (A) and (B) were tested for ISG expression by immunoblotting with anti-viperin antibody. Note that IFN activity is not required for viperin expression mediated by peroxisomal MAVS. All data are the representative of at least 3 independent experiments. Error bars show standard deviation of triplicate infections
The global transcriptional response to reovirus infection is mediated by the collective actions of MAVS-dependent peroxisomal and mitochondrial signaling
Based on the set of candidate genes examined, our data suggests that peroxisomal and mitochondrial MAVS each induce a complementary set of genes that are collectively induced by MAVS-WT. To determine if this is the case, microarrays were performed on reovirus-infected cells. Infections were performed for 3, 9, or 16 h and RNA was collected for genome-wide expression analysis. At all times examined, similar expression profiles were observed when comparing MAVS-WT or MAVS-Mimic cells, confirming that similarities in MAVS localization are predictive of similarities in MAVS function (Figure 5A, B). These cells induced the expression of numerous ISGs, IFNs, and chemokines (Figure 5C). Notably, we were unable to detect the expression of the proinflammatory cytokines TNFα, IL-1β or IL-6 (data not shown). MAVS-Cyto cells were most similar to MAVS-KO cells (Figure 5A, B), further confirming that membrane localization of MAVS is critical for its function in antiviral signaling. Interestingly, MAVS-Pex or -Mito expressing cells displayed a transcriptome that each partially overlapped with that of MAVS-WT cells, but were distinct from one another (Figure 5B, C). For example, at 16 h post-infection, MAVS-Mito cells upregulated genes encoding chemokines, ISGs, IFNβ, and several IFNα family members (Figure 5B, C). MAVS-Pex cells also induced the expression of chemokines and ISGs, but without any detectable changes in IFN expression and with much faster kinetics (within 3 h). Thus, on a global scale, peroxisomal MAVS induces the rapid expression of ISGs without inducing IFN expression, whereas mitochondrial MAVS promotes IFN and ISG expression but with delayed kinetics. We confirmed these results by examining the expression of several candidate IFNs and ISGs using nCounter, which allows for multiplex analysis of gene expression with the sensitivity of quantitative RT-PCR (Geiss et al., 2008) (Figure S4A, B). Overall, at all times examined, most genes expressed by either peroxisomal or mitochondrial MAVS were induced by MAVS-WT or -Mimic (Figure 5B, C). These data therefore support a model whereby the host transcriptional response is the result of MAVS signaling from both mitochondria and peroxisomes. We do note however, that the magnitude of antiviral gene expression induced by cells expressing MAVS-WT or MAVS-Mimic was greater than the magnitude induced by cells where MAVS was restricted to a single organelle, which suggests that signaling from both organelles may be coordinated to ensure maximal antiviral gene expression.
Figure 5. Genome-wide transcriptome analysis reveals a general role for peroxisomal and mitochondrial MAVS in antiviral gene expression.
(A) RNA from MAVS-expressing MEFs and parental MAVS-KO cells after infection with reovirus for 3, 9 or 16 h was subject to microarray analysis. The similarity of the overall gene expression profiles mediated by the indicated MAVS alleles is displayed as Pearson correlation coefficient-based heat map. Samples are clustered along both axes based on their correlation value. Note that at 3 h post infection MAVS-Pex cells display a gene expression pattern that is most similar to MAVS-WT cells. (B) Pairwise comparisons of indicated cell lines based on 4089 significantly regulated genes depicted in a log-log scale scatter plot. Each data point indicates a gene whose expression level exhibited a change of greater than 2-fold.
(C) Heat map of selected genes based on their expression ratios across all 6 cell lines and during all time points upon reovirus infection. Genes are colored according to a log2-based color bar depicted underneath each heat map. See also Figure S4.
Peroxisomal signal transduction creates a transient but functional antiviral state
MAVS-dependent signaling promotes an antiviral state, which is functionally defined as the ability of cells to restrict multiplication of viruses. To determine the significance of mitochondrial or peroxisomal signaling pathways in this regard, we asked if signaling from either organelle is sufficient to restrict viral replication. This was addressed by infecting MAVS-expressing cells with reovirus and measuring production of infectious virions over time. As expected, MAVS-WT and -Mimic cells were most resistant to infection and MAVS-KO and -Cyto cells were most susceptible (Figure 6A). These data indicate that MAVS signaling is required to limit reovirus replication. Interestingly, cells expressing MAVS-Pex or MAVS-Mito exhibited an unusual biphasic behavior. Over the first 24 h, these cells restricted viral replication as well as MAVS-WT, but this capacity diminished, and by 72 h were most similar to the MAVS-KO cells. These data establish that signaling from either peroxisomes or mitochondria is sufficient to induce a functional antiviral response, but signaling from both organelles is necessary for maximal containment of reovirus replication.
Figure 6. Peroxisomal MAVS elicits a functional antiviral response.
(A) MAVS-expressing MEFs and MAVS-KO MEFs were infected with reovirus at an MOI of 3. At indicated times virus titers were determined by plaque assay. (B) MAVS-WT, -Mimic expressing cells and MAVS-KO MEFs were infected with VSV at an MOI of 3. After 8h RNA was isolated and analyzed for ISG and Type I IFN expression using nCounter. (C) Same as (B) except MAVS-Pex, MAVS-Mito and MAVS-Cyto MEFs were analyzed.
(D) MAVS-expressing MEFs and parental MAVS-KO cells were infected with VSV at an MOI of 0.01. At indicated times, virus titers were determined by plaque assay. (E) 293T cells were transiently transfected with an ISRE, IRF1, NF-κB or AP1 luciferase reporters together with empty pMSCV vector (control) or MAVS (WT, Mimic, Pex, Mito or Cyto). Results were normalized to Renilla luciferase activity and are shown as fold increase relative to cells transfected with empty vector. Error bars show standard deviation of triplicate transfections. See also Figure S5.
Vesicular stomatitis virus (VSV) is one of many viruses that interfere with Type I IFN expression as part of their pathogenic lifecycle (Figure 6B, C) and (Ferran and Lucas-Lenard, 1997). Under these conditions, the IFN-independent means of signaling that is induced by peroxisomal MAVS may be particularly important in controlling infection. Consistent with this idea, MAVS-Mito cells were as susceptible to VSV infection as MAVS-KO or -Cyto cells (Figure 6D), suggesting that in the absence of IFN production, the mitochondrial signaling pathway is functionally defective. Most notably, MAVS-Pex cells were nearly as effective at controlling VSV as MAVS-WT cells (Figure 6D). These results suggest that MAVS signaling from peroxisomes is the primary means of controlling viruses that interfere with IFN expression, thus underscoring the importance of this organelle in host defense.
Downstream regulators of MAVS signaling from peroxisomes
To identify downstream signaling regulators of peroxisomal MAVS, each MAVS allele was overexpressed in 293T human kidney epithelial cells. MAVS-WT, -Mimic, -Mito, and -Pex each induced the activation of reporter genes controlled by NF-κB and AP-1 (Figure 6E). In addition, an IRF1-reporter and an ISRE that typically reports IRF3 activity were induced, suggesting a role of these IRFs in MAVS signaling from peroxisomes (Figure 6E). MAVS-Cyto did not activate any reporter. Within these cells, we found that ISRE activation by either MAVS-WT or MAVS-Pex was potentiated by overexpression of TRAF3 and inhibited by expression of a dominant negative allele of TRAF6 (Figure S5A), suggesting the involvement of these known RLR regulators in peroxisomal signaling (Saha et al., 2006; Yoshida et al., 2008). In contrast, expression of a dominant-negative allele of the antiviral factor FADD has a minimal affect on MAVS signaling (Figure S5A), which is consistent with a recent report (Balachandran et al., 2007). Notably, overexpression of NLRX1, a negative regulator that is uniquely located on mitochondria (Figure S5B) (Moore et al., 2008) did not interfere with MAVS-Pex signaling, but did inhibit signaling by MAVS-WT (Figure S5A).
To confirm the roles of IRF1 and IRF3 in peroxisomal signaling, we enlisted VSV to study the IFN-independent means of ISG expression, since only the peroxisomal pathway functions to control replication of this virus, although some ISGs were induced by VSV in cells expressing MAVS-Mito (Figure 6C). MEFs derived from various IRF KO mice were infected with VSV and assessed for their ability to induce ISGs. Whereas WT cells induced the expression of several ISGs (and no IFNs), cells lacking IRF1 or IRF3 were incapable of inducing ISG expression (Figure 7A, B). A few ISGs (e.g., OAS1g) also required another family member, IRF5. These data indicate that IRF1 and IRF3 are central regulators of IFN-independent ISG expression and may act downstream of peroxisomal MAVS.
Figure 7. Transcription factors that control peroxisomal MAVS signaling.
(A,B) WT, IRF1, IRF3 and IRF5 KO MEFs were infected with VSV at an MOI of 3. After 8h RNA was isolated and analyzed for ISG and Type I IFN expression using nCounter.
(C,D) Immortalized MAVS-KO macrophages were retrovirally transduced with MAVS-WT, -Pex, -Mito and-Cyto for 48 h. The transduction efficiency of each cell population was determined to be 20%–30% as assessed by fluorescence microscopy. Cells were infected with reovirus and at the indicated times, were harvested and RNA was analyzed for expression of Type I IFN (C) and other inflammatory genes (D) using nCounter. See also Figure S5.
(E) Model of organelle specific MAVS signaling in fibroblasts. Peroxisomal MAVS is essential for rapid ISG expression independent of Type I IFN, whereas mitochondrial MAVS induces ISGs with delayed kinetics and primarily dependent on Type I IFN secretion. Therefore peroxisomal MAVS mediates immediate and transient antiviral effects, while mitochondrial MAVS promotes a sustained response later during infection. See also Figure S5.
Cell-type specific actions of peroxisomal and mitochondrial MAVS
The prototypical innate-immune adaptor MyD88 regulates TLR signaling and induces different transcriptional responses in different cell types. Whether other adaptor proteins also display this diversity of responses is unclear. To address this for MAVS, we examined the function of peroxisomal and mitochondrial MAVS in macrophages. Each MAVS allele was expressed in immortalized bone marrow-derived macrophages isolated from MAVS-KO mice. The localization of each MAVS protein was similar to that observed in MEFs (compare Figures S2 and S5C). In response to reovirus infection, macrophages that contained mitochondrial MAVS (MAVS-WT or -Mito) induced transcripts encoding IFNs (Figure 7C) and ISGs (Figure S5D). MAVS-Cyto was unable to induce gene expression in response to reovirus infection. Unlike MEFs, reovirus-infected macrophages expressed inflammatory cytokines such as IL-1β (Figure 7D), IL-6, IL-12b, and TNFα (Figure S5D and data not shown). Another difference between MEFs and macrophages was that MAVS-Mito expressing macrophages exhibited no kinetic delay in reovirus-induced gene expression. These results suggest that like the TLR adaptor MyD88, the function of MAVS is controlled in a cell-type specific way.
Peroxisomal MAVS induced the expression of some genes to the same levels observed with the WT allele, such as A20, IL-1β, Cox2, CXCL2 (MIP-2α), CCL4 (MIP-1β) and Fos (Figure 7D), whereas others were induced more than 3-fold but still less than in WT cells e.g.,viperin, IFIT1 and IFIT2 (Figure S5D). Of note, peroxisomal MAVS was unable to induce the expression of any IFN gene in macrophages (Figure 7C). Thus, despite cell-type specific activities of MAVS, a fundamental feature of the RLR signaling network appears to be that peroxisomal MAVS functions to promote an IFN-independent means of gene expression.
Discussion
The best-characterized sensors of cytosolic viruses are members of the RLR family, which enlist the adaptor protein MAVS to initiate antiviral signaling (Kawai and Akira, 2007). MAVS is one of a growing group of tail-anchored membrane proteins, which contain a C-terminal transmembrane domain (Gandre-Babbe and van der Bliek, 2008; Koch et al., 2005; Seth et al., 2005). This anchor was originally reported to promote MAVS recruitment to the mitochondrial outer membrane, providing a landmark of where RLR signaling can occur (Seth et al., 2005). This discovery established that cytosolic detection systems, like extracellular detection systems (e.g., TLRs), use membranes as scaffolds for signal transduction. In the TLR network, however, signaling occurs from a variety of different organelles, not just one (Barton and Kagan, 2009). We report here that in addition to mitochondria, MAVS is located on peroxisomes in several human and murine cell types, and represents the first antiviral signaling protein found on this organelle.
The central finding of this study—that peroxisomes are a site of signal transduction—was established with a complementary set of assays that measured 1) mRNAs encoding ISGs and IFNs, 2) protein levels of ISGs and IFNs, 3) the induction of a functional antiviral state in cells, and 4) infection-induced changes in peroxisome morphology. In each of these assays, we found that peroxisomes are a site of MAVS-dependent signaling. Moreover, these results were obtained by using several unrelated RNA viruses as physiological triggers of RLR signaling, which suggests that peroxisomal signaling is a fundamental component of the RLR network.
The RLR signaling network now joins the TLRs as pattern recognition systems that signal from multiple organelles. Both systems require that a transmembrane protein be positioned on specific organelles—the receptors themselves in the case of TLRs and the MAVS adaptor in the case of RLRs (Akira et al., 2006; Barton and Kagan, 2009). Interestingly, when considering these two networks, the function of diversifying signaling locale appears to be distinct. In the case of TLRs, differential receptor placement diversifies the types of pathogens that can be detected: TLRs found on endosomes recognize viruses, while TLRs found on the plasma membrane typically recognize bacteria. In contrast, differential MAVS placement does not diversify the types of viruses detected by RLRs, but diversifies the types of signaling pathways that are activated. In the case of reovirus and influenza virus infection of fibroblasts, peroxisomal MAVS triggers the rapid expression of ISGs, whereas mitochondrial MAVS triggers delayed ISG and IFN expression. This diversification is functionally important, as our data indicate that MAVS signaling must occur from both organelles to limit reovirus replication.
Our studies revealed another important similarity between the RLR and TLR networks, that of cell-type specific functions for adaptor proteins. In fibroblasts, MAVS functioned to induce expression of IFNs and ISGs, but not inflammatory cytokines. In contrast, IFNs, ISGs and cytokines were all induced by MAVS signaling in macrophages. Thus, MAVS can be grouped with the TLR adaptor MyD88 as immune regulators that induce cell-type specific transcriptional responses. What is the benefit of cell-type specific actions of innate immune signaling pathways? One benefit may lie in the primary functions of the cells responding to a given virus. For example, macrophages are dedicated sentinels of the innate immune system. As such, within these cells, infection triggers MAVS-dependent inflammatory cytokine production and antiviral factors. Fibroblasts, in contrast, are tissue resident cells that are primarily involved in organ homeostasis—a condition that is disrupted under inflammatory conditions. Thus, designing MAVS to induce antiviral factors but not inflammatory cytokines in fibroblasts may aid these cells in maintaining homeostasis under infectious conditions. We speculate that the diversification of adaptor functions in innate immunity may be a general mechanism to tailor signaling pathways to the needs of functionally diverse cell types.
Our finding that peroxisomal localization of MAVS is required for rapid but transient induction of antiviral ISGs, whereas mitochondrial MAVS promotes ISG expression with delayed kinetics in fibroblasts is especially intriguing. The kinetic differences of ISG expression were explained by the observation that peroxisomal MAVS induced a cell-intrinsic means of ISG induction, which occurred in the absence of detectable IFN expression. Mitochondrial MAVS induced cell-intrinsic ISG expression as well, but maximal induction occurred through the actions of secreted IFNs. Our studies did not reveal an obvious difference in the downstream regulators activated by peroxisomal vs. mitochondrial MAVS, but the studies performed in 293T cells suggest that the selective positioning of negative regulators (e.g NLRX1) may contribute to organelle-specific responses. Future work will be required to address this point.
The functional importance of RLR signaling from peroxisomes was best revealed by experiments with VSV, which interferes with IFN expression and renders the mitochondrial pathway ineffective. As a result, even though MAVS is present on mitochondria and peroxisomes in WT cells, a functional antiviral response against VSV is only induced by the peroxisomal pathway. We also exploited VSV infection to dissect the peroxisomal signaling pathway using cells derived from genetically-deficient mice. While we found that IRF3 plays a role in ISG expression, this factor is also involved in the regulation of IFN expression (Sato et al., 2000) and may therefore be considered a more general regulator of antiviral gene expression. Indeed IRF3 is also involved in IFN expression induced by non-RLRs (Kawai and Akira, 2007). IRF1, on the other hand, is needed for expression of all ISGs that we examined in VSV-infected cells and is not required for IFN expression (Tamura et al., 2008). IRF1 may thus uniquely control IFN-independent signaling events that lead to ISG expression and antiviral immunity. Our experiments with VSV also revealed a probable benefit of utilizing both IFN-dependent and IFN- independent mechanisms of ISG induction: for pathogens that disrupt the expression of IFNs, the peroxisomal pathway retains the ability to induce ISGs and create a functional, albeit temporary antiviral state.
In fibroblasts, the cooperative actions of MAVS on peroxisomes and mitochondria are needed for maximal antiviral immunity, and signaling from each organelle occurs independently of the other. As such, it appears that a simple mathematical equation can be proposed to explain antiviral signal transduction: RLR=Pex+Mito (Figure 7E). If either term in this equation is removed, then the RLR signaling network operates inefficiently, and antiviral immunity is compromised. We note however, that maximal ISG and IFN expression requires signaling from both organelles, which likely indicates that crosstalk exists to allow the two pathways to be properly integrated. In closing, our studies establish a new function for peroxisomes, that of a subcellular compartment that promotes a rapid response to viral infection. We speculate that additional organelles may harbor pathogen detection systems, and our work provides a mandate to expand the search for these organelles.
MATERIAL and METHODS
Plasmids and antibodies
pCMV2 Flag-IPS-1, pEF-HA-MAVS, pCDNA3-HA-NLRX1 and the myc-Mff plasmid were gifts from S. Akira, Z. Chen, J.Ting and A. van der Bliek, respectively. The plasmids Dsred-PTS1, bicistronic Pex19 / EGFP-PTS1, Pex19, EGFP-Pex19, Pex13p-EGFP, and pCMV-TIRAP-flag were described (Fransen et al., 2001; Horng et al., 2001; Vastiau et al., 2006). Pex19 was amplified from EGFP-Pex19 and inserted into the retroviral vector pMSCV IRES GFP. All MAVS constructs are based on the allele BC044952. The full length (1–540) and a truncated (1–500) sequence were amplified from pEF-HA-MAVS by PCR. For chimeric MAVS alleles the C-terminal 40 residues were replaced by the following sequences: PEX13 (NM_002618) residues 136–233, FIS1 (NM_016068) residues 128–152, and Omp25 (NM_022599) 109–145 by overlap extension PCR and cloned into pMSCV IRES GFP.
Anti-Pex19 and anti-Pex14, anti-viperin and anti-myc 9E10 were gifts from M. Fransen, P. Cresswell and S. Hansen respectively. Anti-MAVS (Bethyl Laboratories), anti-mtHSP70 (ABR Affinity Reagents), anti-Flag M2 (Sigma), anti-Fis (Santa Cruz), anti-HA (Roche) and anti-β-actin (Sigma) were used according to manufacturers’ recommendations.
Cell lines, retroviral gene transfer and cell fractionation
MEFs, Huh-7, 293T, Vero and MDCK cells were cultured according to standard techniques. MAVS-KO MEFs, PEX19 deficient human skin fibroblasts and L929 cells stably expressing an ISRE-luciferase reporter were provided by Z. Chen, R. Wanders, and B. Beutler, respectively. MEFs lacking IRF1, 3 and 5 were provided by K. Fitzgerald. MAVS and PEX19 alleles were introduced in MAVS-KO MEFs and Pex19 deficient human skin fibroblasts by retroviral gene transfer and then sorted for equal GFP fluorescence to normalize MAVS expression levels. Fractionations of HepG2 cells were performed as described (Fransen et al., 2004).
DNA transfections and immunofluorescence
MEFs and Huh-7 cells were transfected using Fugene-6 (Roche) for 24h at 37°C. Where indicated, cells were incubated with 250nM MitoTracker Deep Red FM (Molecular Probes) for 30min at 37°C prior to fixation. For NLRX1 visualization, Huh-7 cells expressing HA-NLRX1 were incubated with 160 nM MitoTracker for 20min at 37°C and permeabilised with 0.1% saponin in 80 mM PIPES, 5 mM EGTA, 1 mM MgCl2 (pH 6.8) for 10min at 25°C. Cells were fixed with 2% paraformaldehyde for 20min at 25°C and permeabilized for 10min with 0.1% Triton X-100. Samples were treated with block buffer (2% goat serum, 50mM ammonium chloride in PBS) for 30min and the appropriate antibodies diluted in block buffer. Antibody binding was detected using antibodies conjugated with Alexa fluor 488, 594 or 647 (Molecular Probes). Samples were imaged on a Nikon TE-2000 inverted microscope fitted with a video-rate confocal system consisting of a spinning disk confocal head (Yokogawa). Using a 100× oil immersion objective with a numerical aperture of 1.4, confocal images were collected as a 3D stack with a focal step size of 0.27 microns. Micrographs were processed using Adobe Photoshop.
Virus stocks, infections, plaque assay
Reovirus Type 3 Dearing (Cashdollar lab clone) was propagated on L929 cells and plaque purified as described (Furlong et al., 1988). Cells were seeded 12–16h prior to infection on 6-well plates. The day of infection, medium was replaced by addition of 2 ml of fresh medium containing virions at a multiplicity of infection (MOI) of 100. Where indicated cells were preincubated with 20µg/ml brefeldin A (Invitrogen) and infections were carried out in the presence of the drug. Type I IFN activity was blocked by addition of 250 neutralizing units/ml anti-IFNβ and 500 neutralizing units/ml anti-IFNα (PBL InterferonSource) antibodies at the time of infection. To assess reovirus replication, purified virions were diluted in 100 µl of attachment buffer (phosphate-buffered saline with 2 mM MgCl2) and incubated with cell monolayers for 1h at room temperature. After removal of unabsorbed virus by two washes with attachment buffer, cells were incubated for the indicated times. Next, cells were lysed by freeze/thaw and infectious titers were measured by serial dilution onto L929 cells as described (Middleton et al., 2007).
Influenza virus (A/Puerto Rico/8/34, H1N1) lacking the NS1 gene was propagated in Vero cells as described (Garcia-Sastre et al., 1998) and titrated by plaque assay on MDCK cells. For infection, cell monolayers were incubated with ΔNS1 virus at a MOI of 5 for 1hr at 37°C in DMEM supplemented with 0.3% BSA, washed and incubated with growth media.
VSV (Indiana) infections were performed as described (Cureton et al., 2009) and viral titer was determined by plaque assay on Vero cells.
Immunoblotting and Type I IFN bioassay
Protein extracts were prepared by standard techniques, and 40µg cell extract was separated by SDS-PAGE and analyzed by immunoblot. Type I IFN activity was measured as described (Jiang et al., 2005).
Gene arrays and bioinformatics
Cells were infected as described above and RNA was purified using QIAShredder and the RNeasy Mini Kit (Qiagen). Microarrays were performed by the Molecular Genetics Core Facility at Children's Hospital Boston supported by NIH-P50-NS40828, and NIH-P30-HD18655. Quantile normalization was used for signal extraction and normalization. Two criteria were applied to identify differentially regulated genes: i) statistical significance of P<0.05 and ii) fold change of greater than 2 (ratio >2.0 or <0.5). 4089 genes passed both criteria for at least one of the assayed conditions. Samples are clustered based on the Pearson’s correlation coefficient for the profile of those 4089 genes. The Pearson correlation, hierarchical clustering and heat map were generated using R functions ‘cor’, ‘hclust’ and ‘heatmap’, respectively. Signal intensity that reflected mRNA expression was presented on heatmaps or scatterplots on a log scale according to a color-coded intensity scale with R software (The R Project for Statistical Computing). Each data point in the scatterplots presented indicates a gene whose expression level exhibited a change >2-fold. The microarray data discussed in this publication are accessible through GEO Series accession number GSE21215 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE21215)
mRNA detection and analysis with nCounter
nCounter codeSets were constructed to detect genes selected by the GeneSelector algorithm and additional controls as described. 2.4*105 cells were lysed in RLT buffer (Qiagen) supplemented with β-mercaptoethanol. 5% of the lysate was hybridized for 16hr with the codeset and loaded onto the nCounter prep station, followed by quantification using the nCounter Digital Analyzer. To allow for side-by-side comparisons of nCounter experiments, we normalized the nCounter data in two steps. We first controlled for small variations in the efficiency of processing by normalizing measurements from all samples analyzed on a given run to the levels of chosen positive controls provided by the nCounter instrument. Second, we normalized the data obtained for each sample to the expression of nine control genes (Gapdh, Ik, Mea1, Ndufs5, Ndufa7, Rbm6, Shfm1, Tomm7, Ywhaz). These genes were described to be unchanged in cells exposed to a variety of infectious conditions (Amit et al., 2009). For every sample, we computed the weighted average of the mRNA counts of the nine control transcripts and normalized the sample’s values by multiplying each transcript count by the weighted average of the controls.
Supplementary Material
Acknowledgements
We would like to thank B. Boush (FACS), J. Wagner (microscopy) and C. Brees (cell fractionation) for expert help. We would like to thank S. Brubaker, C. Glanemann, L.R. Marek, , M. Schneider I. Zanoni for helpful discussions. This work has been supported by the following sources: JK, Children’s Hospital Boston Career Development Fellowship; ED, Austrian Science Fund (FWF; DK CCHD W1205); MF, Fonds voor Wetenschappelijk Onderzoek – Vlaanderen (G.0754.09) and the Bijzonder Onderzoeksfonds of the K.U.Leuven (OT/09/045).
References
- Ablasser A, Bauernfeind F, Hartmann G, Latz E, Fitzgerald KA, Hornung V. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009;10:1065–1072. doi: 10.1038/ni.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Venkataraman T, Fisher PB, Barber GN. Fas-associated death domain-containing protein-mediated antiviral innate immune signaling involves the regulation of Irf7. J Immunol. 2007;178:2429–2439. doi: 10.4049/jimmunol.178.4.2429. [DOI] [PubMed] [Google Scholar]
- Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanier C, Garcin D, Vazquez A, Arnoult D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 11:133–138. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci U S A. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SE, Noyce RS, Mossman KL. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J Virol. 2004;78:1706–1717. doi: 10.1128/JVI.78.4.1706-1717.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cureton DK, Massol RH, Saffarian S, Kirchhausen TL, Whelan SP. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009;5:e1000394. doi: 10.1371/journal.ppat.1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferran MC, Lucas-Lenard JM. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol. 1997;71:371–377. doi: 10.1128/jvi.71.1.371-377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies CA, Mansell AS, Brady G, Brint E, Dunne A, Gray P, Harte MT, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413:78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- Fransen M, Vastiau I, Brees C, Brys V, Mannaerts GP, Van Veldhoven PP. Potential role for Pex19p in assembly of PTS-receptor docking complexes. J Biol Chem. 2004;279:12615–12624. doi: 10.1074/jbc.M304941200. [DOI] [PubMed] [Google Scholar]
- Fransen M, Wylin T, Brees C, Mannaerts GP, Van Veldhoven PP. Human pex19p binds peroxisomal integral membrane proteins at regions distinct from their sorting sequences. Mol Cell Biol. 2001;21:4413–4424. doi: 10.1128/MCB.21.13.4413-4424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong DB, Nibert ML, Fields BN. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988;62:246–256. doi: 10.1128/jvi.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy DE, Durbin JE, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Motley AM. How peroxisomes multiply. J Cell Sci. 2009;122:2331–2336. doi: 10.1242/jcs.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng T, Barton GM, Medzhitov R. TIRAP: an adapter molecule in the Toll signaling pathway. Nat Immunol. 2001;2:835–841. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- Ingelmo-Torres M, Gonzalez-Moreno E, Kassan A, Hanzal-Bayer M, Tebar F, Herms A, Grewal T, Hancock JF, Enrich C, Bosch M, et al. Hydrophobic and basic domains target proteins to lipid droplets. Traffic. 2009;10:1785–1801. doi: 10.1111/j.1600-0854.2009.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z, Georgel P, Du X, Shamel L, Sovath S, Mudd S, Huber M, Kalis C, Keck S, Galanos C, et al. CD14 is required for MyD88-independent LPS signaling. Nat Immunol. 2005;6:565–570. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Antiviral signaling through pattern recognition receptors. J Biochem (Tokyo) 2007;141:137–145. doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
- Koch A, Yoon Y, Bonekamp NA, McNiven MA, Schrader M. A role for Fis1 in both mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2005;16:5077–5086. doi: 10.1091/mbc.E05-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzono Y, Kinoshita N, Tamura S, Shimozawa N, Hamasaki M, Ghaedi K, Wanders RJ, Suzuki Y, Kondo N, Fujiki Y. Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc Natl Acad Sci U S A. 1999;96:2116–2121. doi: 10.1073/pnas.96.5.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton JK, Agosto MA, Severson TF, Yin J, Nibert ML. Thermostabilizing mutations in reovirus outer-capsid protein mu1 selected by heat inactivation of infectious subvirion particles. Virology. 2007;361:412–425. doi: 10.1016/j.virol.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Mossman KL, Macgregor PF, Rozmus JJ, Goryachev AB, Edwards AM, Smiley JR. Herpes simplex virus triggers and then disarms a host antiviral response. J Virol. 2001;75:750–758. doi: 10.1128/JVI.75.2.750-758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Nemoto Y, De Camilli P. Recruitment of an alternatively spliced form of synaptojanin 2 to mitochondria by the interaction with the PDZ domain of a mitochondrial outer membrane protein. Embo J. 1999;18:2991–3006. doi: 10.1093/emboj/18.11.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol. 2000;148:931–944. doi: 10.1083/jcb.148.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, Cheng G. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. Embo J. 2006;25:3257–3263. doi: 10.1038/sj.emboj.7601220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Severa M, Coccia EM, Fitzgerald KA. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J Biol Chem. 2006;281:26188–26195. doi: 10.1074/jbc.M604516200. [DOI] [PubMed] [Google Scholar]
- Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- Vastiau IM, Anthonio EA, Brams M, Brees C, Young SG, Van de Velde S, Wanders RJ, Mannaerts GP, Baes M, Van Veldhoven PP, Fransen M. Farnesylation of Pex19p is not essential for peroxisome biogenesis in yeast and mammalian cells. Cell Mol Life Sci. 2006;63:1686–1699. doi: 10.1007/s00018-006-6110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJ. Peroxisomes, lipid metabolism, and peroxisomal disorders. Mol Genet Metab. 2004;83:16–27. doi: 10.1016/j.ymgme.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, Seya T, Kawabata S, Koshiba T. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2:ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yoshida R, Takaesu G, Yoshida H, Okamoto F, Yoshioka T, Choi Y, Akira S, Kawai T, Yoshimura A, Kobayashi T. TRAF6 and MEKK1 play a pivotal role in the RIG-I-like helicase antiviral pathway. J Biol Chem. 2008;283:36211–36220. doi: 10.1074/jbc.M806576200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.