Summary
Dedicated AAA+ ATPases help deposit hexameric ring-shaped helicases onto DNA to promote replication in cellular organisms. To understand how loading occurs, we used negative-stain electron microscopy and small-angle X-ray scattering to determine the ATP-bound structure of the intact E. coli DnaB•DnaC helicase/loader complex. The 480 kDa dodecamer forms a three-tiered assembly, in which DnaC adopts a spiral configuration that remodels N-terminal scaffolding and C-terminal motor regions of DnaB to produce a clear break in the helicase ring. Surprisingly, DnaC’s AAA+ fold is dispensable for ring remodeling, as the isolated helicase-binding domain of DnaC can both load DnaB onto DNA and increase the efficiency by which the helicase acts on substrates in vitro. Our data demonstrate that DnaC opens DnaB by a mechanism akin to that of polymerase clamp loaders, and indicate that bacterial replicative helicases, like their eukaryotic counterparts, possess auto-regulatory elements that influence how the hexameric motor domains are loaded onto and unwind DNA.
INTRODUCTION
Precise and reliable transmission of genetic information is critical to the proliferation of all organisms. In cells, copying of the genome is carried out by the replisome, a multi-component macromolecular machine that couples the unwinding of parental DNA duplexes with the synthesis of new daughter strands (Baker and Bell, 1998; MacNeill, 2011; Pomerantz and O'Donnell, 2007; Schaeffer et al., 2005). Replisome progression is driven by hexameric helicases, ring-shaped protein motors that use the energy of ATP hydrolysis to unwind duplex DNA (McGlynn, 2011). Correct recruitment and deposition of replicative helicases is highly regulated, occurring only once per cell cycle to maintain genome integrity (Katayama et al., 2010; Nielsen and Lobner-Olesen, 2008; Soultanas, 2012).
How replicative helicases are loaded onto cellular chromosomes, which lack free DNA ends, has been a long-standing question. At present there appear to be two dominant approaches used by cells to promote hexameric helicase association with DNA, both of which rely on dedicated loading factors (Davey and O'Donnell, 2003). One strategy is to physically open and deposit a preformed hexameric ring onto target substrates. The other is to construct a hexamer around DNA from monomeric subunits. Examples of both approaches have been found in different species (Soultanas, 2012); for instance, pre-assembled MCM-family hexamers are thought to be loaded by the action of the Origin Recognition Complex (ORC) in eukaryotes (Bowers et al., 2004; Evrin et al., 2009; Remus et al., 2009), whereas the replicative helicase of Bacillus subtilis appears to be actively assembled by a protein known as DnaI (Soultanas, 2002; Velten et al., 2003). The physical mechanisms by which specific factors work to facilitate helicase loading are diverse and not well understood.
In E. coli, the replicative helicase DnaB associates with a dedicated loader known as DnaC (Kobori and Kornberg, 1982a, b; Wickner and Hurwitz, 1975). Along with the replication initiator DnaA, DnaC helps chaperone two copies of DnaB onto newly-melted DNA strands during the initiation of replication (Davey et al., 2002; Fang et al., 1999; Marszalek and Kaguni, 1994; Seitz et al., 2000). Notably, DnaC – like many hexameric helicase loading factors – is a member of the AAA+ (ATPases Associated with various cellular Activities) superfamily of nucleotide hydrolases (Koonin, 1992; Mott et al., 2008). As a consequence, appropriate DnaC action on DnaB during replication depends on ATP (Davey et al., 2002; Makowska-Grzyska and Kaguni, 2010). How and whether DnaC opens DnaB hexamers, and how nucleotide is used for productive deposition, is not known.
To address these questions, we determined the 3D structure of DnaB complexed with DnaC (referred hereafter as DnaBC) in a nucleotide-bound state by negative-stain electron microscopy (EM). Fitting of atomic structures into the 25 Å reconstruction reveals a gapped-ring architecture for a dodecameric DnaBC particle in which a helical arrangement of six DnaC subunits can be clearly seen to engage and open a DnaB hexamer. Small angle x-ray scattering confirms the organization of DnaBC seen by EM, indicating that the open ring complex is a dominant form in solution. Interestingly, a set of N-terminal scaffold domains present in DnaB exhibit a new conformational state not seen previously, while the helicase’s motor domains adopt a helical configuration seen only in a structure of a truncated DnaB variant that lacks the N-terminal region. Moreover, we find that in vitro, the isolated DnaB-binding domain of DnaC is sufficient to load the helicase onto a topologically closed DNA substrate, as well as activate helicase function on substrates with free DNA ends that should, in principle, not require a loader. Together, our data indicate that: 1) the DnaB N-terminal collar is a control element that impedes improper DNA loading, 2) DnaC can overcome this auto-repressed state to both load and activate DnaB, and 3) that the ATPase domains of DnaC are not required for DnaB opening, but instead appear to stabilize an open-ring state. These findings not only help explain how DnaC assists DnaB-dependent processes such as replication initiation, but also show that bacterial replicative helicases, like those of eukaryotes, bear auto-regulatory domains that control DNA loading and unwinding functions.
RESULTS AND DISCUSSION
DnaBC adopts a gapped, three-tiered structure
Although a 3D cryo-electron microscopic (EM) reconstruction of DnaBC had been reported previously (Barcena et al., 2001), it exhibited a closed, barrel-shaped toroid that did not immediately explain how DnaC might aid DnaB loading. Analysis of the cryo-EM DnaBC model in light of several subsequently-determined crystal structures of DnaB hexamers showed that it likely derives from a mixture of DnaBC and free DnaB particles (Figure S1) (Bailey et al., 2007b; Lo et al., 2009; Wang et al., 2008). We thus set out to reinvestigate the higher-order organization of DnaBC, using the well-studied E. coli proteins as a model system. To assemble the DnaBC complex, both proteins were independently overexpressed in E. coli, purified, and combined using a 1.2-fold molar excess of the loader. Formation of DnaBC was confirmed by gel filtration chromatography and SDS-PAGE (Figure S2A). To further stabilize the complex, the sample was next dialyzed into buffer containing a non-hydrolyzable ATP mimic, ADP·BeF3. An analytic size-exclusion column equilibrated with nucleotide was then employed to prepare the sample for single particle analysis by negative-stain EM (Experimental Procedures).
Two-dimensional analysis of 21,000 particles shows that, as previously observed (Barcena et al., 2001; San Martin et al., 1998), DnaBC adsorbs to grids with relatively few preferential orientations (Figure S2B and C). Using grids pre-treated with polylysine, we additionally obtained 10,681 particles with an even more uniform angular distribution (Figure S2C and D). Initial 2D alignment and classification of the particles revealed an abundance of the DnaBC complex (~70%) with some free DnaB. Random conical tilt reconstructions were aligned without bias, classified, and averaged into two groups: one showing a cracked ring and the other a closed toroid.
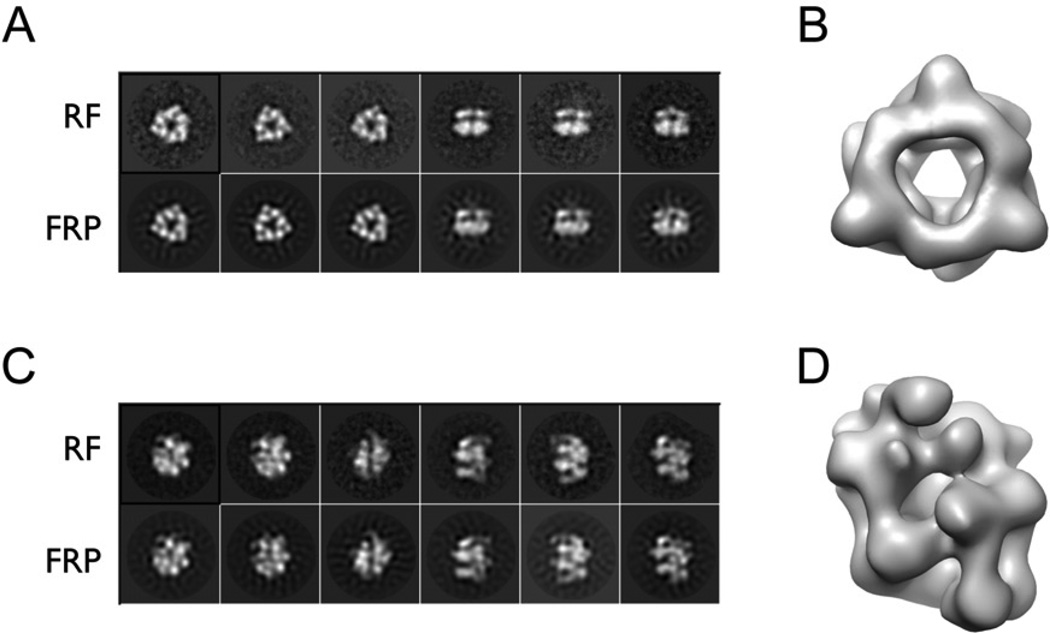
The two models were next used as initial references for multi-model refinement (Figure S3A). 3D reconstructions show that the final closed-ring and cracked-ring structures correspond to DnaB (Figure 1A and 1B) and a DnaBC complex (Figure 1C and 1D), respectively, at 25 Å resolution as measured by Fourier Shell Correlation (Figure S3B). The consistency of the final structures was supported by the agreement between reference-free 2D classes and forward reprojections of the EM reconstructions (Figure 1A and 1C). Although prior DnaB models have displayed three-fold symmetry (San Martin et al., 1998; San Martin et al., 1995; Yang et al., 2002), symmetry restraints were not imposed while generating the DnaBC reconstructions. For DnaB alone, we did perform a symmetry analysis, applying no, three-fold, or six-fold symmetry to the particle reconstructions; only models calculated in the absence of rotational averaging or with three-fold symmetry produced shapes whose projections matched both our reference-free 2D class averages and known atomic-resolution structures (Bailey et al., 2007b; Lo et al., 2009; Wang et al., 2008). By contrast, DnaB particles treated with sixfold symmetry gave rise to highly distinct structures (Figure S4A–C).
Figure 1. EM reconstructions of E.coli DnaB and DnaBC.
(A) Representative reference-free 2D class averages of DnaB (RF) compared to forward reprojections of the 3D structure obtained after multi-reference refinement (FRP).
(B) The 3D reconstruction of DnaB shows a closed ring with a broad inner channel.
(C) Representative reference-free 2D class averages of the DnaBC complex (RF) compared to forward reprojections of the 3D structure obtained after multi-reference refinement (FRP).
(D) The 3D reconstruction for DnaBC shows a 3-tiered, cracked-ring particle.
See Figures S1–S4.
Several recent crystal structures have established that, in the absence of nucleotide, DnaB-family proteins form ring-shaped structures comprising two tiers of domains (Bailey et al., 2007b; Lo et al., 2009; Wang et al., 2008). One tier is composed of a set of N-terminal domains that form a distinctive triangular arrangement of homodimers, while the other derives from six RecA-family ATPase domains arrayed with cyclic, pseudo-six-fold symmetry. A linker helix connects the two modules in a domain-swapped configuration, and further serves to bind adjacent ATPase subunits together. Automated docking showed that a full-length DnaB hexamer structure from B. stearothermophilus fit well with our DnaB EM model (correlation coefficient 0.91) (Figure 2A); the close resemblance is also evident upon filtering the crystal structure to 25 Å (Figure 2B). Hence, the closed-ring species appears to correspond to DnaB alone.
Figure 2. EM analysis of E.coli DnaB and DnaBC.
(A) Closed-ring B. stearothermophilus DnaB crystal structure (PDB ID 2R6A, (Bailey et al., 2007b)) fitted in the E.coli DnaB EM reconstruction.
(B) Closed-ring B. stearothermophilus DnaB crystal structure (PDB ID 2R6A) filtered at 25 Å.
(C) DnaBC forms a three-tiered, right-handed, cracked-ring structure. The upper tier is colored in blue (1), middle in purple (2) and lower in orange (3).
(D) DnaBC view showing the discontinuity in the cracked ring. The orientation is orthogonal to that of panel C.
(E) The DnaBC lower tier exhibits 61 symmetry.
(F) The DnaBC upper tier exhibits 31 symmetry.
(G) Longitudinal DnaBC section, showing the central channel that runs along the three tiers. The orientation is equivalent to that of panel D
(H) DnaBC displayed at lower threshold (5.7 σ contour) to highlight the discontinuity of the ring. This view is equivalent to that of panel D.
In contrast to the free DnaB present in our EM images, the DnaBC complex adopts a right-handed spiral that exhibits three-tiers (Figure 2C and D), along with a central channel that runs through the helical axis of the particle (Figure 2E, F and G). Each tier exhibits a clear recurring pattern, with six spirally-arranged lobes repeated in one outer layer and the center tier. The other (outer) tier is composed of three copies of an elongated, zigzag structure, again arranged in a spiral pattern (Figure 2F). The helical nature of these repeating elements leads to a crack in the DnaBC particle that runs along one side of the entire structure, creating an entryway into the central channel (Figure 2H). This configuration differs markedly from structures of closed-ring DnaB hexamers (Bailey et al., 2007b; Lo et al., 2009; San Martin et al., 1995; Wang et al., 2008; Yang et al., 2002), highlighting the dramatic structural rearrangements accompanied by the binding of DnaC. Our structure also differs from prior EM studies of DnaBC (Barcena et al., 2001; San Martin et al., 1998).
Stoichiometry and organization of DnaBC
To determine which regions of the DnaBC volume correspond to specific elements of the two proteins, we fit domains from available crystal structures into our EM reconstruction (Bailey et al., 2007b; Mott et al., 2008). The N-terminal domains of DnaB form homodimers in which two globular subdomains associate through a pair of helical hairpins, forming a distinctive “S”-shaped architecture (Figure 3A–B) (Bailey et al., 2007a, b; Biswas and Tsodikov, 2008; Lo et al., 2009; Wang et al., 2008). Inspection of the DnaBC model showed that one outer tier of the complex (tier 1) could unambiguously accommodate three N-terminal domain homodimers (Figure 3C). Interestingly, the configuration adopted by the three N-terminal homodimers in the reconstruction constricts one end of the axial pore of the DnaBC complex, but does not obstruct the channel.
Figure 3. Fitting DnaB into the DnaBC model.
(A) Primary structure of E.coli DnaB and E.coli DnaC.
(B) Structure of a typical DnaB N-terminal domain homodimer (PDB ID 2R6A, (Bailey et al., 2007b)).
(C) Docking of DnaB N-terminal domain homodimers into tier 1 of the DnaBC model. Subunits are alternatingly colored as per panel B.
(D) Docking of DnaB RecA folds into tier 2 of the DnaBC model. Subunits are alternatingly colored purple and pink. The density corresponding to the third tier has been removed for clarity.
(E) Side view of both domains of DnaB fitted into the DnaBC complex. The density corresponding to the third tier is not shown.
Having placed the N-terminal domains, the central tier (tier 2) was immediately implicated in housing the C-terminal domains of DnaB. Examination of this region revealed density for six globular lobes, each bearing a distinctive protrusion that gives the central tier a pinwheel-like appearance. Despite the modest resolution of the structure, we were able to computationally fit six individual DnaB-family RecA folds into this region, using a hook-like appendage on the motor domain as a fiducial to verify its position and orientation (Figure 3D). The resulting model produced a clear spiral arrangement for the DnaB C-terminus that, together with the N-terminal domains, readily defined the right-handed chirality of the reconstruction (Figure 3E).
With two segments of the particle accounted for, we next assessed whether the third tier of DnaBC could accommodate DnaC. The loader consists of a small N-terminal domain linked to a C-terminal AAA+ fold (Koonin, 1992) (Figure 3A). Numerous studies have suggested a 6:6 stoichiometry for DnaBC (Galletto et al., 2003; Kobori and Kornberg, 1982b); however, other ratios also have been proposed (Kaguni, 2011; Makowska-Grzyska and Kaguni, 2010). Since no atomic-resolution structure of full-length DnaC is available at present, we turned to a crystal structure of the isolated DnaC AAA+ fold (DnaCAAA+) (Mott et al., 2008). Because this domain is small and globular, we were unable to unambiguously fit single copies of the fold into our reconstructions. However, a previous study of Aquifex aeolicus DnaCAAA+ showed that the protein could assemble into a right-handed spiral filament with 61 crystallographic symmetry that recapitulated appropriate functional interactions in the ATPase active site (Mott et al., 2008) (Figure 4A). Notably, a DnaCAAA+ hexamer in this configuration proved to be an excellent fit to the third tier of the DnaBC EM volume (correlation coefficient 0.87) (Figure 4B). This result, together with previous biochemical and in vivo genetic studies (Mott et al., 2008), strongly indicates that the spiral configuration of DnaC AAA+ domains seen crystallographically represents a physiologic state of the loader.
Figure 4. Molecular architecture of the DnaBC complex.
(A) Detail view of DnaCAAA+ crystal structure (PDB ID 3ECC (Mott et al., 2008)). The DnaC ATPase domains appear to productively associate with nucleotide binding in this complex.
(B) DnaCAAA+ hexamer (reconstituted by imposing the crystallographically-observed 61 packing arrangement on PDB ID 3ECC), fitted into the lower tier of the DnaBC complex.
(C) Composite model for DnaBC. The six columns of unaccounted density connecting the DnaCAAA+ and DnaB RecA domains likely correspond to the DnaC N-terminal regions, which bind DnaB. See Movie S1.
(D) SAXS analysis of DnaBC. Experimental scattering data (1 mM AMPPNP, pink) fits well to a theoretical curve calculated from the DnaBC EM reconstruction (blue), but not to that of a closed-ring DnaB hexamer (purple). Improved fitting is obtained from a multiple ensemble search containing a mixture of DnaBC model and free DnaB (green). See Figure S2.
(E) Clamp-loader AAA+ domains oligomerize into a right-handed spiral to open a processivity clamp (PDB ID 3U5Z (Kelch et al., 2011)).
The 21 kDa AAA+ module of DnaC comprises 75 % of the mass of the loader, and occupies a similar fraction of the third tier within the DnaBC volume. The missing portion of this layer, which corresponds to a set of “legs” that connect DnaCAAA+ protomers with the DnaB RecA folds, likely accommodates the DnaC N-terminal region (Figure 4C). This logic is consistent with genetic and biochemical data indicating that the N-terminal domain of the loader associates with the helicase (Ioannou et al., 2006; Ludlam et al., 2001; Tsai et al., 2009). Together, the fitted structures account for 90 % of the functional elements of DnaB and DnaC, providing a molecular model for nearly the entirety of the assembly (Figure 4C, Movie S1).
Overall, our EM reconstruction both resolves the relative disposition of DnaC with respect to DnaB and firmly establishes DnaBC as a dodecameric complex. To determine if the DnaBC state seen by EM corresponds to a dominant state in solution, we further analyzed the complex in the presence of nucleotide using small-angle x-ray scattering (SAXS), a method that reports on the ensemble distribution of particle shapes and sizes in solution (Putnam et al., 2007). Plotting the average intensity of the scattering data (I) as a function of scattering angle (q) gave rise to a one-dimensional scattering profile of the sample that matched well to a theoretical scattering curve calculated from our fitted DnaBC model (c = 11.8), but not to DnaB alone (c = 46.3) (Figure 4D). We next employed Multiple Ensemble Searches (MES) to assess whether a mixture of structures was present in the solution sample (Schneidman-Duhovny et al., 2010). MES analysis showed that including scattering terms from DnaB along with DnaBC improved the fit of the theoretical models to the data (c = 9.3, with 86 % DnaBC and 14 % DnaB), indicating that, similar to the DnaBC sample used for EM, a portion of free DnaB was present in the preparation (Figure 4D). Together, these data confirm prior studies indicating that the loader engages the ATPase domains of DnaB (Galletto et al., 2003; Soultanas, 2002; Tsai et al., 2009), and further demonstrate that in the presence of nucleotide, this interaction opens a gap in the DnaB ring.
DnaC opens DnaB by a mechanism analogous to, but distinct from that of clamp loader
The structure of E. coli DnaBC firmly establishes DnaC as a ring-breaker for loading DnaB onto DNA. While distinctive, the spiral configuration adopted by both DnaCAAA+, and the coincident mirroring of this state by the target RecA folds of DnaB, is nonetheless reminiscent of another type of AAA+ protein remodeling system, namely the polymerase sliding-clamp loaders (Kelch et al., 2011, 2012). Clamp loaders form pentameric complexes held together in the absence of ATP by a conserved C-terminal collar domain. ATP binding actively remodels clamp loader AAA+ domains, which are loosely connected in the absence of nucleotide (Jeruzalmi et al., 2001), into a right-handed spiral akin to that seen for DnaC (Bowman et al., 2004; Kelch et al., 2011; Miyata et al., 2005; Seybert et al., 2006; Simonetta et al., 2009) (Figure 4E). Extensive interactions between the sliding clamp and the loader ATPase domains transduce this spiral configuration into the clamp, converting it into an open lockwasher state that is competent for binding primer-template DNA substrates (Kelch et al., 2011; Miyata et al., 2005). ATP hydrolysis is triggered upon binding DNA, which in turn leads the loader to dissociate from the clamp (reviewed in (Pomerantz and O'Donnell, 2007)).
Although the DnaC loading mechanism is globally similar to that of clamp loader, there also are key differences. For one, DnaC uses a dedicated N-terminal domain to associate with its target (Ioannou et al., 2006; Ludlam et al., 2001; Tsai et al., 2009), rather than the AAA+ region employed by clamp loaders (Bowman et al., 2004; Kelch et al., 2011; Latham et al., 1997). Moreover, unlike clamp-loader, ATP alone is insufficient to promote productive self-associations between the ATPase domains of full-length DnaC subunits; instead, DnaB, single-stranded DNA, and/or removal of the DnaC N-terminal domain is required for nucleotide-dependent assembly (Biswas et al., 2004; Galletto et al., 2004b; Ioannou et al., 2006; Mott et al., 2008). Finally, whereas clamp loaders depend on their C-terminal collar to remain stably associated (Bowman et al., 2004; Jeruzalmi et al., 2001; Kelch et al., 2011), DnaC exists as a monomer in solution, assembling into a defined hexamer only upon binding DnaB (Galletto et al., 2004b; Kobori and Kornberg, 1982b). Together, these data indicate that DnaC relies on a target-guided, ATP-dependent monomer/oligomer transition for function, a property shared with the AAA+-family replication initiator and DnaC paralog, DnaA (Mott et al., 2008). This mechanism not only distinguishes DnaC from pre-assembled clamp loader systems, but also marks an unusual mechanistic strategy found only in a few other types of AAA+ ATPases.
The DnaB N-terminal collar is a restraining belt that is relieved by DnaC
In comparing our DnaBC reconstruction to other DnaB structures, we noted that DnaC has a more significant effect on DnaB conformation than was previously anticipated. In particular, the N-terminal collar is not just cracked open, but also remodeled into an entirely new organizational state distinct from that seen in closedring DnaB states (Figure 5A): as the N-terminal dimers accompany the ATPase domains in forming a spiral, they also laterally rotate by ~55° from their closed-ring state. This shift maintains the helical hairpin interface seen between N-terminal domain homodimers, but completely alters inter-dimer interactions, producing a more trefoil-shaped arrangement (Figure 5B). The rotation in turn modifies the contacts between the N-terminal collar and the RecA tier; in closed-ring DnaB structures, alternating N-terminal globular heads sit atop the RecA fold of an adjacent protomer, whereas in DnaBC, they rest upon their own C-terminal RecA domains (Movie S2, S3); interestingly, a closed-ring form of the DnaB N-terminal domain packing seen in the DnaBC complex is echoed in a crystal structure recently determined in our group for a nucleotide-bound state of DnaB (Strycharska et al., manuscript in preparation). Overall, the conformational change manifest by DnaB upon binding DnaC aligns the opening within each tier of the DnaBC complex, creating a contiguous cleft with a minimum width of 11–13 Å that allows entry into the central channel from the side of the particle.
Figure 5. The DnaB N-terminal domain collar is remodeled by DnaC.
(A) The N-terminal homodimers of DnaB in the absence of nucleotide form a wide, closed triangular collar (PDB ID 2R6A, (Bailey et al., 2007b)). DnaB RecA domains are displayed as surfaces, whereas the N-terminal domains and linker helices are shown as light blue/orange cylinders.
(B) The N-terminal domains of DnaB within the DnaBC complex undergo a marked positional shift from the closed ring state, forming new packing arrangements between dimers. The DnaB RecA domains form a cracked spiral (boxed inset); DnaC is omitted for clarity. A schematic of the arrangement for the N-terminal domain dimers is shown in the upper left corner of each panel. Arrows in panel A indicate the movement of the DnaB NTDs to the state shown in panel B.
See Figures S5 and S6 and Movies S2 and S3.
In apo crystal structures of full-length DnaB, the motor’s RecA domains are roughly co-planar and do not associate into a catalytically competent state, possibly because nucleotide is not bound in these models (Bailey et al., 2007b; Lo et al., 2009; Wang et al., 2008). By contrast, the DnaB ATPase domains in DnaBC deviate significantly from this pattern, assembling into a clear spiral that positions adjacent RecA folds in a manner consistent with the binding of nucleotide; although the resolution of the reconstruction is insufficient to model specific interactions, the region containing a catalytic arginine on each subunit is closely juxtaposed with the surface containing the Walker A and B nucleotide-binding motifs of a partner protomer (excepting the interface where the ring is broken) (Figure S5). Consistent with previous findings that DnaC inhibits the ATPase activity of DnaB when associated with the helicase (Biswas et al., 1986; Wahle et al., 1989), maintenance of a stable discontinuity between DnaB ATPase domains within the DnaBC complex would be expected to interfere with the helicase’s ATP hydrolysis cycle. The pitch of the helicase ATPase domains in the DnaBC complex also deviates from a recently-published complex between DnaB, nucleotide, and single-stranded DNA that has been proposed to represent a translocation intermediate (Figure S6A) (Itsathitphaisarn et al., 2012); this difference suggests that DnaC can help push DnaB toward attaining a productive ATPase configuration, but that it also prevents the helicase from fully reaching that state when the loader remains bound.
Given that several helicases (e.g., eukaryotic MCM2–7 and the E. coli Rho transcription termination factor) naturally form split-ring, lockwasher structures in the absence of DNA and/or ATP (Costa et al., 2011; Lyubimov et al., 2012; Skordalakes and Berger, 2003), why is DnaC needed to breach the DnaB hexamer? The pitch of the DnaB spiral mirrors that of DnaC, suggesting that DnaC could be responsible for defining the shape of the helicase. However, one DnaB crystal structure, a variant of the G40P helicase, also has been found to adopt a helical configuration (Wang et al., 2008) (Figure 6A). Notably, the pitch of the G40P RecA domains seen in this model closely matches that seen from our docking of individual subunits into the DnaBC EM complex (Figure 6B). The key difference between this G40P model and other, planar DnaB structures is that it lacks the N-terminal domains. This observation suggests that the N-terminal collar of DnaB-family proteins is a restraining belt that prevents DnaB from inadvertently opening, and that the binding of DnaC might not necessarily drive the formation of a spiral helicase structure, but rather exploit the natural propensity of the RecA domains to open on their own. This idea is supported by structural studies of the bacteriophage T7 gp4 protein, a more-distantly related DnaB homolog that also can assemble into a helix when a portion of its N-terminus is removed (Sawaya et al., 1999). The lockwasher-array of motor domains seen together with a split N-terminal collar in the DNA-bound structure of DnaB further reinforces such a concept (Itsathitphaisarn et al., 2012) (Figure S6B).
Figure 6. Intrinsic and extrinsic control of DnaB ring opening and function.
(A) Removal of the N-terminal domain of the DnaB-family G40P protein allows its associated RecA ATPase to assemble into a helical filament (PDB ID 3BH0, (Wang et al., 2008)).
(B) The fitted DnaB RecA domains within DnaBC structure adopt a spiral pitch similar to that seen for N-terminally truncated G40P.
(C) The DnaC NTD can promote the DnaB-dependent unwinding of a topologically-closed DNA substrate that requires helicase ring opening and loading. (Top) Gel showing unwinding of a 3’ tailed oligonucleotide annealed to circular M13mp18 ssDNA in the presence of DnaB with or without DnaC or the DnaC NTD. Lanes N and D indicate the native and boiled substrate, respectively. (Bottom) Quantification of product seen by gel. Concentrations of DnaB hexamers in the reactions are shown; DnaC, when present, was included at a 2-fold molar excess of DnaB monomers. Columns and error bars represent the average and standard deviations, respectively, of at least five measurements.
(D) DnaC and the DnaC NTD can promote the DnaB-dependent unwinding of a topologically-accessible forked DNA substrate. Plot shows unwinding of a fluorophore/quench-labeled DNA by DnaB in the presence or absence of DnaC or the DnaC NTD. Data points and error bars represent the average and standard deviation, respectively, from at least six measurements. See Figure S7.
DnaC not only loads but activates DnaB in an ATP-independent manner
Previous studies have established that a mutant of DnaC incapable of binding ATP can nonetheless facilitate the loading of DnaB onto DNA (Davey et al., 2002). This finding, together with preponderance of spirally-organized RecA domains in various DnaB models suggested to us that DnaC AAA+ ATPase regions might not be required to actively promote the formation of the spiral configuration seen in DnaBC, but to instead stabilize it. To test this consideration, we compared the ability of both full-length DnaC and the isolated DnaB-interaction domain of DnaC to load the helicase onto a closed-circular, single-stranded M13 DNA substrate. In this assay, helicase loading is reflected by the ability of DnaB to displace an annealed radiolabeled oligonucleotide bearing a 3’ overhang (Figure 6C); since DnaB translocates 5’→3’ (LeBowitz and McMacken, 1986), unwinding can only be observed when the helicase is opened and placed around the M13 DNA (Wahle et al., 1989). As expected, reactions containing DnaB and full-length DnaC resulted in unwinding of a significant fraction of the substrate, comparable to previously-observed activity levels for this assay (Davey et al., 2002; Fang et al., 1999; Wahle et al., 1989). However, the isolated helicase interaction domain of DnaC also allowed DnaB to gain access to and unwind the duplex, albeit at a reduced level compared to the full-length loader (Figure 6C). This finding demonstrates that the DnaC N-terminal domain is sufficient for remodeling DnaB into a loading-competent state, but that its AAA+ domain increases loading efficiency, potentially by participating in the binding of single-stranded DNA (Davey et al., 2002; Mott et al., 2008), stabilizing an open-ring state, or both.
A curious biochemical feature of DnaB is the relative inefficiency by which the helicase unwinds DNA duplexes bearing forked single-stranded ends; although the helicase can thread onto such substrates (Jezewska et al., 1998), thereby bypassing an obvious need for a helicase loader, we and others have observed that robust helicase activity requires either DnaC or a large excess of DnaB over DNA (Galletto et al., 2004a; Kaplan, 2000; Kaplan and Steitz, 1999). In light of the dramatic structural effect DnaC has on DnaB (Figure 5), this biochemical behavior suggested that the loader not only assists in depositing the helicase onto DNA, but that the action of DnaC might further help switch the motor from an inactive to an active configuration. To test this idea, we analyzed the effect of both DnaC and the DnaC N-terminal domain on DnaB’s ability to unwind a forked DNA oligonucleotide when the helicase is present at a near stoichiometric ratio compared to substrate (Figures 6D and S7). A suitable substrate was formed by annealing two complementary oligos, one bearing a Cy3 fluorophore and the other a quenching label, which contained either a 5’ or 3’ dT25 overhang. The increase in fluorescence emission in the reaction as DnaB separated the duplex substrate was then monitored as a function of time.
Using a two-fold molar excess of helicase over DNA, we observed negligible unwinding of the fork for reactions containing only DnaB and ATP (Figure 6D). By contrast, addition of full-length DnaC to these reactions resulted in unwinding of most of the duplex present in the reaction. This result suggests that DnaC can help load the helicase onto DNA in a conformation that is distinct from a closed-ring DnaB hexamer that has threaded itself onto a forked end, or that the loader can remodel DnaB that already resides on the substrate; either event would serve to activate the motor for translocation once the loader dissociates. Since productive unwinding by DnaB is known to be fast (291 bp/sec) (Galletto et al., 2004a), the lag in the appearance of product in this assay probably arises as a result of the action of DnaC on the helicase. Interestingly, the DnaC N-terminal region proved nearly as efficient as full-length DnaC in promoting DNA unwinding by DnaB, indicating that it exerts a similar effect on the helicase as the intact loader, despite lacking its ATPase domain (Figure 6D). Together, these observations demonstrate that DnaC not only aids DnaB loading, but that it also can potentiate the ability of the helicase to efficiently unwind substrate, likely by helping to remodel DnaB into a more productive configuration.
Implications for DNA replication associated events
The mechanisms by which DnaC associates with and controls the loading of DnaB have long remained enigmatic. Our findings provide fundamental insights into this process, and further highlight new facets of DnaC and DnaB function that account for the action of the loader during replication (Figure 7A). In this scheme, the DnaC N-terminal domain would first bind to the RecA fold of a closed DnaB ring. As successive loaders continue to bind, the interaction between the N-terminal domain and DnaB would induce a conformational change that alleviates a restraining activity resident within the N-terminal DnaB collar, allowing the helicase ring to open. In the presence of ATP, the AAA+ domains of neighboring DnaC protomers would oligomerize, stabilizing the helicase in an open lockwasher conformation to permit DNA binding and loading of a de-repressed DnaB hexamer. The interaction of DnaB with other factors and/or ATP hydrolysis would in turn promote the release of DnaC from the helicase, allowing translocation and unwinding to ensue.
Figure 7. Mechanism of DnaC action.
(A) DnaC opens and remodels DnaB to facilitate DNA loading and unwinding. Closedring DnaB cannot engage a topologically-closed DNA substrate. DnaC associates with the helicase, remodeling the N-terminal collar and triggering helicase opening. In the presence of ATP, DnaC AAA+ domains further assemble into a helical conformation that stabilizes the open-ring complex and assists with DNA binding. ATP hydrolysis by DnaC and/or DnaG help disengage the loader (Davey et al., 2002; Makowska-Grzyska and Kaguni, 2010) leaving an active helicase encircled around DNA. See also Figure S7.
(B) Hypothetical model showing how DnaC is free to associate with a DnaA filament even when bound to DnaB (as proposed in (Mott et al., 2008)). The model was generated by aligning a DnaA filament bound to single-stranded DNA (PDB ID 3R8F (Duderstadt et al., 2011)) – which bears an exposed arginine-finger at the 5’ end of the complex – with the solvent accessible nucleotide-binding face of the terminal DnaC protomer in DnaBC in a manner consistent with typical AAA+/AAA+ interactions. The superposition co-aligns the pores of the all three proteins, positioning DNA bound by the central channel of DnaA to enter into the helicase/loader complex.
An interesting feature of this model is that, unlike clamp loaders (Kelch et al., 2011; Miyata et al., 2005; Pietroni et al., 1997; Simonetta et al., 2009; Turner et al., 1999; Zhuang et al., 2006), DnaC does not appear to require ATP (or even its ATPase domain) to actively open and load a helicase ring (Davey et al., 2002) (Figure 6C). This behavior raises the question of why a functional DnaC ATPase is needed for replication initiation both in vitro and in vivo (Davey et al., 2002; Ludlam et al., 2001; Mott et al., 2008). A plausible explanation is that the ATPase elements in DnaC provide a switch to trigger the timely release of DnaC from the helicase after loading has occurred (Davey et al., 2002). This situation is analogous to clamp loader systems, where ATP hydrolysis, induced by the binding of DNA, reduces the affinity of the loader for both the clamp and DNA (Ason et al., 2000; Goedken et al., 2005; Hingorani et al., 1999; Jarvis et al., 1989). In a similar manner, the binding of both DnaB and ssDNA stimulates ATP hydrolysis by DnaC (Davey et al., 2002), which may allow the loader to be ejected more efficiently from the helicase. The DnaG primase protein also binds to DnaB (Chang and Marians, 2000; Tougu et al., 1994) and together with primer synthesis, appears to promote DnaC release from DnaB (Makowska-Grzyska and Kaguni, 2010). Interestingly, DnaG binds to a different organizational state of the N-terminal DnaB collar than is observed here for the DnaBC complex (Bailey et al., 2007b); this behavior suggests that the binding of DnaG and DnaC to DnaB may mutually antagonize one another at certain points in the DnaB functional cycle.
Further implications for other replication-associated events can be drawn from these studies. For example, in A. aeolicus, the ATPase domains of DnaA and DnaC have been shown to associate in a nucleotide-dependent manner, an interaction that may assist with orienting one copy of DnaB onto a replication origin (Mott et al., 2008). The disposition of DnaC ATPase domains in the DnaBC complex leaves one loader subunit sufficiently accessible to readily accommodate the binding of DnaA oligomer through AAA+/AAA+ interactions. As the central channel of a DnaA oligomer binds to an extended single-stranded DNA substrate (Duderstadt et al., 2011), this association would provide a means for orienting and threading a nucleic acid chain into the pore of the helicase/loader complex; the polarity by which DnaA would feed single-stranded DNA into DnaBC in such a scheme is consistent with the known 5’→3’ directionality of the helicase (Figure 7B). Our analyses additionally demonstrate that DnaC can directly activate the DnaB helicase. Besides promoting efficient transition from replication initiation to elongation, this latter action also could assist with other events, such as the restart of stalled replication forks in which the helicase remains encircled around DNA. Future studies will be needed to probe these concepts further.
EXPERIMENTAL PROCEDURES
A more detailed description of the methods used in this work can be found in the Supplemental Material.
Protein Expression and Purification
E. coli DnaB and DnaC were independently expressed in E. coli using a pET-based plasmid system. Following harvesting and lysis, proteins were purified by ammonium sulfate precipitation (for DnaB) and column chromatography (affinity, ion-exchange, and gel-filtration), after which they were concentrated and stored at − 80 °C for subsequent studies. DnaBC complexes prepared for structural analyses were passed over an analytic gel filtration column just prior to spotting on EM grids.
Electron microscopy and image analysis
DnaBC samples were stained on EM grids with 2% (w/v) uranyl formate. All electron microscopy data were collected using LEGINON (Suloway et al., 2005), and preprocessed with EMAN2 (Lander et al., 2009; Tang et al., 2007) . Initial random conical tilt reconstructions were obtained using XMIPP (Scheres et al., 2008), and refined with EMAN2 and SPARX (Hohn et al., 2007; Tang et al., 2007). UCSF Chimera was used for volume segmentation, docking of the atomic structures and figure generation (Pettersen et al., 2004).
Small Angle X-ray Scattering
SAXS data were collected at beamline 12.3.1 at the Advanced Light Source of Lawrence Berkeley National Laboratory. Samples contained 6 mg/mL DnaBC and 1 mM AMPPNP. Data were processed and analyzed using PRIMUS (Konarev et al., 2003). Theoretical scattering curves from the DnaBC EM model fitting and DnaB crystal structures were generated by FOXS and compared to DnaBC experimental data using MES (Pelikan et al., 2009; Schneidman-Duhovny et al., 2010).
DNA Unwinding Assays
M13 loading and unwinding assays was performed as previously described (Davey et al., 2002) with minor modifications. A 37-mer oligonucleotide (5’-TGTACCCCGGTTGATAATCAGAAAAGCCCCTTGCTAC 3’) was 32P-end labeled and annealed to single-stranded M13mp18 DNA, leaving a 7-nt 3’-overhang. 25 mL reactions containing 50 fmol substrate DNA, varying amounts of DnaB, DnaC and/or the DnaC NTD, and reaction buffer (40 mM HEPES-NaOH pH 7.5, 10 mM MgCl2, 5 mM DTT, 0.1 mg/mL BSA, and 5 mM ATP) were incubated at 37 °C for 15 minutes, and then quenched with 20 mM EDTA and 0.2 % SDS. Reactions were resolved on a 15 % acrylamide native gel at room temperature and quantified by autoradiography.
Unwinding of forked DNA substrates was carried out by first annealing a 5’-Cy3-labeled oligonucleotide (5’-Cy3-TACGTAACGAGCCTGC(dT)25-3’) to a 1.2 molar excess of a 3’-black hole quencher-labeled strand (5’-(dT)25-GCAGGCTCGTTACGTA-BHQ2–3’). A capture oligo (5’-GCAGGCTCGTTACGTA-3’) complementary to the base-paired region of the Cy3-labeled strand was added to all reactions to prevent re-annealing of the unwound substrate and recycling of DnaB by the loader. Reactions containing 200 nM DnaB hexamer (with or without 1.2 mM loader monomer), 100 nM fork substrate, and 200 nM capture oligo were monitored at 37 °C for 15 minutes in assay buffer (20 mM HEPES-KOH pH 7.5, 5 mM magnesium acetate, 50 mM potassium glutamate, 5 % glycerol, 4 mM DTT, 0.2 mg/mL BSA and 1 mM ATP).
Supplementary Material
Highlights.
The DnaB-DnaC complex forms a topologically open, three-tiered toroid.
DnaC remodels DnaB to produce a cleft in the helicase ring suitable for DNA passage.
DnaC’s AAA+ fold is dispensable for DnaB loading and activation.
DnaB possesses auto-regulatory elements that control helicase loading and unwinding.
ACKNOWLEDGEMENTS
The authors are grateful to the Nogales lab and the staff at beamlines 8.3.1 and 12.3.1 of the Advanced Light Source for help with data collection and analysis, and to the Berger lab for helpful discussions and aid in preparing the manuscript. 3D EM models for E. coli DnaBC and DnaB have been deposited to the EMDB (www.emdatabank.org) under accession number EMD-2321 and EMD-2322, respectively. This work was supported by an NIH NRSA post-doctoral fellowship (F32GM090803 ) to VLO, a postdoctoral fellowship from the Programa Nacional de Movilidad de Recursos Humanos del Plan Nacional de I+D+i 2008–2011 from the Spanish Ministry of Education to EA, a Department of Energy Office of Science Graduate Fellowship (DOE SCGF) to IVH, and by the NIGMS to JMB (RO1-GM071747).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ason B, Bertram JG, Hingorani MM, Beechem JM, O’Donnell M, Goodman MF, Bloom LB. A model for Escherichia coli DNA polymerase III holoenzyme assembly at primer/template ends. DNA triggers a change in binding specificity of the gamma complex clamp loader. J Biol Chem. 2000;275:3006–3015. doi: 10.1074/jbc.275.4.3006. [DOI] [PubMed] [Google Scholar]
- Bailey S, Eliason WK, Steitz TA. The crystal structure of the Thermus aquaticus DnaB helicase monomer. Nucleic Acids Res. 2007a;35:4728–4736. doi: 10.1093/nar/gkm507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Eliason WK, Steitz TA. Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science. 2007b;318:459–463. doi: 10.1126/science.1147353. [DOI] [PubMed] [Google Scholar]
- Baker TA, Bell SP. Polymerases and the replisome: machines within machines. Cell. 1998;92:295–305. doi: 10.1016/s0092-8674(00)80923-x. [DOI] [PubMed] [Google Scholar]
- Barcena M, Ruiz T, Donate LE, Brown SE, Dixon NE, Radermacher M, Carazo JM. The DnaB.DnaC complex: a structure based on dimers assembled around an occluded channel. Embo J. 2001;20:1462–1468. doi: 10.1093/emboj/20.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas EE, Biswas SB, Bishop JE. The dnaB protein of Escherichia coli: mechanism of nucleotide binding, hydrolysis, and modulation by dnaC protein. Biochemistry. 1986;25:7368–7374. doi: 10.1021/bi00371a019. [DOI] [PubMed] [Google Scholar]
- Biswas SB, Flowers S, Biswas-Fiss EE. Quantitative analysis of nucleotide modulation of DNA binding by DnaC protein of Escherichia coli. Biochem J. 2004;379:553–562. doi: 10.1042/BJ20031255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas T, Tsodikov OV. Hexameric ring structure of the N-terminal domain of Mycobacterium tuberculosis DnaB helicase. Febs J. 2008;275:3064–3071. doi: 10.1111/j.1742-4658.2008.06460.x. [DOI] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2–7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- Chang P, Marians KJ. Identification of a region of Escherichia coli DnaB required for functional interaction with DnaG at the replication fork. J Biol Chem. 2000;275:26187–26195. doi: 10.1074/jbc.M001800200. [DOI] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–477. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, Fang L, McInerney P, Georgescu RE, O’Donnell M. The DnaC helicase loader is a dual ATP/ADP switch protein. Embo J. 2002;21:3148–3159. doi: 10.1093/emboj/cdf308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, O'Donnell M. Replicative helicase loaders: ring breakers and ring makers. Curr Biol. 2003;13:R594–R596. doi: 10.1016/s0960-9822(03)00523-2. [DOI] [PubMed] [Google Scholar]
- Duderstadt KE, Chuang K, Berger JM. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011;478:209–213. doi: 10.1038/nature10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2–7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci U S A. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Davey MJ, O’Donnell M. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- Galletto R, Jezewska MJ, Bujalowski W. Interactions of the Escherichia coli DnaB helicase hexamer with the replication factor the DnaC protein. Effect of nucleotide cofactors and the ssDNA on protein-protein interactions and the topology of the complex. J Mol Biol. 2003;329:441–465. doi: 10.1016/s0022-2836(03)00435-2. [DOI] [PubMed] [Google Scholar]
- Galletto R, Jezewska MJ, Bujalowski W. Unzipping mechanism of the double-stranded DNA unwinding by a hexameric helicase: quantitative analysis of the rate of the dsDNA unwinding, processivity and kinetic step-size of the Escherichia coli DnaB helicase using rapid quench-flow method. J Mol Biol. 2004a;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Galletto R, Maillard R, Jezewska MJ, Bujalowski W. Global conformation of the Escherichia coli replication factor DnaC protein in absence and presence of nucleotide cofactors. Biochemistry. 2004b;43:10988–11001. doi: 10.1021/bi049377y. [DOI] [PubMed] [Google Scholar]
- Goedken ER, Kazmirski SL, Bowman GD, O’Donnell M, Kuriyan J. Mapping the interaction of DNA with the Escherichia coli DNA polymerase clamp loader complex. Nat Struct Mol Biol. 2005;12:183–190. doi: 10.1038/nsmb889. [DOI] [PubMed] [Google Scholar]
- Hingorani MM, Bloom LB, Goodman MF, O’Donnell M. Division of labor--sequential ATP hydrolysis drives assembly of a DNA polymerase sliding clamp around DNA. Embo J. 1999;18:5131–5144. doi: 10.1093/emboj/18.18.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Ioannou C, Schaeffer PM, Dixon NE, Soultanas P. Helicase binding to DnaI exposes a cryptic DNA-binding site during helicase loading in Bacillus subtilis. Nucleic Acids Res. 2006;34:5247–5258. doi: 10.1093/nar/gkl690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsathitphaisarn O, Wing RA, Eliason WK, Wang J, Steitz TA. The Hexameric Helicase DnaB Adopts a Nonplanar Conformation during Translocation. Cell. 2012;151:267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis TC, Paul LS, Hockensmith JW, von Hippel PH. Structural and enzymatic studies of the T4 DNA replication system. II. ATPase properties of the polymerase accessory protein complex. J Biol Chem. 1989;264:12717–12729. [PubMed] [Google Scholar]
- Jeruzalmi D, O’Donnell M, Kuriyan J. Crystal structure of the processivity clamp loader gamma complex of E. coli DNA polymerase III. Cell. 2001;106:429–441. doi: 10.1016/s0092-8674(01)00463-9. [DOI] [PubMed] [Google Scholar]
- Jezewska MJ, Rajendran S, Bujalowska D, Bujalowski W. Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB Helicase? Fluorescence energy transfer studies. J Biol Chem. 1998;273:10515–10529. doi: 10.1074/jbc.273.17.10515. [DOI] [PubMed] [Google Scholar]
- Kaguni JM. Replication initiation at the Escherichia coli chromosomal origin. Curr Opin Chem Biol. 2011;15:606–613. doi: 10.1016/j.cbpa.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DL. The 3'-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J Mol Biol. 2000;301:285–299. doi: 10.1006/jmbi.2000.3965. [DOI] [PubMed] [Google Scholar]
- Kaplan DL, Steitz TA. DnaB from Thermus aquaticus unwinds forked duplex DNA with an asymmetric tail length dependence. J Biol Chem. 1999;274:6889–6897. doi: 10.1074/jbc.274.11.6889. [DOI] [PubMed] [Google Scholar]
- Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8:163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- Kelch BA, Makino DL, O’Donnell M, Kuriyan J. How a DNA polymerase clamp loader opens a sliding clamp. Science. 2011;334:1675–1680. doi: 10.1126/science.1211884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelch BA, Makino DL, O’Donnell M, Kuriyan J. Clamp loader ATPases and the evolution of DNA replication machinery. BMC Biol. 2012;10:34. doi: 10.1186/1741-7007-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori JA, Kornberg A. The Escherichia coli dnaC gene product. II. Purification, physical properties, and role in replication. J Biol Chem. 1982a;257:13763–13769. [PubMed] [Google Scholar]
- Kobori JA, Kornberg A. The Escherichia coli dnaC gene product. III. Properties of the dnaB-dnaC protein complex. J Biol Chem. 1982b;257:13770–13775. [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- Koonin EV. DnaC protein contains a modified ATP-binding motif and belongs to a novel family of ATPases including also DnaA. Nucleic Acids Res. 1992;20:1997. doi: 10.1093/nar/20.8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham GJ, Bacheller DJ, Pietroni P, von Hippel PH. Structural analyses of gp45 sliding clamp interactions during assembly of the bacteriophage T4 DNA polymerase holoenzyme. II. The Gp44/62 clamp loader interacts with a single defined face of the sliding clamp ring. J Biol Chem. 1997;272:31677–31684. doi: 10.1074/jbc.272.50.31677. [DOI] [PubMed] [Google Scholar]
- LeBowitz JH, McMacken R. The Escherichia coli dnaB replication protein is a DNA helicase. J Biol Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- Lo YH, Tsai KL, Sun YJ, Chen WT, Huang CY, Hsiao CD. The crystal structure of a replicative hexameric helicase DnaC and its complex with single-stranded DNA. Nucleic Acids Res. 2009;37:804–814. doi: 10.1093/nar/gkn999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlam AV, McNatt MW, Carr KM, Kaguni JM. Essential amino acids of Escherichia coli DnaC protein in an N-terminal domain interact with DnaB helicase. J Biol Chem. 2001;276:27345–27353. doi: 10.1074/jbc.M101940200. [DOI] [PubMed] [Google Scholar]
- Lyubimov AY, Costa A, Bleichert F, Botchan MR, Berger JM. ATP-dependent conformational dynamics underlie the functional asymmetry of the replicative helicase from a minimalist eukaryote. Proc Natl Acad Sci U S A. 2012;109:11999–12004. doi: 10.1073/pnas.1209406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill SA. Protein-protein interactions in the archaeal core replisome. Biochem Soc Trans. 2011;39:163–168. doi: 10.1042/BST0390163. [DOI] [PubMed] [Google Scholar]
- Makowska-Grzyska M, Kaguni JM. Primase directs the release of DnaC from DnaB. Mol Cell. 2010;37:90–101. doi: 10.1016/j.molcel.2009.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek J, Kaguni JM. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J Biol Chem. 1994;269:4883–4890. [PubMed] [Google Scholar]
- McGlynn P. Helicases that underpin replication of protein-bound DNA in Escherichia coli. Biochem Soc Trans. 2011;39:606–610. doi: 10.1042/BST0390606. [DOI] [PubMed] [Google Scholar]
- Miyata T, Suzuki H, Oyama T, Mayanagi K, Ishino Y, Morikawa K. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc Natl Acad Sci U S A. 2005;102:13795–13800. doi: 10.1073/pnas.0506447102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mott ML, Erzberger JP, Coons MM, Berger JM. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008;135:623–634. doi: 10.1016/j.cell.2008.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen O, Lobner-Olesen A. Once in a lifetime: strategies for preventing re-replication in prokaryotic and eukaryotic cells. EMBO Rep. 2008;9:151–156. doi: 10.1038/sj.embor.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelikan M, Hura GL, Hammel M. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pietroni P, Young MC, Latham GJ, von Hippel PH. Structural analyses of gp45 sliding clamp interactions during assembly of the bacteriophage T4 DNA polymerase holoenzyme. I. Conformational changes within the gp44/62-gp45-ATP complex during clamp loading. J Biol Chem. 1997;272:31666–31676. doi: 10.1074/jbc.272.50.31666. [DOI] [PubMed] [Google Scholar]
- Pomerantz RT, O'Donnell M. Replisome mechanics: insights into a twin DNA polymerase machine. Trends Microbiol. 2007;15:156–164. doi: 10.1016/j.tim.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JF. Concerted loading of Mcm2–7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martin C, Radermacher M, Wolpensinger B, Engel A, Miles CS, Dixon NE, Carazo JM. Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC. Structure. 1998;6:501–509. doi: 10.1016/s0969-2126(98)00051-3. [DOI] [PubMed] [Google Scholar]
- San Martin MC, Stamford NP, Dammerova N, Dixon NE, Carazo JM. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J Struct Biol. 1995;114:167–176. doi: 10.1006/jsbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- Schaeffer PM, Headlam MJ, Dixon NE. Protein--protein interactions in the eubacterial replisome. IUBMB Life. 2005;57:5–12. doi: 10.1080/15216540500058956. [DOI] [PubMed] [Google Scholar]
- Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, Marabini R. Image processing for electron microscopy single-particle analysis using XMIPP. Nat Protoc. 2008;3:977–990. doi: 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Weigel C, Messer W. The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol Microbiol. 2000;37:1270–1279. doi: 10.1046/j.1365-2958.2000.02096.x. [DOI] [PubMed] [Google Scholar]
- Seybert A, Singleton MR, Cook N, Hall DR, Wigley DB. Communication between subunits within an archaeal clamp-loader complex. Embo J. 2006;25:2209–2218. doi: 10.1038/sj.emboj.7601093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O’Donnell M, Kuriyan J. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM. Structure of the Rho transcription terminator: mechanism of mRNA recognition and helicase loading. Cell. 2003;114:135–146. doi: 10.1016/s0092-8674(03)00512-9. [DOI] [PubMed] [Google Scholar]
- Soultanas P. A functional interaction between the putative primosomal protein DnaI and the main replicative DNA helicase DnaB in Bacillus. Nucleic Acids Res. 2002;30:966–974. doi: 10.1093/nar/30.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soultanas P. Loading mechanisms of ring helicases at replication origins. Mol Microbiol. 2012;84:6–16. doi: 10.1111/j.1365-2958.2012.08012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tougu K, Peng H, Marians KJ. Identification of a domain of Escherichia coli primase required for functional interaction with the DnaB helicase at the replication fork. J Biol Chem. 1994;269:4675–4682. [PubMed] [Google Scholar]
- Tsai KL, Lo YH, Sun YJ, Hsiao CD. Molecular interplay between the replicative helicase DnaC and its loader protein DnaI from Geobacillus kaustophilus. J Mol Biol. 2009;393:1056–1069. doi: 10.1016/j.jmb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- Turner J, Hingorani MM, Kelman Z, O'Donnell M. The internal workings of a DNA polymerase clamp-loading machine. Embo J. 1999;18:771–783. doi: 10.1093/emboj/18.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velten M, McGovern S, Marsin S, Ehrlich SD, Noirot P, Polard P. A two-protein strategy for the functional loading of a cellular replicative DNA helicase. Mol Cell. 2003;11:1009–1020. doi: 10.1016/s1097-2765(03)00130-8. [DOI] [PubMed] [Google Scholar]
- Wahle E, Lasken RS, Kornberg A. The dnaB-dnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing dnaB functions. J Biol Chem. 1989;264:2469–2475. [PubMed] [Google Scholar]
- Wang G, Klein MG, Tokonzaba E, Zhang Y, Holden LG, Chen XS. The structure of a DnaB-family replicative helicase and its interactions with primase. Nat Struct Mol Biol. 2008;15:94–100. doi: 10.1038/nsmb1356. [DOI] [PubMed] [Google Scholar]
- Wickner S, Hurwitz J. Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc Natl Acad Sci U S A. 1975;72:921–925. doi: 10.1073/pnas.72.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yu X, VanLoock MS, Jezewska MJ, Bujalowski W, Egelman EH. Flexibility of the rings: structural asymmetry in the DnaB hexameric helicase. J Mol Biol. 2002;321:839–849. doi: 10.1016/s0022-2836(02)00711-8. [DOI] [PubMed] [Google Scholar]
- Zhuang Z, Yoder BL, Burgers PM, Benkovic SJ. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc Natl Acad Sci U S A. 2006;103:2546–2551. doi: 10.1073/pnas.0511263103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.