Abstract
Animal development requires temporal coordination between recurrent processes and sequential events, but the underlying timing mechanisms are not yet understood. The molting cycle of C. elegans provides an ideal system to study this basic problem. We recently characterized LIN-42, which is related to the circadian clock protein PERIOD, as a key component of the developmental timer underlying rhythmic molting cycles. In this context, LIN-42 coordinates epithelial stem cell dynamics with progression of the molting cycle. Repeated actions of LIN-42 may enable the reprogramming of seam cell temporal fates, while stage-specific actions of LIN-42 and other heterochronic genes select fates appropriate for upcoming, rather than passing, life stages. Here, we discuss the possible configuration of the molting timer, which may include interconnected positive and negative regulatory loops among lin-42, conserved nuclear hormone receptors such as NHR-23 and -25, and the let-7 family of microRNAs. Physiological and environmental conditions may modulate the activities of particular components of this molting timer. Finding that LIN-42 regulates both a sleep-like behavioral state and epidermal stem cell dynamics further supports the model of functional conservation between LIN-42 and mammalian PERIOD proteins. The molting timer may therefore represent a primitive form of a central biological clock and provide a general paradigm for the integration of rhythmic and developmental processes.
Keywords: lin-42, let-7 microRNAs, Period, circadian clocks, developmental timing, heterochrony, lethargus, molting, nuclear hormone receptors
Introduction
Once-in-a-lifetime changes, such as metamorphosis, and repeated processes, such as cell division, are both essential for animal development. However, the timing mechanisms that coordinate sequential and recurrent events in developing organisms are not well understood.
Studies of developmental timing have focused on hormones and gene regulatory cascades that trigger specific chronological events. One prominent example is the regulation of insect metamorphosis by the steroid hormone 20-hydroxy-ecydsone (20-E) and related transcriptional cascades.1 Specific cytokines and transcription factors that promote the stepwise maturation of mammalian B-cells from pluripotent hematopoietic stem cells have also been well-characterized.2 The use of C. elegans as a model system led to the seminal discovery of conserved microRNAs (miRNAs) and protein-coding genes that program the successive temporal fates of the stem cell-like lateral epithelial seam cells.3-5 These cells divide asymmetrically early in every larval stage; undergo one additional symmetric division in the L2 stage; and ultimately terminally differentiate, by fusing with one another and exiting the cell cycle, at the larval-to-adult transition.5,6 Collectively, the genes that control these stage-specific patterns of seam cell division and differentiation make up the heterochronic gene regulatory network.7,8
Relatively little is known about biological timers that drive temporally reiterated processes in the context of metazoan development. One of the best-characterized developmental oscillators is the segmentation clock of vertebrates, which is composed of interconnected positive and negative regulatory interactions among components of the Notch and Wnt signaling pathways. The segmentation clock drives the rhythmic expression of master transcription factors, which in turn program spatial and temporal cell fates.9 The clock operates in somite precursors but not mature tissues. In C. elegans, most epithelial cells and synctyia of juveniles exhibit rhythmic gene expression profiles associated with the four larval molts, as we shall describe.10,11
The best-characterized biological timers are the circadian clocks of mature animals. These clocks synchronize daily behavioral, hormonal, and metabolic rhythms with predictable fluctuations in environmental and physiological conditions.12 The mammalian clock consists of interlocked positive and negative feedback loops among the transcription co-factor PERIOD (PER), the basic helix-loop-helix (HLH) transcription factors CLOCK and BMAL, the nuclear hormone receptors (NRs) RORα and REV-ERB, and other ancillary components.12 PER and other core clock proteins also regulate the cell cycle, and disruption or misalignment of the circadian clock leads to tumor progression in mouse models.13,14 However, the extent to which the canonical circadian clock or any other PER-based oscillator coordinates cell cycles with developmental transitions had not been examined.
To address these basic questions, we investigated the molecular mechanism that times the molting cycles of C. elegans.15 The larval molts involve distinctive cellular and organism behaviors that together enable the rapid and repeated reconstruction of cuticle, which is a collagen-rich extracellular matrix (ECM) (Fig. 1).16,17 Briefly, the underlying hypoderm detaches from the preexisting cuticle (apolysis), and generates a new cuticle underneath the old one. Various cell-ECM adhesive complexes that collectively tether the cuticle to the hypoderm, the underlying basement membrane (BM), and body wall muscles are also remade during the molts.11,18 Notably, the seam cells repeatedly switch between proliferative and quiescent states in phase with the periodic molts. As described, these cells undergo stem cell-like asymmetric divisions early in every larval stage. The posterior daughters retain pluripotency and proliferative potential. In contrast, the anterior daughters fuse with the hypodermal syncytium (hyp7), increasing the size of the body.19,20 The pluripotent seam cells are largely quiescent and contribute to the synthesis of cuticles during the molts.6 The mechanisms that coordinate stem cell and ECM dynamics in this context are not well understood, but are essential for viability. Indeed, both genetic mutations that prevent seam cell specification and drugs that delay seam cell division cause aberrant, fatal molts.21-24
The rhythmic behaviors associated with molting cycles include lethargy and idiosyncratic movements used to escape the old cuticle (ecdyse). Lethargus has been used to model mammalian sleep, as this reversible quiescent state is characterized by the cessation of food-intake and locomotion as well as decreased responsiveness to external stimuli.25,26 Communication among neurons, muscles, and epithelial cells likely coordinates these behavioral routines with progression of the molting cycle.15,25-27 Larvae molt four times, once every 8–10 h under favorable culture conditions. Although this periodicity resembles a harmonic of the circadian clock, the anticipated pacemaker had not been characterized.
Using a candidate gene approach, we identified lin-42, which is related to PERIOD,28,29 as a key component of the molting clock that operates in the epithelium of juveniles.15 This LIN-42-based timer sustains the rhythm of the molting cycle and synchronizes seam cell dynamics with the larval molts.15 In addition, LIN-42 acts in the heterochronic pathway to program the L3-stage, and possibly other, seam cell temporal fates.15,30,31
Here, we summarize our recent findings and further discuss the possible molecular configurations, environmental and physiological inputs, and systemic outputs of the molting clock. Several lines of evidence now support the model of functional conservation between C. elegans LIN-42 and mammalian PER proteins. The use of both reiterated and consecutive functions of LIN-42/PER to regulate developmental timing in C. elegans may therefore provide a general paradigm for the integration of rhythmic and sequential processes in biology (Fig. 1). Our ongoing studies of the molting timer will likely uncover novel mechanisms by which conventional and unconventional PER-based clocks regulate biological rhythms throughout the life of metazoans.
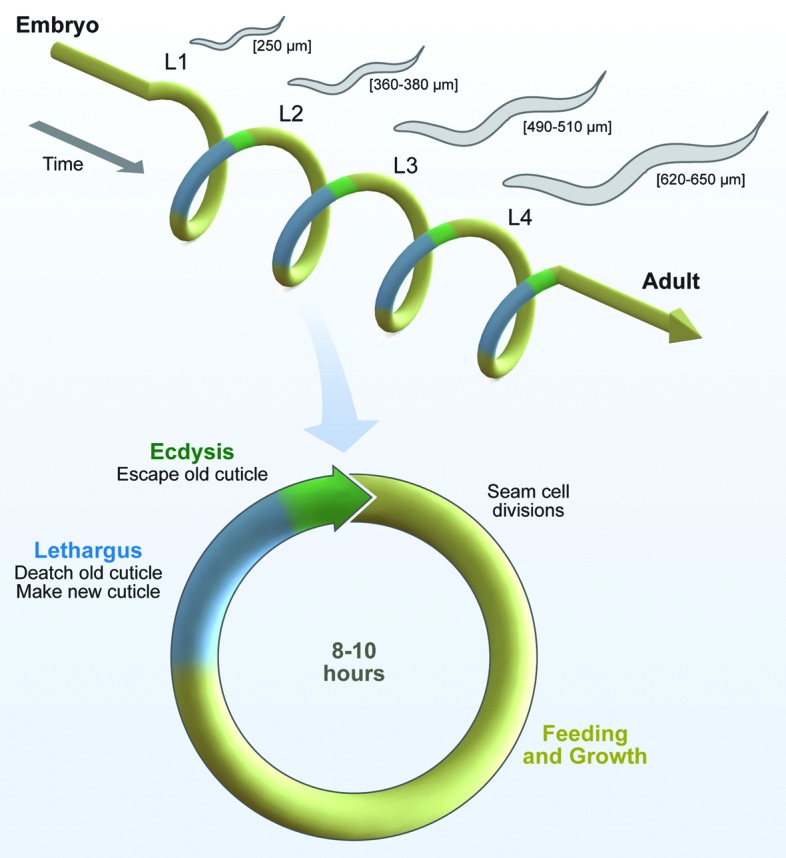
Figure 1. Molting is a reiterated process in development. Each molt involves specialized cellular programs and animal behaviors. Epithelial cells and syncytia detach from the preexisting cuticle and secrete a new cuticle underneath the old one. Larvae are quiescent for approximately 2 h while the cuticle is remade (lethargus). Larvae then execute a series of idiosyncratic movements to escape the old cuticle (ecdysis); this step takes about 20 min. The entire process is repeated four times, every 8–10 h under favorable culture conditions. The lateral epithelial seam cells undergo stem cell-like asymmetric divisions early in every larval stage, but contribute to the synthesis of new cuticles during the molts. Reprogramming of the successive, stage-specific temporal fates of the seam cells occurs around the time of the molts (not shown).
Identifying Components of the Molting Timer
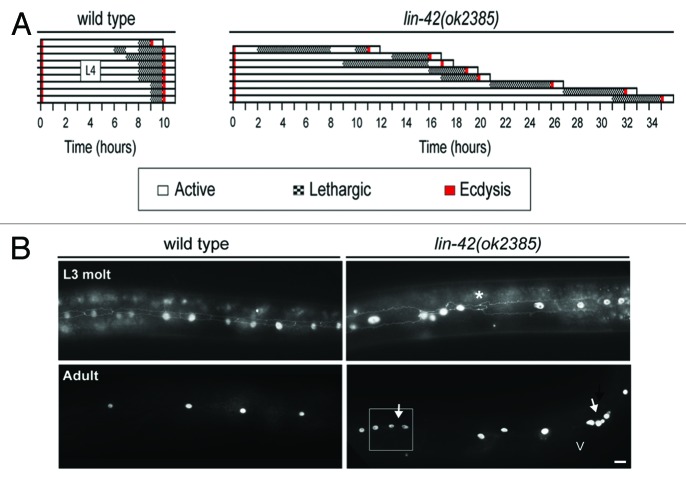
The genetic basis of the circadian clock was established by the characterization of three distinct alleles of Drosophila melanogaster period that shortened, lengthened, or abolished daily rhythms in locomoter activity.32 Two hallmarks of core clock components have since emerged. The first is that either inactivation or constitutive expression of the corresponding genes disrupts the clock-controlled biological rhythm.12 We found that lin-42(ok2385) mutants molted at unpredictable times.15 Strikingly, individual lin-42(ok2385) mutants completed the fourth molt up to 24 h apart, whereas wild-type animals completed the molts virtually in synchrony (Fig. 2A). Unusually long periods of time typically elapsed between ecdyses in lin-42(ok2385) mutants. However, new molts occasionally began prior to the completion of earlier ecdyses. These unnaturally short cycles became evident when larvae shed two cuticles within an hour, or became trapped within two incompletely shed cuticles. Further, forced expression of lin-42a from an inducible promoter led to anachronistic and fatal molts, in which the hypoderm detached from the cuticle prior to stage-specific developmental benchmarks.15
Figure 2. Molting cycles and seam cell dynamics are temporally misaligned in lin-42 mutants. (A) Chart shows the asynchronous execution of the fourth molt in lin-42(ok2385) mutants, compared with wild-type larvae. Each bar represents the behavior of a single animal monitored from the third to the fourth molt. Larvae were observed for 30 sec of every hour and behavior scored by visual inspection. The average duration of the L4 stage was 21.8 ± 3.1 (Cν = 0.4) hours in lin-42(ok2385) mutants, compared with 9.9 ± 0.1 (Cν = 0.05) in wild-type animals. (B) Seam cells were visualized in lin-42(ok2385) mutants and wild-type animals by fluorescence microscopy, using the AJM-1::GFP marker for adherens junctions and the scm::gfp reporter for seam nuclei. Seam cells of lin-42 mutants often underwent precocious homotypic fusion during the third molt (asterisk). However, some seam nuclei underwent supernumerary divisions in reproductively mature animals (arrows). The white box indicates one region where alae were detected on the cuticle. “V” indicates the position of the vulva. Scale bar corresponds to 10 μM. This figure was adapted from Monsalve et al., LIN-42/PERIOD Controls Cyclical and Developmental Progression of C. elegans Molts, Current Biology21, 2033–2045 (2011).
We also found that lin-42 was essential for proper seam cell dynamics throughout larval development.15 The seam cells were inappropriately detached from one another and misshapen in lin-42(ok2385) mutants undergoing molts. At the L3-to-L4 transition, some seam cells fused prematurely and secreted alae, a phenotype that was previously observed in other lin-42 mutants.30,31 However, at the L4-to-adult transition, some seam cells failed to exit the cell cycle and subsequently underwent supernumerary divisions (Fig. 2B). Thus, lin-42(ok2385) animals exhibited defining features of both “precocious” and “retarded” heterochronic mutants. Taken together, our findings suggest that a LIN-42-based timer synchronizes seam cell dynamics with larval molting cycles. The lin-42 gene regulates both the periodicity and the quality of the molts, as lin-42 also functions in the heterochronic pathway to program the L3, and possibly other, seam cell temporal fates.28 In this context, reiterated functions of LIN-42 may enable the reprogramming of seam cell temporal fates, while sequential actions of LIN-42 and other heterochronic genes select the fates appropriate for upcoming, rather than passing, life stages.
Oscillatory activity is the second distinctive feature of core clock components. Both transcriptional and post-transcriptional regulatory mechanisms contribute to these oscillations, which occur in phase with the clock-controlled biological rhythm.12 The lin-42 locus is complex and encodes three major transcripts with both shared and unique cis-acting regulatory elements.29 The overall abundance of lin-42 transcripts and proteins rises and falls in phase with the larval molts.28 To better define the temporal expression profiles of lin-42a, b and c, we separately combined both the unique promoter of lin-42a and the shared promoter of lin-42b and c with the gfp-pest reporter. Expression of the corresponding fusion genes in hyp7 showed that activity of the lin-42b promoter peaked during the intermolts, as previously reported.29 However, the lin-42a promoter was most active toward the end of every larval stage; GFP was robustly expressed in lethargic larvae undergoing molts, and was barely detectable about two hours after ecdysis.15 Expression of lin-42a restored both rhythmic molts and proper epidermal development to lin-42 mutants, underscoring the significance of the independent transcription of lin-42a.15,29
Taken together, our findings provide substantial evidence that LIN-42A functions as a central component of the molting clock. The isolation of missense alleles of lin-42 associated with consistently shorter or longer, rather than unpredictable, molting cycles would further support this model. In theory, the molecular identities of related substitutions might identify motifs essential for the activity of LIN-42A, and possibly sites of post-translational modifications that affect the abundance or intracellular distribution of LIN-42A. Similar approaches uncovered pivotal roles for the phosphorylation and O-glycosylation of PER in the circadian clock.33-36 Identifying the transcriptional regulators and targets of LIN-42 would also improve our understanding of the molting timer. This task is challenging, in part, because the genome of C. elegans encodes scores of HLH transcription factors, none of which are clearly homologous to CLOCK or BMAL.
Particular conserved NRs are excellent candidates for additional components of the molting clock: namely, NHR-23, which is homologous to both mammalian RORα and Drosophila DHR3, NHR-25, which is homologous to both mammalian SF-1 and Drosophila FTZ-F1, and DAF-12, which is related to the Vitamin D receptor.37 Inactivation of either nhr-23 or -25 prevents completion of the molts, and overexpression of either gene causes larval lethality.38-41 NHR-23 and -25 also directly or indirectly activate the expression of many genes involved in the process of molting.11,39,42-45 These genes encode various intercellular signaling molecules, matrix modification enzymes, and ECM proteins, including collagens and MLT-10.46 NHR-25 also regulates seam cell dynamics.23,24 In addition, we found that RNAi of either nhr-23 or -25 exacerbated the molting defects of lin-42(ok2385) mutants.15 DAF-12 regulates the decision to undergo rapid development or facultative diapause in L2 larvae.37 In this context, lin-42 interacts with daf-12 and ligand-activated DAF-12 induces the expression of lin-42a when conditions favor rapid development.47,48 However, daf-12 is expressed throughout development and may regulate the progression of additional life stages.37
The abundance of nhr-23 and -25 transcripts also oscillates in phase with the molting cycle.39,49 Overall levels of nhr-23 transcripts peak midway through the intermolt, but the temporal expression patterns of the six distinct isoforms of nhr-23 curated in Wormbase229 have not yet been examined. Levels of nhr-25α transcripts, which encode the full-length receptor, peak during the molts, whereas levels of nhr-25β transcripts, which encode a truncated receptor, peak after ecdysis.49 Once made, NHR-25β might interfere with transcriptional activation by NHR-25α, as the former isoform lacks a DNA binding domain. Based on the timing of peak nhr-23 expression and the annotations of confirmed targets of the receptor, we hypothesize that transcriptional activation by NHR-23 enforces a commitment to molt. NHR-23 might directly promote the expression of NHR-25, given that Drosophila DHR3 activates the transcription of FTZ-F1.50 In theory, the expression of various targets of NHR-25 may then promote completion of the molt and the start of the next life stage. The precise temporal expression pattern of daf-12 has not yet been characterized.37
The conserved let-7 family of miRNAs has been predicted to target and downregulate both lin-42 and nhr-25,4,51 and may thereby fine-tune the periodicity of larval molting cycles. The best-characterized function of let-7 is to promote terminal differentiation of the lateral hypoderm by downregulating additional protein-coding genes in the heterochronic pathway.4 The seam cells fail to terminally differentiate at the larval-to-adult transition in let-7 mutants, and instead undergo extra divisions in reproductively mature animals.4 Mutations in let-7 are also associated with supernumerary molts, which can be suppressed by knocking-down either nhr-23 or -25.4,51 The fact that let-7 mutants undergo supernumerary molts at predictable, rather than sporadic, times indicates that the core molting timer operates in this background. However, the extent to which let-7 or related miRNAs affect the duration of specific larval stages or the synchronicity of the molts has not yet been determined. Notably, mutations in lin-42 partially suppress the phenotypes of let-7 mutants and vice-versa,29,31 but the mechanism of this co-suppression has not yet been defined. One interesting possibility is that lin-42 negatively regulates the expression of mature let-7 miRNAs.
Primary transcripts of let-7 are also expressed during each of the four molts, even though mature let-7 miRNAs are only produced in late stage larvae and adults.52 However, additional members of the let-7 family act during earlier larval stages,53,54 and the expression of some, if not all, paralogs of let-7 might oscillate in phase with the molts. Although the cyclical expression of lin-42, nhr-23, nhr-25, and let-7 is critical for larval development,15,55 the transcriptional and post-transcriptional regulatory mechanisms that account for these oscillations have not yet been described.
Modeling the Molting Timer
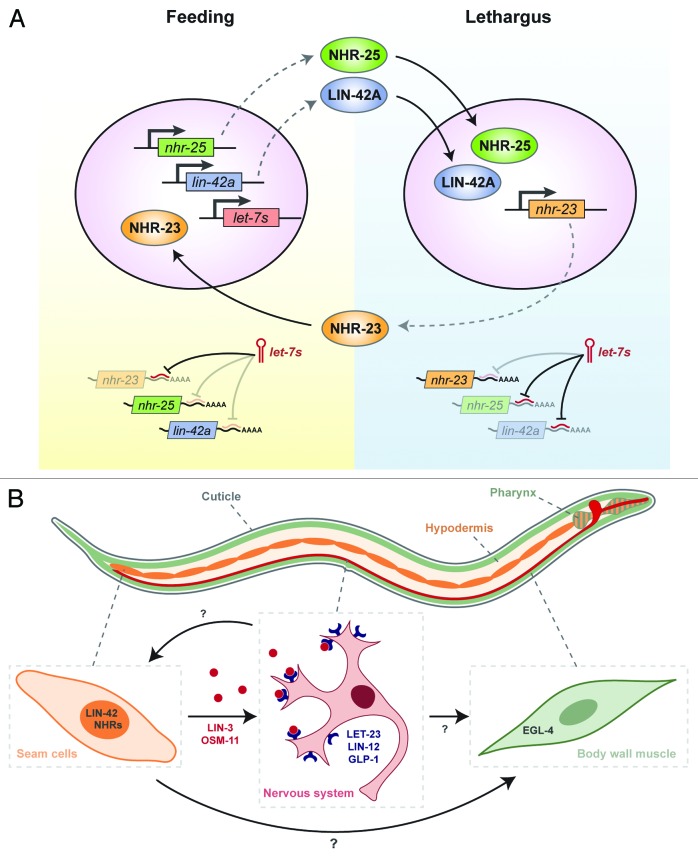
As depicted in Figure 3A, we postulate that interlocked positive and negative regulatory loops involving LIN-42, conserved NRs, and the let-7 family of miRNAs compose a developmental oscillator that drives rapid molting cycles. In the positive limb of the proposed timer, specific NRs activate the expression of lin-42a and let-7 family miRNAs, and levels of the corresponding gene products begin to rise. The precise time of gene activation within each larval stage may be determined by the abundance of the receptors, affiliated co-factors, and small-molecule ligands. Consistent with this hypothesis, there are several half-sites for NR binding56 in the experimentally defined promoters of lin-42a and let-7.15,29,52,57 Further, DAF-12 is known to induce the expression of lin-42a and let-7 under favorable conditions.47,48,58,59 In addition, the mammalian NR NF-κB activates expression of let-7 in tissue culture.60 Although a high-throughput screen for transcriptional regulators of the let-7 family did not uncover NHR-23 or -25,61 the possibility that one or both of these receptors periodically activate the expression of let-7 family miRNAs in larvae has not been evaluated. In the negative limb of the proposed timer, let-7 family miRNAs target lin-42 and nr transcripts for translational inhibition or degradation, contributing to a decline in levels of the corresponding proteins. Consistent with this model, let-7 is known to directly target daf-12 transcripts.58,59 The extent, if any, to which mammalian let-7 miRNAs downregulate per or other components of the circadian clock is not known.
Figure 3. Modeling the Molting Timer. (A) We propose that interconnected positive and negative regulatory interactions among LIN-42, certain NRs, and let-7 family miRNAs compose a developmental oscillator that operates in the epidermis of juveniles and drives rhythmic molting cycles. This diagram depicts hypothetical molecular events that might occur in seam cells as animals enter and exit the larval molts. (B) The proposed molting timer may also regulate the rhythmic production of endocrine cues that affect animal behavior. Additional signals among the epidermis (orange), the nervous system (red), and body wall muscles (green) are expected to regulate lethargus and ecdysis. See text for additional information about this model.
The molecular function of LIN-42A has not yet been defined, but the protein is thought to act as a transcriptional co-factor, similar to larger PER proteins. Consistent with this model, several sequence motifs are conserved between LIN-42A and mammalian PER proteins, although LIN-42A lacks a Period-Ant-Sim (PAS) domain.29 Further, LIN-42A was detected in the nuclei of epithelial cells.15 Per2 was recently found to bind the NR REV-ERB through an LXXLL motif as part of the circadian clock.62 Various co-activators of Drosophila FTZ-F1 and Mammalian SF-1 also use LXXLL motifs to bind the receptors.63,64 LIN-42A and B contain two LXXLL motifs, one or both of which might enable physical interactions with NHR-23 or -25. Consistent with this general idea, initial studies have suggested that LIN-42B interacts with DAF-12 in a manner that does not depend on the PAS domain.48 If LIN-42 does in fact associate with these particular NRs, then the abundance of related complexes in epithelial cells might fluctuate during every larval stage and contribute to the rhythmic expression of downstream target genes. Protein-protein interactions between LIN-42 and these particular NRs may therefore play a role in timing molting cycles.
In summary, LIN-42A functions in a developmental oscillator underlying rhythmic molting cycles. In this context, high levels of LIN-42A coincide with lethargy, ECM remodeling, and stem cell quiescence, whereas low levels of LIN-42A coincide with physical activity, stable matrices, and stem cell divisions. Rising levels of LIN-42 may allow the reprogramming of seam cell temporal fates; falling levels of LIN-42 may license the subsequent round of cell division. Reprogramming of seam cell temporal fates may be achieved, in part, by repressing the expression of preexisting miRNAs, including lin-4 in L1 larvae and the let-7 family in older larvae. The timer would start to tick in the epithelium of late-stage embryos, around the time of synthesis of the L1-stage cuticle, and cease to operate when the epithelium terminally differentiates at the larval-to-adult transition. Anticipated clock-controlled genes include some components of the heterochronic network. In theory, the 8-h rhythm of the molting cycle may represent a harmonic of a circadian oscillator adapted to support the rapid growth of nematode larvae.20
Finding that individual lin-42 mutants complete the larval stages at variable times underscores the need to account for progression of the molts, in addition to the passage of time, when characterizing mutations that affect developmental timing and the expression profiles of corresponding genes. The heterochronic mutants of C. elegans have been classified based on the relative timing of either the symmetric divisions or the terminal differentiation of the seam cells, compared with development of the reproductive system.65 It has since become standard practice in the field to synchronize hatchlings by starvation-induced L1-stage diapause, cultivate larvae on food for pre-selected periods of time, and then collect animals for visual inspection or the extraction of nucleic acids. Interpretations of related data often assume that all larvae of a given genotype developed at a consistent pace, similar to wild-type animals. Some heterochronic phenotypes may therefore be attributable to slower, faster, or arrhythmic molting cycles, rather than the misspecification of temporal cell fates.
Potential Inputs to the Molting Timer
The circadian clocks of mammals beneficially synchronize rhythms in behavior and metabolism with daily fluctuations in sunlight, temperature, and the availability of food.12,66,67 These environmental factors are typically held constant when nematodes are cultivated in the lab, but almost certainly vary in the natural habitat of the soil, and may impact the periodicity of molting cycles in that context. Indeed, cooler cultivation temperatures reduce the speed of larval development, and rhythmic fluctuations in ambient temperature produce oscillations in the expression of many C. elegans genes.68 Further, both the quality and abundance of food affect the pace of larval development.69
Fluctuating levels of specific nutrients or metabolites and thresholds in body size may also provide physiologic inputs to the molting timer. In general, energy reserves should increase as larvae feed. However, because the volume of the body increases continuously, whereas the volume of the mouth only increases during molts, the ratio of food intake to energy expenditure likely drops when larval reach a critical size.20 The process of molting then requires substantial work, at a time when larvae are unable to feed. Consistent with the idea that metabolic states influence the progression of molting cycles, signaling through the insulin pathway promotes rapid larval development.70,71 Mechanotransduction pathways coupled to stress on the cuticle may also promote molting when larvae attain critical sizes. These concepts are consistent with the observation that the average size of shed cuticles increases saltationally from one larval stage to the next.69 We further anticipate that “checkpoints” on the status of the new cuticle regulate the transition from lethargus to ecdysis, although the corresponding circuits have not yet been described.
Fluctuations in the abundance of particular steroid hormones may also affect the periodicity of the molts, by modulating the activity of NRs including NHR-23, -25, and DAF-12. C. elegans cannot synthesize cholesterol de novo, and an exogenous supply of cholesterol is required for completion of the molts.72,73 Cholesterol is generally thought to serve as a precursor for the biosynthesis of essential steroid hormones,74 although 20-E has not been detected in any free-living nematode.75 The natural ligand of human RORα is a derivative of cholesterol,76 and a related molecule might serve as a physiologic ligand for NHR-23. Recent findings have shown that FTZ-F1 functions as a constitutive transcriptional activator without a small molecule ligand,63 and NHR-25 might operate in a similar manner. The endogenous ligand for DAF-12 was the first steroid hormone characterized in C. elegans.77 Both insulin and TGF-β signaling promote production of this hormone.37
Endocrine and neuroendocrine cues coupled to any or all of the aforementioned environmental and physiological factors may converge on the regulation of LIN-42 and other components of the molting timer. In natural habitats, such interactions might align the molts with favorable extrinsic and intrinsic conditions, and thereby decrease the possibility of injury or death during the molting process.
Systemic Outputs of the Molting Timer
As described, molting cycles include the distinctive behaviors of lethargy and ecdysis, which are regulated by endocrine and neuroendocrine circuits.25,27 Both the onset and duration of lethargus were irregular in lin-42(ok2385) mutants, and ecdyses were often aberrant.15 Remarkably, seam cell-specific expression of lin-42a restored normal cycles of physical activity and quiescence to lin-42(ok2385) mutants, in addition to successful ecdyses.15 This particular finding implies that activity of LIN-42 in the seam cells promotes the rhythmic production or release of intercellular signals that directly or indirectly modulate the functions of various neurons and muscles throughout the body (Fig. 3B). Several recent reports have confirmed that neuroendocrine cues produced by epithelial cells either initiate or maintain the behavioral quiescence associated with larval molts. Secretion of the DOS protein OSM-11 from the seam cells induces quiescence by activating the Notch receptors LIN-12 and GLP-1 in the nervous system.27 In a parallel pathway, the EGF-like molecule LIN-3 induces quiescence by activating the EGF receptor LET-23 in the ALA interneuron.25 The relevant source of endogenous LIN-3 has not yet been defined. However, the lin-3 gene is expressed in the pharynx, the hypoderm, and the intestine, and expression of lin-3 in some epithelial cells is activated by NHR-25.78,79 The ectodomain of the Amyloid Precursor-Like protein APL-1, which may be released from both seam cells and neurons, might also regulate behavior during the molts.80-82 Moreover, the expression of an apl-1 fusion gene in the seam cells of L4 larvae is induced by nhr-25, and indirectly repressed by the let-7 family of miRNAs.45,83 Another key component of the signaling network that regulates lethargus is the conserved cyclic GMP protein kinase EGL-4, which is expressed in body wall muscles, the hypodermis, and head neurons.26,84,85 Loss-of-function mutations in egl-4 are associated with increased physical activity and partly suppress the behavioral quiescence triggered by forced expression of lin-42a, osm-11, or lin-3.15,25,27 Although the events downstream of EGFR and Notch Receptor activation have not yet been characterized, these pathways likely converge on the regulated secretion of neurotransmitters at neuromuscular junctions. In theory, the molting timer might control behavioral rhythms by directly or indirectly modulating the activities of OSM-11, LIN-3, or APL-1. Additional outputs of the molting timer may affect osmoregulation, metabolism, and other systemic processes that differ between molting and non-molting larvae.
The Molting Timer as a Primitive Biological Clock
In mammals, a master circadian clock operates in the suprachiasmatic nucleus (SCN) of the brain. The central clock resets and synchronizes peripheral clocks in vital organs, and responds to feedback from peripheral oscillators.12 The ontogeny of this system is not well understood.86 When viewed in this context, the LIN-42/PER-based molting timer, which operates in epithelial cells and affects rhythmic behaviors of several other tissues, may represent a primitive form of a central clock. Interestingly, the eclosion of insects and the rhythmic growth of plants are also regulated by the circadian clock.87-89 The use of PER-based clocks as developmental timers may therefore represent an ancient adaptation of multicellular organisms.
The regulation of sleep-wake cycles is perhaps the best-characterized function of human PER proteins, and mutations in the corresponding genes are associated with various inherited disorders of sleep.90-93 Finding that lin-42 regulates the timing of lethargus further supports the view that nematode lethargy and human sleep share an ancient evolutionary origin. Homologs of several additional molecules that regulate lethargus also affect the sleep-wake cycles of mammals. RORα, which is related to NHR-23, regulates sleep as a component of the circadian clock.94 The mammalian EGF receptor and the EGL-4 homolog PRKG1 also influence the timing and quality of sleep.95,96 Further, levels of let-7 increase in the hippocampus of sleep-deprived rats, suggesting that the activity of let-7 correlates with sleep-wake cycles.97,98
Another striking similarity between the function of C. elegans LIN-42 and mammalian PER is that both proteins regulate epidermal stem cell dynamics.15,99 The epidermal stem cells of mammalian hair follicles also undergo cycles of proliferation and quiescence,100 and PER was recently been found to regulate the overall size of this stem cell population and the propensity of cells therein to enter the cell cycle.99,101 This particular function of human PER may be analogous to the role of LIN-42 in regulating seam cell proliferation and temporal specification.15
Recent studies have uncovered the clinical significance of clock misalignment or dysfunction in modern epidemics including obesity, diabetes and age-related declines in bodily function.12,,102-105 Further basic research on the molting timer may therefore uncover novel but conserved aspects of PER-based oscillators relevant to human development as well as metabolic syndromes and sleep disorders.
Acknowledgments
This work was supported by the UCLA School of Medicine and graduate research fellowships to G.C.M. from the National Science Foundation and the Ford Foundation. We are grateful to Chris Colwell, Ann Rougvie, Cathy Clarke, Feng Guo, Eddy De Robertis, Chris Gissendanner and John Kim for helpful discussions. We thank Ryan Smit for initial bioinformatic analyses, and Rusty Howson for scientific illustrations.
Glossary
Abbreviations:
- NR
nuclear hormone receptor
- RNAi
RNA-interference
- GFP
Green Fluorescent Protein
- UTR
untranslated region
- let-7s
let-7 family of microRNAs
- 20-E
20-hydroxy-ecydsone
- HLH
helix-loop-helix
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/20874
References
- 1.Thummel CS. Flies on steroids--Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 1996;12:306–10. doi: 10.1016/0168-9525(96)10032-9. [DOI] [PubMed] [Google Scholar]
- 2.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 3.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 6.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol. 1977;56:110–56. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 7.Rougvie AE. Intrinsic and extrinsic regulators of developmental timing: from miRNAs to nutritional cues. Development. 2005;132:3787–98. doi: 10.1242/dev.01972. [DOI] [PubMed] [Google Scholar]
- 8.Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–34. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- 9.Pourquié O. The segmentation clock: converting embryonic time into spatial pattern. Science. 2003;301:328–30. doi: 10.1126/science.1085887. [DOI] [PubMed] [Google Scholar]
- 10.Johnstone IL, Barry JD. Temporal reiteration of a precise gene expression pattern during nematode development. EMBO J. 1996;15:3633–9. [PMC free article] [PubMed] [Google Scholar]
- 11.Frand AR, Russel S, Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3:e312. doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–54. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borgs L, Beukelaers P, Vandenbosch R, Belachew S, Nguyen L, Malgrange B. Cell “circadian” cycle: new role for mammalian core clock genes. Cell Cycle. 2009;8:832–7. doi: 10.4161/cc.8.6.7869. [DOI] [PubMed] [Google Scholar]
- 14.Greene MW. Circadian rhythms and tumor growth. Cancer Lett. 2012;318:115–23. doi: 10.1016/j.canlet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 15.Monsalve GC, Van Buskirk C, Frand AR. LIN-42/PERIOD controls cyclical and developmental progression of C. elegans molts. Curr Biol. 2011;21:2033–45. doi: 10.1016/j.cub.2011.10.054. [DOI] [PubMed] [Google Scholar]
- 16.Singh RN, Sulston JE. Some observations on moulting in Caenorhabditis elegans. Nematologica. 1978;24:63–71. doi: 10.1163/187529278X00074. [DOI] [Google Scholar]
- 17.Page AP, Johnstone IL. The cuticle. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaidel-Bar R, Miller S, Kaminsky R, Broday L. Molting-specific downregulation of C. elegans body-wall muscle attachment sites: the role of RNF-5 E3 ligase. Biochem Biophys Res Commun. 2010;395:509–14. doi: 10.1016/j.bbrc.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 19.Podbilewicz B, White JG. Cell fusions in the developing epithelial of C. elegans. Dev Biol. 1994;161:408–24. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- 20.Knight CG, Patel MN, Azevedo RB, Leroi AM. A novel mode of ecdysozoan growth in Caenorhabditis elegans. Evol Dev. 2002;4:16–27. doi: 10.1046/j.1525-142x.2002.01058.x. [DOI] [PubMed] [Google Scholar]
- 21.Koh K, Rothman JH. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 2001;128:2867–80. doi: 10.1242/dev.128.15.2867. [DOI] [PubMed] [Google Scholar]
- 22.Ruaud AF, Bessereau JL. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development. 2006;133:2211–22. doi: 10.1242/dev.02392. [DOI] [PubMed] [Google Scholar]
- 23.Chen Z, Eastburn DJ, Han M. The Caenorhabditis elegans nuclear receptor gene nhr-25 regulates epidermal cell development. Mol Cell Biol. 2004;24:7345–58. doi: 10.1128/MCB.24.17.7345-7358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silhánková M, Jindra M, Asahina M. Nuclear receptor NHR-25 is required for cell-shape dynamics during epidermal differentiation in Caenorhabditis elegans. J Cell Sci. 2005;118:223–32. doi: 10.1242/jcs.01609. [DOI] [PubMed] [Google Scholar]
- 25.Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–7. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- 26.Raizen DM, Zimmerman JE, Maycock MH, Ta UD, You YJ, Sundaram MV, et al. Lethargus is a Caenorhabditis elegans sleep-like state. Nature. 2008;451:569–72. doi: 10.1038/nature06535. [DOI] [PubMed] [Google Scholar]
- 27.Singh K, Chao MY, Somers GA, Komatsu H, Corkins ME, Larkins-Ford J, et al. C. elegans Notch signaling regulates adult chemosensory response and larval molting quiescence. Curr Biol. 2011;21:825–34. doi: 10.1016/j.cub.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE. Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science. 1999;286:1141–6. doi: 10.1126/science.286.5442.1141. [DOI] [PubMed] [Google Scholar]
- 29.Tennessen JM, Gardner HF, Volk ML, Rougvie AE. Novel heterochronic functions of the Caenorhabditis elegans period-related protein LIN-42. Dev Biol. 2006;289:30–43. doi: 10.1016/j.ydbio.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 30.Lui Z. Thesis, Harvard University. 1990. [Google Scholar]
- 31.Abrahante JE, Miller EA, Rougvie AE. Identification of heterochronic mutants in Caenorhabditis elegans. Temporal misexpression of a collagen:green fluorescent protein fusion gene. Genetics. 1998;149:1335–51. doi: 10.1093/genetics/149.3.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konopka RJ, Benzer S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1971;68:2112–6. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kivimäe S, Saez L, Young MW. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 2008;6:e183. doi: 10.1371/journal.pbio.0060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3beta/SGG in circadian clocks. J Neurosci. 2010;30:12664–75. doi: 10.1523/JNEUROSCI.1586-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu JC, Ko HW, Edery I. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell. 2011;145:357–70. doi: 10.1016/j.cell.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim EY, Jeong EH, Park S, Jeong HJ, Edery I, Cho JW. A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 2012;26:490–502. doi: 10.1101/gad.182378.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antebi A. Nuclear hormone receptors in C. elegans. WormBook. 2006:1–13. doi: 10.1895/wormbook.1.64.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kostrouchova M, Krause M, Kostrouch Z, Rall JE. CHR3: a Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development. 1998;125:1617–26. doi: 10.1242/dev.125.9.1617. [DOI] [PubMed] [Google Scholar]
- 39.Kostrouchova M, Krause M, Kostrouch Z, Rall JE. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2001;98:7360–5. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asahina M, Ishihara T, Jindra M, Kohara Y, Katsura I, Hirose S. The conserved nuclear receptor Ftz-F1 is required for embryogenesis, moulting and reproduction in Caenorhabditis elegans. Genes Cells. 2000;5:711–23. doi: 10.1046/j.1365-2443.2000.00361.x. [DOI] [PubMed] [Google Scholar]
- 41.Gissendanner CR, Sluder AE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol. 2000;221:259–72. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- 42.Kouns NA, Nakielna J, Behensky F, Krause MW, Kostrouch Z, Kostrouchova M. NHR-23 dependent collagen and hedgehog-related genes required for molting. Biochem Biophys Res Commun. 2011;413:515–20. doi: 10.1016/j.bbrc.2011.08.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks DR, Appleford PJ, Murray L, Isaac RE. An essential role in molting and morphogenesis of Caenorhabditis elegans for ACN-1, a novel member of the angiotensin-converting enzyme family that lacks a metallopeptidase active site. J Biol Chem. 2003;278:52340–6. doi: 10.1074/jbc.M308858200. [DOI] [PubMed] [Google Scholar]
- 44.Davis MW, Birnie AJ, Chan AC, Page AP, Jorgensen EM. A conserved metalloprotease mediates ecdysis in Caenorhabditis elegans. Development. 2004;131:6001–8. doi: 10.1242/dev.01454. [DOI] [PubMed] [Google Scholar]
- 45.Hada K, Asahina M, Hasegawa H, Kanaho Y, Slack FJ, Niwa R. The nuclear receptor gene nhr-25 plays multiple roles in the Caenorhabditis elegans heterochronic gene network to control the larva-to-adult transition. Dev Biol. 2010;344:1100–9. doi: 10.1016/j.ydbio.2010.05.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meli V, Osuna B, Ruvkun G, Frand AR. MLT-10 defines a family of DUF644 and proline-rich repeat proteins involved in the molting cycle of C. elegans MBOC 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hochbaum D, Zhang Y, Stuckenholz C, Labhart P, Alexiadis V, Martin R, et al. DAF-12 regulates a connected network of genes to ensure robust developmental decisions. PLoS Genet. 2011;7:e1002179. doi: 10.1371/journal.pgen.1002179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tennessen JM, Opperman KJ, Rougvie AE. The C. elegans developmental timing protein LIN-42 regulates diapause in response to environmental cues. Development. 2010;137:3501–11. doi: 10.1242/dev.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gissendanner CR, Crossgrove K, Kraus KA, Maina CV, Sluder AE. Expression and function of conserved nuclear receptor genes in Caenorhabditis elegans. Dev Biol. 2004;266:399–416. doi: 10.1016/j.ydbio.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 50.Kageyama Y, Masuda S, Hirose S, Ueda H. Temporal regulation of the mid-prepupal gene FTZ-F1: DHR3 early late gene product is one of the plural positive regulators. Genes Cells. 1997;2:559–69. doi: 10.1046/j.1365-2443.1997.1460344.x. [DOI] [PubMed] [Google Scholar]
- 51.Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–41. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- 52.Van Wynsberghe PM, Kai ZS, Massirer KB, Burton VH, Yeo GW, Pasquinelli AE. LIN-28 co-transcriptionally binds primary let-7 to regulate miRNA maturation in Caenorhabditis elegans. Nat Struct Mol Biol. 2011;18:302–8. doi: 10.1038/nsmb.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, et al. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–14. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 55.Hayes GD, Ruvkun G. Misexpression of the Caenorhabditis elegans miRNA let-7 is sufficient to drive developmental programs. Cold Spring Harb Symp Quant Biol. 2006;71:21–7. doi: 10.1101/sqb.2006.71.018. [DOI] [PubMed] [Google Scholar]
- 56.Sandelin A, Wasserman WW. Prediction of nuclear hormone receptor response elements. Mol Endocrinol. 2005;19:595–606. doi: 10.1210/me.2004-0101. [DOI] [PubMed] [Google Scholar]
- 57.Turner MJ, Slack FJ. Transcriptional control of microRNA expression in C. elegans: promoting better understanding. RNA Biol. 2009;6:49–53. doi: 10.4161/rna.6.1.7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bethke A, Fielenbach N, Wang Z, Mangelsdorf DJ, Antebi A. Nuclear hormone receptor regulation of microRNAs controls developmental progression. Science. 2009;324:95–8. doi: 10.1126/science.1164899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hammell CM, Karp X, Ambros V. A feedback circuit involving let-7-family miRNAs and DAF-12 integrates environmental signals and developmental timing in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18668–73. doi: 10.1073/pnas.0908131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang DJ, Legesse-Miller A, Johnson EL, Coller HA. Regulation of the let-7a-3 promoter by NF-κB. PLoS One. 2012;7:e31240. doi: 10.1371/journal.pone.0031240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, et al. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–49. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev. 2010;24:345–57. doi: 10.1101/gad.564110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoo J, Ko S, Kim H, Sampson H, Yun JH, Choe KM, et al. Crystal structure of Fushi tarazu factor 1 ligand binding domain/Fushi tarazu peptide complex identifies new class of nuclear receptors. J Biol Chem. 2011;286:31225–31. doi: 10.1074/jbc.M111.252916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bernardo TJ, Dubrovsky EB. The Drosophila juvenile hormone receptor candidates methoprene-tolerant (MET) and germ cell-expressed (GCE) utilize a conserved LIXXL motif to bind the FTZ-F1 nuclear receptor. J Biol Chem. 2012;287:7821–33. doi: 10.1074/jbc.M111.327254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–16. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 66.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–85. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fuller PM, Lu J, Saper CB. Differential rescue of light- and food-entrainable circadian rhythms. Science. 2008;320:1074–7. doi: 10.1126/science.1153277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Linden AM, Beverly M, Kadener S, Rodriguez J, Wasserman S, Rosbash M, et al. Genome-wide analysis of light- and temperature-entrained circadian transcripts in Caenorhabditis elegans. PLoS Biol. 2010;8:e1000503. doi: 10.1371/journal.pbio.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szewczyk NJ, Udranszky IA, Kozak E, Sunga J, Kim SK, Jacobson LA, et al. Delayed development and lifespan extension as features of metabolic lifestyle alteration in C. elegans under dietary restriction. J Exp Biol. 2006;209:4129–39. doi: 10.1242/jeb.02492. [DOI] [PubMed] [Google Scholar]
- 70.Ruaud AF, Katic I, Bessereau JL. Insulin/Insulin-like growth factor signaling controls non-Dauer developmental speed in the nematode Caenorhabditis elegans. Genetics. 2011;187:337–43. doi: 10.1534/genetics.110.123323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baugh LR, Sternberg PW. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr Biol. 2006;16:780–5. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 72.Hieb WF, Rothstein M. Sterol requirement for reproduction of a free-living nematode. Science. 1968;160:778–80. doi: 10.1126/science.160.3829.778. [DOI] [PubMed] [Google Scholar]
- 73.Yochem J, Tuck S, Greenwald I, Han M. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development. 1999;126:597–606. doi: 10.1242/dev.126.3.597. [DOI] [PubMed] [Google Scholar]
- 74.Kurzchalia TV, Ward S. Why do worms need cholesterol? Nat Cell Biol. 2003;5:684–8. doi: 10.1038/ncb0803-684. [DOI] [PubMed] [Google Scholar]
- 75.Chitwood DJ. Biochemistry and function of nematode steroids. Crit Rev Biochem Mol Biol. 1999;34:273–84. doi: 10.1080/10409239991209309. [DOI] [PubMed] [Google Scholar]
- 76.Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, et al. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure. 2002;10:1697–707. doi: 10.1016/S0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- 77.Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–23. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 78.Hwang BJ, Sternberg PW. A cell-specific enhancer that specifies lin-3 expression in the C. elegans anchor cell for vulval development. Development. 2004;131:143–51. doi: 10.1242/dev.00924. [DOI] [PubMed] [Google Scholar]
- 79.Liu G, Rogers J, Murphy CT, Rongo C. EGF signalling activates the ubiquitin proteasome system to modulate C. elegans lifespan. EMBO J. 2011;30:2990–3003. doi: 10.1038/emboj.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daigle I, Li C. apl-1, a Caenorhabditis elegans gene encoding a protein related to the human beta-amyloid protein precursor. Proc Natl Acad Sci U S A. 1993;90:12045–9. doi: 10.1073/pnas.90.24.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wiese M, Antebi A, Zheng H. Intracellular trafficking and synaptic function of APL-1 in Caenorhabditis elegans. PLoS One. 2010;5:5. doi: 10.1371/journal.pone.0012790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ewald CY, Raps DA, Li C. APL-1, the Alzheimer’s Amyloid Precursor Protein in Caenorhabditis elegans, Modulates Multiple Metabolic Pathways Throughout Development. Genetics. 2012:in press. doi: 10.1534/genetics.112.138768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Niwa R, Zhou F, Li C, Slack FJ. The expression of the Alzheimer’s amyloid precursor protein-like gene is regulated by developmental timing microRNAs and their targets in Caenorhabditis elegans. Dev Biol. 2008;315:418–25. doi: 10.1016/j.ydbio.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raizen DM, Cullison KM, Pack AI, Sundaram MV. A novel gain-of-function mutant of the cyclic GMP-dependent protein kinase egl-4 affects multiple physiological processes in Caenorhabditis elegans. Genetics. 2006;173:177–87. doi: 10.1534/genetics.106.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stansberry J, Baude EJ, Taylor MK, Chen PJ, Jin SW, Ellis RE, et al. A cGMP-dependent protein kinase is implicated in wild-type motility in C. elegans. J Neurochem. 2001;76:1177–87. doi: 10.1046/j.1471-4159.2001.00131.x. [DOI] [PubMed] [Google Scholar]
- 86.Yamazaki S, Yoshikawa T, Biscoe EW, Numano R, Gallaspy LM, Soulsby S, et al. Ontogeny of circadian organization in the rat. J Biol Rhythms. 2009;24:55–63. doi: 10.1177/0748730408328438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Myers EM, Yu J, Sehgal A. Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr Biol. 2003;13:526–33. doi: 10.1016/S0960-9822(03)00167-2. [DOI] [PubMed] [Google Scholar]
- 88.Kyriacou CP, Oldroyd M, Wood J, Sharp M, Hill M. Clock mutations alter developmental timing in Drosophila. Heredity (Edinb) 1990;64:395–401. doi: 10.1038/hdy.1990.50. [DOI] [PubMed] [Google Scholar]
- 89.Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, et al. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–61. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- 90.Carpen JD, Archer SN, Skene DJ, Smits M, von Schantz M. A single-nucleotide polymorphism in the 5′-untranslated region of the hPER2 gene is associated with diurnal preference. J Sleep Res. 2005;14:293–7. doi: 10.1111/j.1365-2869.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- 91.Carpen JD, von Schantz M, Smits M, Skene DJ, Archer SN. A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J Hum Genet. 2006;51:1122–5. doi: 10.1007/s10038-006-0060-y. [DOI] [PubMed] [Google Scholar]
- 92.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, et al. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–3. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 93.Reid KJ, Zee PC. Circadian rhythm sleep disorders. Handb Clin Neurol. 2011;99:963–77. doi: 10.1016/B978-0-444-52007-4.00017-5. [DOI] [PubMed] [Google Scholar]
- 94.Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–8. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- 95.Feil R, Hölter SM, Weindl K, Wurst W, Langmesser S, Gerling A, et al. cGMP-dependent protein kinase I, the circadian clock, sleep and learning. Commun Integr Biol. 2009;2:298–301. doi: 10.4161/cib.2.4.8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–5. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 97.Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, et al. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–91. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Davis CJ, Bohnet SG, Meyerson JM, Krueger JM. Sleep loss changes microRNA levels in the brain: a possible mechanism for state-dependent translational regulation. Neurosci Lett. 2007;422:68–73. doi: 10.1016/j.neulet.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Janich P, Pascual G, Merlos-Súrez A, Batlle E, Ripperger J, Albrecht U, et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–14. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 100.Hsu YC, Fuchs E. A family business: stem cell progeny join the niche to regulate homeostasis. Nat Rev Mol Cell Biol. 2012;13:103–14. doi: 10.1038/nrm3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin KK, Kumar V, Geyfman M, Chudova D, Ihler AT, Smyth P, et al. Circadian clock genes contribute to the regulation of hair follicle cycling. PLoS Genet. 2009;5:e1000573. doi: 10.1371/journal.pgen.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nakamura TJ, Nakamura W, Yamazaki S, Kudo T, Cutler T, Colwell CS, et al. Age-related decline in circadian output. J Neurosci. 2011;31:10201–5. doi: 10.1523/JNEUROSCI.0451-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Halbout B, Perreau-Lenz S, Dixon CI, Stephens DN, Spanagel R. Per1(Brdm1) mice self-administer cocaine and reinstate cocaine-seeking behaviour following extinction. Behav Pharmacol. 2011;22:76–80. doi: 10.1097/FBP.0b013e328341e9ca. [DOI] [PubMed] [Google Scholar]
- 105.Gale JE, Cox HI, Qian J, Block GD, Colwell CS, Matveyenko AV. Disruption of circadian rhythms accelerates development of diabetes through pancreatic beta-cell loss and dysfunction. J Biol Rhythms. 2011;26:423–33. doi: 10.1177/0748730411416341. [DOI] [PMC free article] [PubMed] [Google Scholar]