Abstract
In preparation for meiotic chromosome segregation, homologous chromosomes need to pair, synapse (i.e., assemble the synaptonemal complex, SC), and then recombine to generate a physical linkage (i.e., chiasma) between them. In many organisms meiotic pairing capacity distributed along the entire chromosome length supports presynaptic alignment. In contrast, the prevailing model for C. elegans proposes that presynaptic homologous pairing is performed solely by a master pairing-site, the pairing center (PC). In this model, the remaining chromosomal regions (the non-PC regions) are not actively involved in presynaptic pairing, and the SC assembling from the PC aligns the homologous chromosomes along non-PC regions and holds them together. Our recent work, however, demonstrates that C. elegans chromosomes establish presynaptic alignment along the entire chromosome length, suggesting that the non-PC regions are also actively involved in the presynaptic pairing process. Furthermore, we have also discovered that the chromodomain protein MRG-1 facilitates this presynaptic non-PC pairing. The phenotype of the mrg-1 mutant indicates that the PC and the non-PC collaborate in successful pairing and synapsis. Therefore, homologous pairing mechanisms in C. elegans possibly share more similarity with those in other organisms than previously thought. Here, we elaborate on these observations and discuss a hypothetical model for presynaptic pairing in C. elegans based on our novel findings.
Keywords: meiosis, homologous pairing, presynaptic alignment, pairing center, non-homologous synapsis, MRG-1
Meiosis is the specialized cell division that generates a haploid genome of gametes (sperm and eggs) from a diploid genome of germ cells. A unique feature of meiosis is segregation of homologous chromosomes during meiosis I, while meiosis II functions to segregate sister chromatids analogous to mitosis. Successful meiosis I is ensured by crossover formation (reciprocal chromatid exchange) between homologous chromosomes,1 which requires accurate pairing and assembly of the synaptonemal complex (SC)2 between them. Because no pairing between homologous chromosomes usually exists before meiotic entry, establishment of homologous pairing at the beginning of meiotic prophase serves as an essential basis for meiosis (Fig. 1). In recent years there has been rapid progress in our understanding of the molecular mechanisms for later steps such as SC formation and meiotic homologous recombination. However, the molecular mechanisms for homologous pairing, particularly homology recognition, remain poorly understood.3

Figure 1. Schematic diagram of meiosis. Faithful segregation of homologous chromosomes is supported by multiple events in meiotic prophase. Red and magenta lines show homologous chromosomes. The orange line is the chromosome axis, and the green lines are the SC central structure. Blue arrowheads show the direction of chromosome or chromatid segregation.
Although many aspects of homologous pairing remain a mystery, it is now well established that a conserved chromosome movement during meiosis contributes to homologous pairing: a part of a chromosome (usually telomeres) is tethered to the nuclear envelope (NE), and the mechanical forces generated by the cytoskeleton outside the nucleus are transmitted to chromosomes via NE-associated and other proteins to move chromosomes.4,5 Such chromosome movement is proposed to facilitate both chromosome encounters and dissociations, thus promoting the homology search. This model implies that there is a mechanism that selectively stabilizes the association between homologous chromosomes, but not between non-homologous ones. However, how such selective stabilization operates is poorly understood. Considering the fact that the SC assembles even between non-homologous chromosomes, it is unlikely that the SC is the primary means of distinguishing homologous vs. non-homologous association. Consistently, cytological observations support the presence of presynaptic stabilization of homolog association in the absence of, or prior to, SC formation.6,7 What molecular mechanism supports presynaptic stabilization? Meiotic DSB formation and its repair by homologous recombination (HR) may stabilize the interaction between homologous chromosomes through physical linkage created by DNA strand exchange, although the exact mechanism has not yet been elucidated. In addition, as evidenced by the capacity for successful homologous pairing and synapsis even in the complete absence of meiotic DSB formation in C. elegans8 and the fruit fly Drosophila,9 there is clearly a DSB-independent, and thus HR-independent, mechanism for homologous chromosome pairing. Although the molecular nature of this mechanism is poorly understood, a DSB-independent mechanism apparently exists in a wide variety of organisms, because some level of homologous pairing is reported in mutants defective in DSB formation and/or HR even in yeast, which primary relies on the DSB-dependent mechanism.10-13 In this regard, C. elegans, which primarily relies on the DSB-independent pairing, is an excellent model to study this mysterious pairing mechanism.
During meiotic prophase in C. elegans, the pairing center (PC), a cis-acting element unique to this organism, functions as an equivalent of telomeres of other organisms and tethers the chromosome end to the NE.5 Each C. elegans chromosome has one PC near one end of the chromosome. During the presynaptic stage, the outer NE protein ZYG-12 and the inner NE protein SUN-1 interact with each other, forming patches on the NE. These ZYG-12/SUN-1 patches colocalize with PC-binding proteins (ZIM-1, -2, -3 and HIM-8) that are bound to specific repeat sequences within the PC.14 ZYG-12/SUN-1 patches move dynamically as a result of forces dependent on cytoplasmic dynein,15 presumably promoting chromosome encounters and dissociations. Although presynaptic pairing (i.e., SC-independent pairing) is strongly stabilized at the PC loci,16 it appears that the PC proteins by themselves do not define the pairing specificity because two PC proteins (ZIM-1 and ZIM-3) are shared between two chromosomes (II and III, and I and IV, respectively) but these chromosomes do not show non-homologous pairing.17 This observation suggests that the PC target sequence is not sufficient and that PC neighboring loci unique to each chromosome are also necessary to define chromosome identity in pairing. It should, however, be noted that artificial recruitment of the PC proteins ZIM-2 and HIM-8 to a high-copy target sequence array integrated into two chromosomes (V and X) leads to highly efficient presynaptic non-homologous association between them.14 Therefore, the PC proteins have a certain potency to stabilize presynaptic interaction, at least in this artificial overloading condition. In addition, even in the non-PC regions PC target sequence are present14 and PC protein loading occurs albeit at a lower density,18 suggesting that the PC protein might directly stabilize presynaptic pairing in the non-PC regions. As a separate function from stabilization of presynaptic pairing, the PC also promotes SC formation.19
Based on this dual function of the PC, the following model has been proposed (Fig. 2A, master pairing-site model): (1) the PC is the master pairing-site for presynaptic pairing, and successful homologous pairing triggers SC assembly at the PC; (2) as the SC assembly extends from the PC to non-PC region, a pair of homologous chromosomes are zipped up to establish full alignment of chromosomes.19 In this model, non-PC chromosomal regions do not actively participate in the presynaptic pairing process, and thus full presynaptic alignment along a chromosome does not occur in C. elegans. However, a few previous studies suggested that non-PC regions could potentially participate in presynaptic pairing, albeit in a minor role.19,20 If this is the case, pairing activity intrinsic to the non-PC region would exist, and there could be the capacity to support presynaptic full alignment (Fig. 2B, a collaborative pairing model). Careful evaluation of the validity of these two models is critical, because presynaptic pairing performed by a solo master pairing-site is rather exceptional and many organisms show a more collaborative mode of pairing in which the pairing activity is distributed along the entire length of a chromosome.6,7,21 However, the role of pairing of the non-PC regions in C. elegans has been often disregarded due to the dominant role of the PC. Moreover, investigation of presynaptic full alignment in C. elegans has been technically difficult because locus-specific fluorescent in situ hybridization (FISH), which has been conventionally used to assess pairing status, is not suitable for demonstrating homologous pairing along the entire length of a chromosome. Therefore we have recently developed a method to apply chromosome paint to a whole mount gonad of C. elegans in order to analyze chromosome alignment.18 We examined a three-dimensional (3D) rendering image of chromosome paint that was reconstructed from optical sections of the syp-1 mutant that is capable of forming a chromosome axis but completely defective in SC central region assembly. From this analysis, it became clear that presynaptic full alignment exists in C. elegans (Fig. 3). Moreover, this full alignment was not merely a rare observation, but was observed in a significant fraction (20–30%) of nuclei in mid meiotic prophase. This finding strongly suggests the presence of pairing activity intrinsic to the non-PC regions in C. elegans.

Figure 2. Alternative models of homologous chromosome association and SC formation. PC, pairing center; SC, synaptonemal complex; MT, microtubule; NE, nuclear envelop.
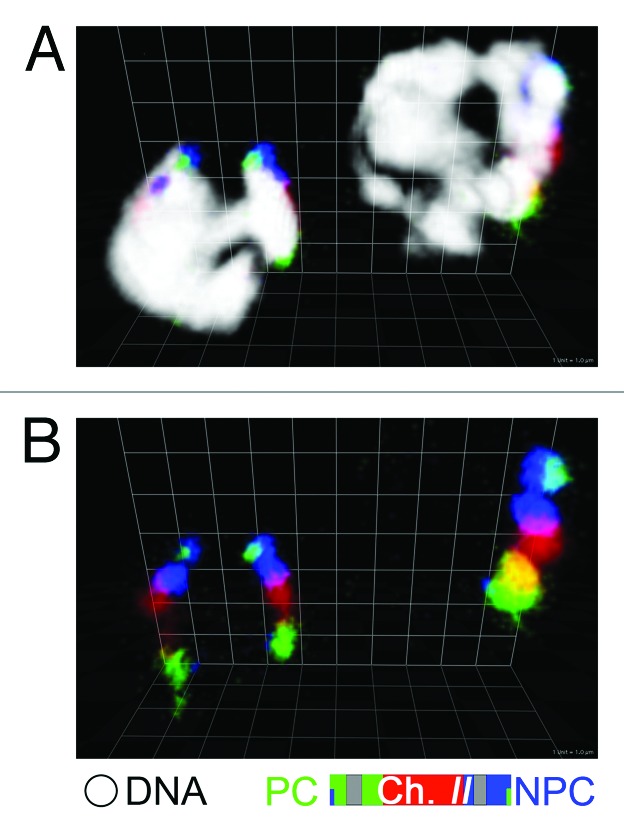
Figure 3. Presynaptic full alignment. Chromosome II in syp-1 mutant is painted with three fluorophores as in the diagram at the bottom, with (A) or without (B) DAPI staining in white. Three-dimensional reconstruction of two nuclei is shown; no pairing (left) and full alignment (right). Scale grid: 1 µm unit square length.
What molecular mechanism supports non-PC pairing activity? A clue to this question came from our recent study,22 in which we found that a chromodomain protein, MRG-1, facilitates presynaptic full alignment activity, possibly in a direct manner. This molecule was discovered in a RNAi screen that targeted the pairing process using a GFP reporter system23 to monitor in vivo pairing status and utilizing the whole genome RNAi library of C. elegans. In loss-of-function mutants of mrg-1 (RNAi or qa6200, a putative null allele), homologous pairing is decreased specifically in the non-PC regions (but not at the PCs) of autosomes. The absence of a pairing defect for the X chromosome in the mrg-1 mutants correlates with the absence of MRG-1 protein on the X chromosome and suggests direct involvement of MRG-1 in the homologous pairing process. Presynaptic full alignment is significantly decreased, but presynaptic PC pairing is not affected in the mrg-1 mutant, indicating that MRG-1 facilitates presynaptic pairing activity intrinsic to the non-PC regions. The phenotype of mrg-1 also points to the importance of presynaptic pairing in non-PC regions for proper SC assembly. Since the PCs successfully pair and synapse between homologous partners in the mrg-1 mutant, the prevailing master pairing-site model predicts that the homologous non-PC regions would be brought together as the SC assembles from the PC toward the non-PC, and thus establish proper homologous synapsis along the entire length. This is clearly not the case in the mrg-1 mutant, in which homologous synapsis at the PC and non-homologous synapsis in the non-PC regions coexist. Therefore, proper homologous pairing supported by pairing activity intrinsic to non-PC regions is necessary to ensure proper synapsis along the entire length of a chromosome. This observation further strengthens the collaborative pairing model: C. elegans chromosomes are not relying on just a single locus (the PC), but instead both the PC and the non-PC regions collaborate to ensure proper homologous interaction during meiotic prophase.
How does MRG-1 facilitate presynaptic homologous association? Regarding a molecular function, the orthologs of MRG-1 in other organisms (yeasts24,25 and humans26,27) are known to be a component of complexes containing multiple proteins, many of which are histone-modifying enzymes. These MRG-1 orthologs bind to di- or tri-methylated histone H3K36 (H3K36me2/3) via their chromodomain.28,29 MRG-1 of C. elegans likely functions in pairing in a similar manner, since it is known that H3K36me2/3 modification is enriched on autosomes.30 Therefore, it is possible that histone modification could be involved in selective stabilization of homologous chromosome association. As for a cellular mechanism, because our knowledge of the mechanism for homology recognition is very limited, we cannot simply place MRG-1 in a known context. Thus, we need to first develop a conceptual framework of homology recognition that incorporates our cytological observations both in the wild type and the mrg-1 mutant, as well as previously reported observations.
Our speculative model hypothesizes two types of force for presynaptic homologous pairing: one that mechanically moves chromosomes (Fm) and one that holds chromosomes (Fh). The Fm would randomly move chromosomes and consequently facilitate both chromosome encounters and dissociations. An example of the Fm is that driven by the PC-SUN-1-ZYG-12-dynein-based mechanism, which applies equally to all chromosomes. Theoretically, any forces that move chromosomes directly or indirectly, even Brownian motion, could contribute to the Fm, and the Fm would be the summation of multiple forces produced by different mechanisms. At the beginning of presynaptic pairing, the Fm could promote chromosome interaction by increasing the chance of their encounter. Once chromosomes encounter each other and start to transiently interact, the Fh kicks in and stabilizes their association, while the Fm now could function to discourage chromosome interaction due to its random nature. At this stage the Fm and the Fh could be opposing each other, and the power balance between the Fm and the Fh determines the stability of presynaptic chromosome interaction. Although it is purely hypothetical, the presence of the Fh, at least in C. elegans, is strongly suggested by the observations that chromosomes establish presynaptic full alignment18 in the presence of their dynamic movement driven by SUN-KASH-dynein.15,31,32 The exact mechanism generating the Fh is not known, particularly in C. elegans in which the DSB-independent mechanism is primarily operating.8 In DSB-dependent pairing, DNA strand invasion during HR repair between homologous chromosomes could generate the Fh. For DSB-independent pairing, there are many candidates that could theoretically generate the Fh. For example, the Fh could be generated through DNA-DNA direct interaction,33 protein-mediated interaction,34 or a transcription coupled RNA-mediated mechanism.35 Like the Fm, the Fh could be the summation of the forces generated by multiple mechanisms. According to this model, if Fm < Fh, chromosome interaction is stabilized and chromosomes would stay associated. If Fm > Fh, the chromosomes would dissociate from each other. Successful homologous pairing would be supported by conditions where the Fh between homologous chromosomes (Fh-hom) is stronger than the Fm, while the Fh between non-homologous (heterologous) chromosomes (Fh-het) is weaker than the Fm (i.e., Fh-het < Fm < Fh-hom). Although Fm and Fh-hom should be positive values, Fh-het could be any value, including a negative value (i.e., repulsion between non-homologous chromosomes) or zero in the wild type. However, at least in mutants that exhibit non-homologous synapsis, Fh-het would be a positive value, otherwise it would be difficult to initiate SC assembly between non-homologous chromosomes.
In this model, the mechanism ensuring the proper power balance among these forces represents the mechanism that selectively stabilizes homologous association, and thus “recognizes” homology between chromosomes. Therefore, any changes that disturb the balance between these forces could result in defective homologous pairing. Among these, the condition of Fm < Fh-het would lead to erroneously stable non-homologous association, which could further lead to non-homologous synapsis. Therefore, we speculate that the mrg-1 mutation creates a condition in which Fm < Fh-het, resulting in a decrease in presynaptic homologous pairing and the presence of non-homologous synapsis.22 This condition can result from either (1) a decrease in the Fm, or (2) an increase in the Fh-het (or Fh in general), or both (1) and (2). In the mrg-1 mutant, a general decrease in the Fm appears to be unlikely, because the Fm not only dissociates chromosomes but also promotes homolog encounters. If the Fm is decreased in the mrg-1 mutant, homolog encounters would be less efficient even at the PC, as seen in the loss-of-function mutant of dynein.15 However, PC pairing and synapsis is not affected in the mrg-1 mutant,22 indicating that an intact Fm is applied, at least to the PC. Although a general decrease in the Fm applied to the PC is unlikely, there could be irregular local decreases in the Fm in the mrg-1 mutant. A partially condensed chromosome in meiotic prophase could be regarded as a flexible and elastic rod-shaped object with a certain stiffness, analogous to a condensed mitotic chromosome.36 When such an object moves in a viscoelastic fluid, the nucleoplasm, it could receive significant viscous resistance as shown with the interphase nucleoplasm.37 In such a case, the Fm applied to one end (i.e., the PC) of a chromosome would move that end very efficiently, but not the other (non-PC) end depending on the stiffness of chromosomes and the viscoelasticity of the nucleoplasm. MRG-1 might support proper transmission of the Fm to the non-PC regions through the regulation of chromosomal properties (e.g., chromosome stiffness). In this scenario, even when an intact Fm is applied to the PC, the non-PC region might receive an irregularly smaller Fm in mrg-1 than in the wild type, which could lead to Fm < Fh(-het) in the non-PC regions. As mentioned above, the other scenario is an increase in Fh(-het) that overwhelms Fm. In this scenario, MRG-1 might be involved in modulation of Fh (either Fh-het selectively or the Fh in general), and in the mrg-1 mutant the Fh(-het) might be increased compared with the wild type and exceeds the Fm, making association between non-homologous chromosomes more stable (i.e., making chromosomes stickier). These two scenarios are not mutually exclusive. Consequently, either one or both of them would create the situation in which it is more difficult to dissociate non-homologous chromosomes in non-PC regions than at the PCs in the mrg-1 mutant (Fig. 4). Future investigation that carefully examines the presence of presynaptic non-homologous association in the mrg-1 mutant and deciphers these two opposing putative forces (Fm and Fh), in addition to studies addressing the molecular basis of these forces, particularly the Fh, will reveal the exact mechanism of action of MRG-1 in homologous pairing and provide a framework to understand the mechanism for homology recognition.

Figure 4. A model hypothesizing the correct balance between the force moving chromosomes (Fm) and the one holding chromosomes (Fh) for selective stabilization of homologous- chromosome interaction.
Acknowledgments
The author is grateful to all current and previous Nabeshima lab members for generating data and developing ideas, to Raymond Chan and Gyorgyi Csankovszki for critical reading of the manuscript and valuable comments, and to Yukiko Yamashita for enormous contribution in improvement of the manuscript and conceptual development. This work was supported by a research grant (5-FY07-666) from the March of Dimes Birth Defects Foundation to K.N.
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/19528
References
- 1.Neale MJ, Keeney S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature. 2006;442:153–8. doi: 10.1038/nature04885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–9. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 3.Barzel A, Kupiec M. Finding a match: how do homologous sequences get together for recombination? Nat Rev Genet. 2008;9:27–37. doi: 10.1038/nrg2224. [DOI] [PubMed] [Google Scholar]
- 4.Chikashige Y, Haraguchi T, Hiraoka Y. Another way to move chromosomes. Chromosoma. 2007;116:497–505. doi: 10.1007/s00412-007-0114-8. [DOI] [PubMed] [Google Scholar]
- 5.Bhalla N, Dernburg AF. Prelude to a division. Annu Rev Cell Dev Biol. 2008;24:397–424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loidl J. The initiation of meiotic chromosome pairing: the cytological view. Genome. 1990;33:759–78. doi: 10.1139/g90-115. [DOI] [PubMed] [Google Scholar]
- 7.Zickler D, Kleckner N. Meiotic chromosomes: integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 8.Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–98. doi: 10.1016/S0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 9.McKim KS, Hayashi-Hagihara A. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 1998;12:2932–42. doi: 10.1101/gad.12.18.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loidl J, Klein F, Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994;125:1191–200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–91. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 12.Nag DK, Scherthan H, Rockmill B, Bhargava J, Roeder GS. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 1995;141:75–86. doi: 10.1093/genetics/141.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips CM, Meng X, Zhang L, Chretien JH, Urnov FD, Dernburg AF. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat Cell Biol. 2009;11:934–42. doi: 10.1038/ncb1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato A, Isaac B, Phillips CM, Rillo R, Carlton PM, Wynne DJ, et al. Cytoskeletal forces span the nuclear envelope to coordinate meiotic chromosome pairing and synapsis. Cell. 2009;139:907–19. doi: 10.1016/j.cell.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacQueen AJ, Colaícovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16:2428–42. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips CM, Dernburg AF. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev Cell. 2006;11:817–29. doi: 10.1016/j.devcel.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 18.Nabeshima K, Mlynarczyk-Evans S, Villeneuve AM. Chromosome painting reveals asynaptic full alignment of homologs and HIM-8-dependent remodeling of X chromosome territories during Caenorhabditis elegans meiosis. PLoS Genet. 2011;7:e1002231. doi: 10.1371/journal.pgen.1002231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–50. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loidl J, Pasierbek P, Rose AM. Conservation and variability of meiotic processes—Lessons from the unconventional meiosis of C. elegans. Chromosomes Today. 2004;14:93–101. [Google Scholar]
- 21.McKee BD. Homolog pairing and segregation in Drosophila meiosis. Genome Dyn. 2009;5:56–68. doi: 10.1159/000166619. [DOI] [PubMed] [Google Scholar]
- 22.Dombecki CR, Chiang ACY, Kang H-J, Bilgir C, Stefanski NA, Neva BJ, et al. The chromodomain protein MRG-1 facilitates SC-independent homologous pairing during meiosis in Caenorhabditis elegans. Dev Cell. 2011;21:1092–103. doi: 10.1016/j.devcel.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Yuen KW, Nabeshima K, Oegema K, Desai A. Rapid de novo centromere formation occurs independently of heterochromatin protein 1 in C. elegans embryos. Curr Biol. 2011;21:1800–7. doi: 10.1016/j.cub.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayama J, Xiao G, Noma K, Malikzay A, Bjerling P, Ekwall K, et al. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. EMBO J. 2003;22:2776–87. doi: 10.1093/emboj/cdg248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Pardo PS, Leung JK, Lucchesi JC, Pereira-Smith OM. MRG15, a novel chromodomain protein, is present in two distinct multiprotein complexes involved in transcriptional activation. J Biol Chem. 2002;277:50860–6. doi: 10.1074/jbc.M203839200. [DOI] [PubMed] [Google Scholar]
- 27.Yochum GS, Ayer DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol Cell Biol. 2002;22:7868–76. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–8. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 29.Zhang P, Du J, Sun B, Dong X, Xu G, Zhou J, et al. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–8. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rechtsteiner A, Ercan S, Takasaki T, Phippen TM, Egelhofer TA, Wang W, et al. The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet. 2010;6:e1001091. doi: 10.1371/journal.pgen.1001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baudrimont A, Penkner A, Woglar A, Machacek T, Wegrostek C, Gloggnitzer J, et al. Leptotene/zygotene chromosome movement via the SUN/KASH protein bridge in Caenorhabditis elegans. PLoS Genet. 2010;6:e1001219. doi: 10.1371/journal.pgen.1001219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wynne DJ, Rog O, Carlton PM, Dernburg AF. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol. 2012;196:47–64. doi: 10.1083/jcb.201106022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danilowicz C, Lee CH, Kim K, Hatch K, Coljee VW, Kleckner N, et al. Single molecule detection of direct, homologous, DNA/DNA pairing. Proc Natl Acad Sci U S A. 2009;106:19824–9. doi: 10.1073/pnas.0911214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas SE, Soltani-Bejnood M, Roth P, Dorn R, Logsdon JM, Jr., McKee BD. Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell. 2005;123:555–68. doi: 10.1016/j.cell.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 35.Ding DQ, Haraguchi T, Hiraoka Y. From meiosis to postmeiotic events: alignment and recognition of homologous chromosomes in meiosis. FEBS J. 2010;277:565–70. doi: 10.1111/j.1742-4658.2009.07501.x. [DOI] [PubMed] [Google Scholar]
- 36.Marko JF. Micromechanical studies of mitotic chromosomes. Chromosome Res. 2008;16:469–97. doi: 10.1007/s10577-008-1233-7. [DOI] [PubMed] [Google Scholar]
- 37.Tseng Y, Lee JSH, Kole TP, Jiang I, Wirtz D. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J Cell Sci. 2004;117:2159–67. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]


