Abstract
MicroRNAs (miRNAs) are endogenously expressed small non-coding RNAs acting at the post-transcriptional level where they promote mRNA degradation and block protein translation. Recent findings suggest that complex transcriptional and post-transcriptional circuits control miRNAs. STAT3 has emerged as an important regulator of their expression and biogenesis and, in turn, STAT3 signaling pathways are controlled by distinct miRNAs. We summarize the current knowledge on STAT3 mediated processing of individual miRNAs and contrariwise, the modulation of the STAT3 pathway by miRNAs in development and in pathophysiological conditions such as immune processes, infection, cancer, cardiovascular disease and pulmonary hypertension.
Keywords: STAT3, miRNA, cancer, cardiovascular disease, lung, immunity
Introduction
A decade ago the discovery of a novel class of evolutionarily conserved small (18–24 nucleotides) non-coding RNA molecules, miRNAs, has revolutionized our view of gene regulation in almost all biological processes.1 MiRNAs emerged as counterparts of transcription factors acting at the post-transcriptional level where they target mostly the 3′ untranslated region (UTR) of gene transcripts (mRNAs), promoting their degradation or suppressing their translation into proteins, thereby silencing genes.2
MiRNA genes are located either as individual or clustered genes in intergenic or intronic regions. They are transcribed by RNA polymerase II as primary transcripts, the so-called pri-miRs. These pri-miRs are cleaved from a length of hundreds to thousands of nucleotides to hairpin-shaped precursors, the pre-miRNAs—a process that is mediated by a ribonuclease III called Drosha and the double-stranded DNA binding protein DGCR8/Pasha.3
The pre-miRNAs are transported to the cytoplasm via the nuclear export factor exportin 5 and further processed into ~22 nucleotide miRNA duplexes by the ribonuclease III Dicer and its cofactors (PACT and TRBP). After dissociation one strand of the RNA molecule is incorporated into the RNA induced silencing complex (RISC).4
The RISC-linked miRNA is capable of binding to the target mRNA leading either to degradation or to translational repression. The pairing specificity of the miRNA to the 3′ UTR of a target mRNA is considered to be based on the 5′ proximal “seed” region (nucleotides 2–8) and the secondary structure of the surrounding region 4–8. Apart from the Watson-Crick base pairing, the number and configuration of mismatches between the miRNA and the target mRNA as well as the number of target sequences on the mRNA determine the efficiency of transcriptional repression.5
Moreover, the ability of a single miRNA to regulate multiple functionally related mRNAs potentiates the strength of miRNA-based regulation. This has been repeatedly demonstrated for the liver-specific miR-122, which regulates numerous metabolic genes.6 Notably, an inhibitor of miR-122 (miravirsen or SPC3649) is currently tested in a phase 2A clinical trial to assess safety and tolerability in treatment-naive patients with chronic hepatitis C.7
Although remarkable progress has been made in our understanding of miRNA biogenesis and function over the past years, the regulatory mechanisms that orchestrate the complex network of miRNAs and the mechanisms miRNAs use to regulate gene expression are not fully understood.8 Likewise, information on upstream regulators of miRNAs that manage their transcriptional and post-transcriptional control is scarce. Genes encoding miRNAs are transcribed by RNA polymerase II similar to most of their target mRNAs suggesting that transcription factors such as STAT3 fine-tune miRNA expression resulting in complex regulatory circuits involving positive and negative feedback loops.9
STAT3 belongs to the family of STAT proteins, which are activated in response to extracellular signaling proteins including the interleukin (IL)-6 family [IL-5, IL-6, IL-11, leukemia inhibitory factor (LIF), oncostatin M (OSM), ciliary neurotrophic factor (CNTF) and cardiotrophin-1 (CT-1)], erythropoietin and leptin, prolactin and angiotensin II (AngII).10-13 Upon activation, i.e., phosphorylation of tyrosine and serine residues, STAT3 forms homo- or heterodimers with other STAT proteins and translocates to the nucleus where it activates the transcription of downstream target genes by binding to specific DNA elements (gas-motives) in their promoter.14 In addition, and as outlined in the present review several miRNAs are also transcriptionally regulated by STAT3.15,16
The goal of the present review is to highlight regulatory circuits involving STAT3 and miRNAs in the context of development and pathophysiological conditions related to inflammation, infection, cancer, cardiovascular disease and pulmonary hypertension (Table 1). A better understanding of the complex regulatory networks between STAT3 and miRNAs may lead to novel specific therapeutic approaches in various disease settings.
Table 1. Function of miRNAs involved in STAT3-dependent circuits in various fields of development and disease.
| Field | miRNA | Function | References |
|---|---|---|---|
|
Embryonic development |
miR-93, miR-17-5p |
Germ layer differentiation |
18 |
| miR-17, miR-20a, miR-106b |
Lung branching morphogenesis |
21 |
|
| miR-124a, miR-9 |
Neural lineage differentiation |
19, 20 |
|
|
Immunity and infection disease |
miR-125b |
Myeloid cell proliferation |
22, 23 |
| miR-17-5p, miR-20a |
Proliferation of myeloid derived suppressor cells |
24 |
|
| miR-155 |
Innate antiviral immunity in HBV-infection |
27 |
|
| let-7a |
Progression of hepatitis B to hepatocellular carcinoma |
23 |
|
|
Cancer disease |
miR-21, miR-19a/b |
Involvement in multiple myeloma |
34, 15, 36, 37 |
| miR-21, miR-181b |
Involvement in colon adenocarcinoma |
38 |
|
| miR-155, miR-20b |
Involvement in breast cancer |
35 |
|
| miR-9, miR-17, miR-20a |
Involvement in glioma |
32 |
|
|
Cardiovascular system |
miR-199a |
Sarcomere protein ubiquitination and turnover sarcomere integration |
16 |
| Myocardial vascular function |
16 |
||
| miR-21 |
Regeneration through MSCs |
48 |
|
| Pulmonary hypertension | miR-17-5p, miR-20a |
Fibrous matrix production |
50 |
| miR-204 | Vascular remodeling | 52 |
STAT3-miRNAs Circuits in Mammalian Development
Soon after the recognition of miRNAs as fundamental regulators of gene expression, it became clear that miRNAs have important regulatory functions during embryonic development.17 For example, Foshay et al. provided evidence that miRNA (miR)-17 family members, miR-17-5p, miR-20a, miR-93 and miR-106a, are differentially expressed in developing mouse embryos and function to control differentiation of stem cells.18 Particularly, miR-93 appears to promote the differentiation of primitive endoderm and trophectoderm in the blastocyst. High expression of miR-93 and miR-17-5p were also found within the mesoderm of gastrulating embryos acting to delay or enhance differentiation into the different germ layers. Notably, one major target gene responsible for the effects of these miRNAs on cellular differentiation was STAT3 that is known to act as an embryonic stem (ES) cell regulator.18 Using STAT3-3′UTR-Luciferase constructs, a regulatory property was particularly shown for miR-93 and to a lesser extent for miR-20a but not for the other members of the miR-17 family.
Regulatory circuits of STAT3 and miRNAs play important roles in the neural lineage differentiation of ES cells. Neural stem cells differentiate into the three main neural lineages: neurons, astrocytes and oligodendrocytes.19 In the developing central nervous system, activation of STAT3 is known to direct the differentiation of neural stem cells toward astrocytes and to suppress neurogenesis.20 Two putative brain-specific miRNAs, miR-124a and miR-9, appear to be essential for this process since they appear to modulate neuronal differentiation by downregulating canonical STAT3 signaling by targeting the gp130, the LIFR and the CNTFR. This miRNA-mediated suppression of STAT3-mediated effects may promote differentiation of neural stem cells toward neuronal lineage a feature that could be of therapeutic value for potential stem cell therapy in neurodegenerative diseases such as Parkinson or Alzheimer disease.
The branching morphogenesis of the lung is a third example where regulatory circuits between STAT3 and miRNA may participate in embryonic development. Here, particularly the miR-17 family seems essential for maintaining the homeostasis of epithelial structures in the developing lung since miR-17, miR-20a, and miR-106b were identified as fine-regulators of both mitogen-activated protein kinase-14 (MAPK14) and STAT3, which in turn regulate cadherin-1 (CDH1) expression.21 As a key adhesion receptor CDH1 is essential for epithelial cell identification and grouping during development and mediates the interaction between extracellular matrix, cytoplasmic plaques, and other adhesion molecules. However, detailed molecular links between STAT3 and the observed alteration of CDH1 expression remain to be further evaluated.
In summary, regulatory circuits involving STAT3 and miRNAs play important roles during stem cell differentiation and early organogenesis. A better understanding of these interactions may help to improve stem cell based regenerative therapy concepts in the future.
STAT3-miRNAs Circuits in Differentiation of Myeloid Cells and Innate Immunity
In the past decade, numerous miRNA-based key regulatory functions in the innate and adaptive immunity have been identified including immune cell lineage commitment, differentiation, maturation and maintenance of immune homeostasis.
For example, in primary lineage-negative cells, miR-125b overexpression enhances colony-formation in vitro and promotes myelopoiesis in mouse bone marrow chimeras. In this process miR-125b coordinated the regulation of several signaling pathways involving also STAT3 to direct distinct phenotypes in myeloid cells, i.e., blocking granulocyte colony-stimulating factor (G-CSF)-dependent differentiation of primary lineage-negative cells and at the same time enhancing colony formation in the presence of G-CSF indicating that miR-125b confers a proliferative or survival advantage to myeloid cells.22,23 MiR-125b overexpression reduced DNA-binding, and transcriptional activity but not induction of tyrosine-phosphorylation and nuclear translocation of STAT3 suggesting that miR125b is not interfering with canonical STAT3 activation pathways but seems to affect one or more STAT3 cofactors.22 The activator protein 1 (AP-1) transcription factors c-Jun and JunD were identified as novel miR-125b targets, but only gene-specific silencing of JunD and not c-Jun mimicked to some extend the miR-125b overexpression phenotype with regard to myelopoiesis and STAT3 silencing.22 Thus, miR-125b, STAT3 and JunD are interconnected in a novel circuit regulating the differentiation and survival of specific inflammatory cell types from myeloid precursors.
Additional miRNAs, such as miR-17-5p and miR-20a, exert also suppressive potential on so-called myeloid-derived suppressor cells (MDSCs) by modulating STAT3 expression.24 MDSCs are considered as important components of the immune suppressive network that particularly affects T cell function and are critical factors for tumor-associated immune suppression.25 Importantly, MDSCs transfected with miR-17-5p or miR-20a display reduced ability to suppress antigen-specific CD4 and CD8 T cells. Similar to miR-125b, miR-17-5p and miR-20a are directly regulating STAT3 via seed sequence binding thereby affecting the activity of MDSCs.24 Interestingly, the expression of both miR-17-5p and miR-20a in MDSCs was found to be lower in tumor bearing mice than in disease-free animals, which may trigger the suppression of T cell mediated immunity under cancerous condition.
It becomes increasingly evident that many miRNAs play important roles for the innate immune response. In the context of viral infections, miRNAs are regulators of both viral and host cell gene expression, and therefore, can benefit either the virus or the host.26 Consequently, the particular interaction determines the degree to which hosts are able to restrict viral replication and infection and finally the viral pathogenesis and outcome. Notably, viruses may use cellular miRNAs for their replication machinery, and furthermore, may influence the expression of cellular miRNAs and thereby the cellular gene expression for the benefit of their own survival.
Su et al. reported on a protective role of miR-155 for the innate antiviral immunity through targeting suppressor of cytokine signaling 1 (SOCS-1), and thus, promoting Janus kinase (JAK)-STAT signaling pathway in response to current anti-hepatitis B virus (HBV) treatment with interferon (IFN)-α, which consequently inhibits HBV replication in human hepatoma cells.27 The contrary situation was shown in a study demonstrating how miRNA expression in host cells is dysregulated by viral proteins.23 The authors showed that the pleiotropic HBV x protein (HBx), which is associated with hepatocellular carcinoma (HCC), induces an alteration of the miRNA expression pattern in hepatoma cells. Among others, the let-7 family, in particular let-7a, was downregulated. Further characterization of let-7a in hepatoma cells implied that it negatively regulates cellular proliferation through direct targeting of STAT3. Thus, HBx-mediated downregulation of let-7a and upregulation of STAT3 would promote cell proliferation in HBx transfected cells and induce tumor formation and development of HCC.
Thus, these studies demonstrate a potential miRNA-controlled STAT3 axis in the regulation of the innate immunity. In this regard, modulating miR-17-5p and miR-20a may lead to immunotherapeutic approaches in the treatment of cancer diseases. In addition, miRNA modulation of the STAT3 pathway may have the potential to be incorporated into therapeutic control of inflammation and infectious diseases affecting the immunity ranging from viral infections to virus-induced cancer diseases.
STAT3-miRNAs Circuits in Cancer Diseases
Enhanced activation of STAT3 is present in many human cancer types and numerous tumor cell lines and is therefore considered a molecular abnormality that supports the tumor phenotype.28,29
Recently, the analysis of gene expression data in glioblastoma in combination with matched miRNA profiles, uncovered a post-transcriptional regulation layer of surprising magnitude, comprising more than 248,000 miRNA-mediated interactions. These include about 7,000 genes, whose transcripts act as miRNA “sponges” and 148 genes that act through alternative, non-sponge interactions.30 Biochemical analyses in cell lines confirmed that this network regulates established drivers of tumor initiation and subtype implementation, including phosphatase and tensin homolog (PTEN), platelet derived growth factor receptor A (PDGFRA), retinoblastoma protein 1 (RB1), vascular endothelial growth factor A (VEGFA), STAT3 and runt-related transcription factor 1 (RUNX1), suggesting that these interactions mediate crosstalk between canonical oncogenic pathways.
In line with this observation, it has been reported that STAT3 is constitutively activated in 60% of primary high-grade malignant gliomas and the extent of activation correlates positively with the glioma grade.31 Interestingly, several miRNAs such as miR-9, miR-17 and miR-20a that assumedly target STAT3 were found to be dysregulated.32 It was suggested that these miRNAs serve as potent modulators of glioma subclass-specific gene expression networks and may be useful for subclassification of gliomas such as oligodendrogliomas and glioblastomas.32 This is an important notion since these disease types have different clinical outcomes and require distinct treatments.32
The negative regulation of STAT3 by miR-20a via binding to the 3′-UTR of STAT3 mRNA was also demonstrated in pancreatic carcinoma cells, which reveal reduced expression of miR-20a and in turn enhanced de-repressed STAT3 expression and activation, boosting proliferation and invasion pathways in pancreatic carcinomas.33 Therefore, applying miRNA-20a is discussed as a novel approach to treat various carcinomas as an alternative or additional option to current cancer therapies targeting STAT3.
MiRNAs also regulate the activity state of STAT3 in tumors. For example, the upregulation of miR-19a/b in multiple myeloma negatively regulates SOCS-1, which is a factor that terminates STAT3 activation.34 Likewise, miR-155 exerts its oncogenic effects in breast cancer, at least in part, by negatively regulating SOCS-1. Notably, a point mutation in the miR-155 binding site of the SOCS-1 3′-UTR was identified in a breast tumor that affects miR-155 mediated repression and, thus, promotes the proliferation of breast cancer cells.35 Conclusively, by de-suppression of STAT3 miR-19a/b and miR-155 contribute to oncogenic processes in multiple myeloma.34,35
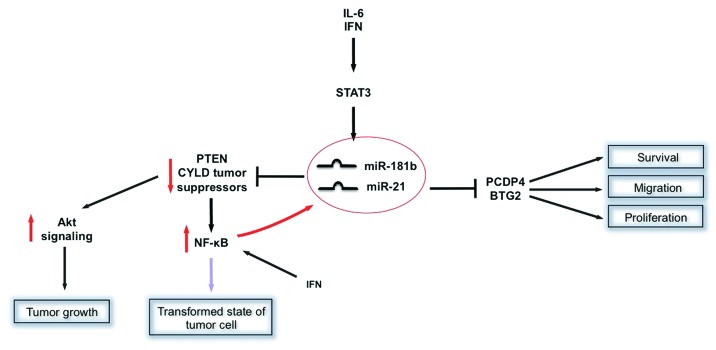
In turn, STAT3-induced miRNAs promote the oncogenic potential of STAT3 by downregulating tumor suppressor genes. For example, PTEN has been long known as a suppressor of tumor growth by de-activating Akt signaling. A key feature of many glioma samples is a loss of PTEN expression. Interestingly, PTEN expression variability induced by miRNA-mediated regulators was predictive for tumor cell growth rates.30 MiR-21 has been identified as a negative regulator of PTEN expression and it has been demonstrated that STAT3 acts as a transcription factor for miR-21 expression in various tumor cell lines (myeloma, prostate cancer cells) in response to IL-6 and IFN-γ.15,36 Meanwhile, miR-21 is considered as an oncomir with numerous anti-apoptotic features in various carcinomas.37 Interestingly, miR-21 initiates the transformation of a non-neoplastic cell into a cancer cell by inducing a complex epigenetic switch.38 This mechanism involves a direct upregulation of miR-21 and also miR-181b in an IL-6/STAT3-dependent manner. MiR-21 and miR-181b inhibit PTEN and cylindromatosis (CYLD) tumor suppressors, leading to increase nuclear factor κB (NFκB) activity that is required to maintain the transformed state. These STAT3-mediated regulatory circuits appear to be crucial for the neoplastic transformation in tumor cells such as in colon adenocarcinomas.38 The role of the STAT3-miR-21 axis in cancerous cells was further demonstrated by Yang et al. who suggested a STAT3 and NFκB co-regulation of miR-21 at the level of the miR-21 promoter in response to IFN, thereby counteracting as a key feedback regulator of IFN-induced apoptotic mechanisms.36 Furthermore, evidence exists that the STAT3-miR-21 circuit also plays a role in tumor metastasis since miR-21 downregulates beside PTEN and IFN-induced apoptosis additional tumor suppressors (programmed cell death protein 4, PCDP4) and anti-proliferative proteins (B cell translocation gene 2, BTG2) thereby promoting cell proliferation, migration and survival in metastatic tumor cells such as B16 melanoma cells.39
Thus, STAT3 is interconnected with miRNAs at multiple levels in complex regulatory circuits (Fig. 1) that are involved in positive feedback loops of oncogenic transformation mechanisms, proliferation, survival and migration of tumor cells and the epigenetic switch that links inflammation to cancer.
Figure 1. Examples of STAT3 involvement in miRNA based regulatory mechanisms in cancer disease. Activated STAT3 acts as a transcription factor for miR-181b and miR-21 expression in various tumor cells. In turn, miR-21 reduces expression of tumor suppressor genes such as PTEN and CYLD, which ultimately promote tumor transformation and growth through activation of Akt and NFκB signaling. Note the positive feedback loop of NFκB as a co-regulator of miR-21 at the level of the miR-21 promoter in response to IFN. Other tumor suppressors regulated by miR-21 are PCDP4 and anti-proliferative proteins like BTG2, which, when downregulated, lead to enhanced cell proliferation, migration and survival in metastatic tumor cells.
STAT3-miRNAs Circuits in Cardiovascular Diseases
In recent years, miRNAs have emerged as fundamental regulators of cardiovascular development, physiology and pathology, such as myocardial infarction, fibrosis, hypertrophy and vascular dysfunction.40-44
This recognition has emerged from numerous studies with genetically modified animal models, by systemic administration of miRNA silencers, the so-called antagomirs, in small animal models, as well as from studies on circulating extracellular miRNAs in the serum or plasma of patients with cardiovascular diseases.45-47
We recently demonstrated a pathophysiological circuit in the heart between reduced STAT3 protein levels, increased miR-199a expression and subsequent impairment of the ubiquitin-proteasome system (UPS) that disrupts the sarcomere structure of cardiomyocytes and impacts on the cardiomyocyte secretome impairing endothelial cell function.16 We found that failure-prone hearts of mice with a cardiomyocyte-specific deletion of STAT3 (STAT3-KO) displayed upregulated cardiac expression of miR-199a before the onset of heart failure. Further analysis revealed that STAT3 protein acts as a potent suppressor of miR-199a transcription in postnatal cardiomyocytes. In turn, upregulated mir-199a expression in STAT3-KO resulted in disturbance of the UPS because miR-199a suppressed the ubiquitin-conjugating enzymes (Ube) Ube2i and Ube2g1. Suppression of Ube2i and Ube2g1 in cardiomyocytes in vivo and in vitro was associated with marked downregulation of specific sarcomeric proteins, i.e., α- and β-myosin heavy chain, derangement of the sarcomeric ultrastructure and an eccentric hypertrophy phenotype of the cardiomyocytes. In addition, the miR-199a-mediated impairment of the UPS caused an accumulation of asymmetric dimethylarginine (ADMA) that was released by cardiomyocytes in concentrations able to impair endothelial cell function suggesting that the regulatory circuit between STAT3 and miR-199a impacts on cardiomyocyte and endothelial function in the heart. The potential clinical importance of our findings were underscored by observations in the terminal failing human heart, where low STAT3 protein levels were associated with increased miR-199a levels and reduced Ube2g1 expression.16
Another study reported on a potential role of STAT3 for cardiac stem cell therapy using a rodent model of myocardial infarction. Here it was demonstrated that ischemic preconditioning would improve the survival of bone marrow-derived mesenchymal stem cells (MSCs) prior to engraftment and promote their paracrine activity as well as their differentiation in a way that involves a STAT3-dependent upregulation of miR-21.48
Thus, regulatory circuits involving STAT3 and miRNAs play important roles in normal cardiac function by maintaining sarcomere homeostasis and endothelial function and seem to be important for protective and regenerative mechanisms during pathophysiological insults such as myocardial ischemia.
STAT3-miRNAs Circuits in Pulmonary Hypertension
Pulmonary hypertension (PH) is a severe disease with high morbidity and mortality that results from an increased pulmonary arterial pressure. Symptoms include shortness of breath, dizziness, fainting, markedly decreased exercise tolerance and right ventricular heart failure. Mutations in the bone morphogenetic protein (BMP) type II receptor (BMPR2) downregulated expression of BMPR2 in idiopathic PH suggest that dysfunctional BMP signaling plays a crucial role in the pathophysiology of PH.49 BMPR2 is known to be crucial for differentiation, proliferation, and the fibrous matrix production of both endothelial and smooth muscle cells. Brock et al. identified BMPR2 as a direct target of miR-17-5p and miR-20a, two miRNAs in the miR-17/92 cluster.50 Furthermore they demonstrated that IL-6 upregulates the expression of the miR-17/92 cluster in human pulmonary arterial endothelial cells via the gp130-STAT3 signaling cascade and showed that a highly conserved STAT3-binding site is present in the promoter region of the miR-17/92 gene (C13orf25).50 In particular, they showed that persistent activation of STAT3 via miR-17-5p and miR-20a reduces the expression of BMPR2 protein through conserved seed matches within the 3′-UTR of its mRNA.50 Thereby, chronic activation of IL-6-gp130-STAT3 signaling leads to a downregulation of BMPR2, which in turn could promote vascular remodeling in the arterial vessels of patients with PH.51
The pathophysiology of PH appears to include enhanced proliferation and reduced apoptosis of pulmonary artery smooth muscle cells (PASMCs). STAT3 seems to promote proliferation and survival of PASMCs by downregulating the expression of miR-204.52 MiR-204 directly targets the expression of protein-tyrosine phosphatase SH2 domain-containing cytoplasmic protein (SHP2), therefore STAT3-dependent downregulation of miR-204 subsequently leads to SHP2 upregulation that via activation of the Src kinase and the nuclear factor of activated T cells promotes PASMCs proliferation and resistance to apoptosis, a feature that may promote PH progression.52
Taken together, these studies uncover novel regulatory pathway involving STAT3 as transcriptional activator or repressor of miRNAs that are critically involved in the etiology of PH and indicate that targeting miRNAs should be explored as a potential new therapeutic strategy for this disease.
Conclusion and Outlook
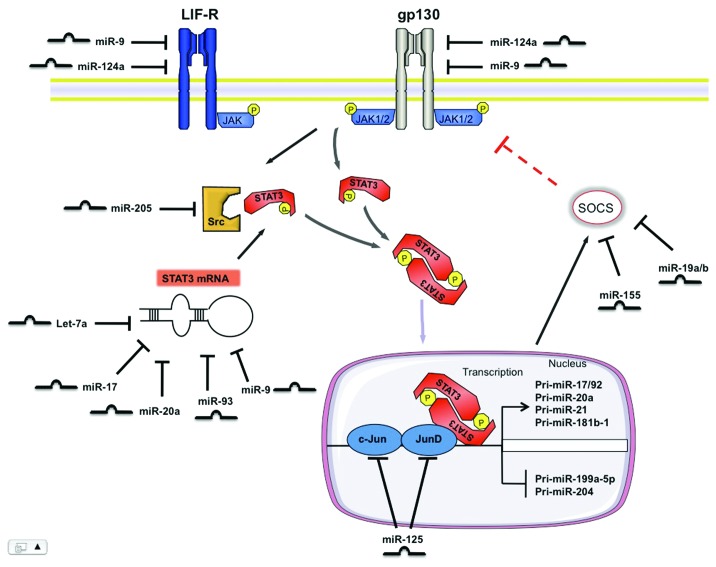
While at the beginning of the miRNA era the main research focus was put on genomic alterations of miRNA expression pattern that would affect the respective target gene or a functional group of genes, recent studies have identified upstream regulators of miRNAs, such as STAT3, and broadened our understanding of how these upstream regulators are interconnected with miRNAs to regulate many physiological and pathophysiological processes (Fig. 2). MiRNA-mediated targeting of STAT3 as well as key steps in the STAT3 signaling pathways illustrate new positive and negative feedback loops that can control the outcome of STAT3 mediated actions and opens up an exciting new avenue in STAT3 research (Fig. 2).
Figure 2. Scheme illustrating STAT3 mediated processing of individual miRNAs and contrariwise, the modulation of the STAT3 pathway by miRNAs at different levels. STAT3 mediates positive and negative regulation of various miRNAs at the transcriptional level. On the other hand, this signaling pathway is controlled by numerous miRNAs at the receptor level, by modulating its activators and suppressors and by direct regulation of STAT3 mRNA.
Approximately 50 miRNAs are predicted to bind the 3′-UTR of STAT3, of which let-7, miR-20a and miR-93 were directly validated using STAT3-3′-UTR-Reporter constructs.18,23
In view of the versatility of the studies illustrated here, it becomes apparent that a miRNA-STAT3 axis plays a major role in development, in adult organ systems and in various pathophysiologies (Table 1). Consequently, a strict tissue-specificity for targeting the miRNA-STAT3 interaction has to be provided since persistent modification of STAT3 in other organs could evoke off-target effects with severe complications.53 A miRNA-based approach to modify the STAT3 pathway may offer novel options to confine the off-target effects, as some miRNAs seem to be predominantly expressed in a tissue-specific manner. At the same time this approach would imply new challenges, as modification of a given miRNA would affect other targets beyond the STAT3 signaling pathway.
In conclusion, with miRNAs as new players in the complex biological networks it remains to be carefully evaluated whether future investigatory efforts will implement sustained translation of experimental miRNA data into the clinical arena or whether it gets lost in translation.
Glossary
Abbreviations:
- ADMA
asymmetric dimethylarginine
- AngII
angiotensin II
- AP-1
activator protein 1
- Bcl-2
B cell lymphoma 2
- BMP
bone morphogenic protein
- BMPR2
bone morphogenetic protein receptor type II
- BTG2
B cell translocation gene 2
- CD
cluster of differentiation
- CDH1
cadherin-1
- CNTF
ciliary neurotrophic factor
- CNTFR
ciliary neurotrophic factor receptor
- CT-1
cardiotrophin-1
- CYLD
cylindromatosis
- ES cell
embryonic stem cell
- G-CSF
granulocyte colony-stimulating factor
- gp130
glycoprotein-130
- HBV
hepatitis B virus
- HBx
hepatitis B virus x protein
- HCC
hepatocellular carcinoma
- IFN
interferon
- IL
interleukin
- JAK
Janus kinase
- LIF
leukemia inhibitory factor
- LIFR
leukemia inhibitory factor receptor
- Mapk14
mitogen-activated protein kinase-14
- MDSCs
myeloid-derived suppressor cells
- miR
miRNA
- miRNA
microRNA
- mRNA
messenger ribonucleic acid
- MSCs
mesenchymal stem cells
- NFκB
nuclear factor κB
- OSM
oncostatin M
- PASMCs
pulmonary artery smooth muscle cells
- PCDP4
programmed cell death protein 4
- PDGFR
platelet derived growth factor receptor
- PH
pulmonary hypertension
- PTEN
phosphatase and tensin homolog
- RB1
retinoblastoma protein 1
- RISC
RNA induced silencing complex
- RUNX
Runt-related transcription factor
- PACT
protein activator of the interferon induced protein kinase
- SHP2
SH2 domain-containing cytoplasmatic protein
- SOCS-1
suppressor of cytokine signaling-1
- Src
proto-oncogene tyrosine-protein kinase Src (sarcoma)
- STAT3
signal transducer and activator of transcription 3
- TRBP
HIV-1 transactivating response RNA-binding protein
- Ube
ubiquitin-conjugating enzyme
- UTR
untranslated region
- VEGFA
vascular endothelial growth factor A
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/19573
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–6. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 3.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 4.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 5.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–26. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 7.Nunnari G, Schnell MJ. MicroRNA-122: a therapeutic target for hepatitis C virus (HCV) infection. Front Biosci (Schol Ed) 2011;3:1032–7. doi: 10.2741/207. [Schol Ed] [DOI] [PubMed] [Google Scholar]
- 8.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 9.Bauersachs J, Thum T. Biogenesis and regulation of cardiovascular microRNAs. Circ Res. 2011;109:334–47. doi: 10.1161/CIRCRESAHA.110.228676. [DOI] [PubMed] [Google Scholar]
- 10.Kirito K, Nakajima K, Watanabe T, Uchida M, Tanaka M, Ozawa K, et al. Identification of the human erythropoietin receptor region required for Stat1 and Stat3 activation. Blood. 2002;99:102–10. doi: 10.1182/blood.V99.1.102. [DOI] [PubMed] [Google Scholar]
- 11.Banks AS, Davis SM, Bates SH, Myers MG., Jr Activation of downstream signals by the long form of the leptin receptor. J Biol Chem. 2000;275:14563–72. doi: 10.1074/jbc.275.19.14563. [DOI] [PubMed] [Google Scholar]
- 12.Hilfiker-Kleiner D, Kaminski K, Podewski E, Bonda T, Schaefer A, Sliwa K, et al. A cathepsin D-cleaved 16 kDa form of prolactin mediates postpartum cardiomyopathy. Cell. 2007;128:589–600. doi: 10.1016/j.cell.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 13.Kodama H, Fukuda K, Pan J, Makino S, Sano M, Takahashi T, et al. Biphasic activation of the JAK/STAT pathway by angiotensin II in rat cardiomyocytes. Circ Res. 1998;82:244–50. doi: 10.1161/01.res.82.2.244. [DOI] [PubMed] [Google Scholar]
- 14.Fischer P, Hilfiker-Kleiner D. Survival pathways in hypertrophy and heart failure: the gp130-STAT axis. Basic Res Cardiol. 2007;102:393–411. doi: 10.1007/s00395-007-0674-z. [DOI] [PubMed] [Google Scholar]
- 15.Löffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermüller J, Kretzschmar AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 16.Haghikia A, Missol-Kolka E, Tsikas D, Venturini L, Brundiers S, Castoldi M, et al. Signal transducer and activator of transcription 3-mediated regulation of miR-199a-5p links cardiomyocyte and endothelial cell function in the heart: a key role for ubiquitin-conjugating enzymes. Eur Heart J. 2011;32:1287–97. doi: 10.1093/eurheartj/ehq369. [DOI] [PubMed] [Google Scholar]
- 17.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foshay KM, Gallicano GI. miR-17 family miRNAs are expressed during early mammalian development and regulate stem cell differentiation. Dev Biol. 2009;326:431–43. doi: 10.1016/j.ydbio.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 19.Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, et al. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–83. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- 20.Gu F, Hata R, Ma YJ, Tanaka J, Mitsuda N, Kumon Y, et al. Suppression of Stat3 promotes neurogenesis in cultured neural stem cells. J Neurosci Res. 2005;81:163–71. doi: 10.1002/jnr.20561. [DOI] [PubMed] [Google Scholar]
- 21.Carraro G, El-Hashash A, Guidolin D, Tiozzo C, Turcatel G, Young BM, et al. miR-17 family of microRNAs controls FGF10-mediated embryonic lung epithelial branching morphogenesis through MAPK14 and STAT3 regulation of E-Cadherin distribution. Dev Biol. 2009;333:238–50. doi: 10.1016/j.ydbio.2009.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Surdziel E, Cabanski M, Dallmann I, Lyszkiewicz M, Krueger A, Ganser A, et al. Enforced expression of miR-125b affects myelopoiesis by targeting multiple signaling pathways. Blood. 2011;117:4338–48. doi: 10.1182/blood-2010-06-289058. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow P, et al. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M, Liu Q, Mi S, Liang X, Zhang Z, Su X, et al. Both miR-17-5p and miR-20a alleviate suppressive potential of myeloid-derived suppressor cells by modulating STAT3 expression. J Immunol. 2011;186:4716–24. doi: 10.4049/jimmunol.1002989. [DOI] [PubMed] [Google Scholar]
- 25.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. 2009;182:4499–506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375–87. doi: 10.1016/j.chom.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Su C, Hou Z, Zhang C, Tian Z, Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J. 2011;8:354. doi: 10.1186/1743-422X-8-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raptis L, Arulanandam R, Geletu M, Turkson J. The R(h)oads to Stat3: Stat3 activation by the Rho GTPases. Exp Cell Res. 2011;317:1787–95. doi: 10.1016/j.yexcr.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lavecchia A, Di Giovanni C, Novellino E. STAT-3 inhibitors: state of the art and new horizons for cancer treatment. Curr Med Chem. 2011;18:2359–75. doi: 10.2174/092986711795843218. [DOI] [PubMed] [Google Scholar]
- 30.Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–81. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natsume A, Kinjo S, Yuki K, Kato T, Ohno M, Motomura K, et al. Glioma-initiating cells and molecular pathology: implications for therapy. Brain Tumor Pathol. 2011;28:1–12. doi: 10.1007/s10014-010-0011-3. [DOI] [PubMed] [Google Scholar]
- 32.Lages E, Guttin A, El Atifi M, Ramus C, Ipas H, Dupré I, et al. MicroRNA and target protein patterns reveal physiopathological features of glioma subtypes. PLoS One. 2011;6:e20600. doi: 10.1371/journal.pone.0020600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan H, Wu J, Liu W, Zuo Y, Chen S, Zhang S, et al. MicroRNA-20a overexpression inhibited proliferation and metastasis of pancreatic carcinoma cells. Hum Gene Ther. 2010;21:1723–34. doi: 10.1089/hum.2010.061. [DOI] [PubMed] [Google Scholar]
- 34.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A. 2008;105:12885–90. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–27. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 36.Yang CH, Yue J, Fan M, Pfeffer LM. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70:8108–16. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 38.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang CH, Yue J, Pfeffer SR, Handorf CR, Pfeffer LM. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem. 2011;286:39172–8. doi: 10.1074/jbc.M111.285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–8. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 41.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, et al. Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation. 2008;118:1567–76. doi: 10.1161/CIRCULATIONAHA.108.769984. [DOI] [PubMed] [Google Scholar]
- 43.Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 44.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, et al. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324:1710–3. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 45.Ji X, Takahashi R, Hiura Y, Hirokawa G, Fukushima Y, Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin Chem. 2009;55:1944–9. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 46.Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–9. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 47.Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol. 2011;31:2383–90. doi: 10.1161/ATVBAHA.111.226696. [DOI] [PubMed] [Google Scholar]
- 48.Haider KH, Idris NM, Kim HW, Ahmed RP, Shujia J, Ashraf M. MicroRNA-21 is a key determinant in IL-11/Stat3 anti-apoptotic signalling pathway in preconditioning of skeletal myoblasts. Cardiovasc Res. 2010;88:168–78. doi: 10.1093/cvr/cvq151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morrell NW. Role of bone morphogenetic protein receptors in the development of pulmonary arterial hypertension. Adv Exp Med Biol. 2010;661:251–64. doi: 10.1007/978-1-60761-500-2_16. [DOI] [PubMed] [Google Scholar]
- 50.Brock M, Trenkmann M, Gay RE, Michel BA, Gay S, Fischler M, et al. Interleukin-6 modulates the expression of the bone morphogenic protein receptor type II through a novel STAT3-microRNA cluster 17/92 pathway. Circ Res. 2009;104:1184–91. doi: 10.1161/CIRCRESAHA.109.197491. [DOI] [PubMed] [Google Scholar]
- 51.Eickelberg O, Morty RE. Transforming growth factor beta/bone morphogenic protein signaling in pulmonary arterial hypertension: remodeling revisited. Trends Cardiovasc Med. 2007;17:263–9. doi: 10.1016/j.tcm.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Courboulin A, Paulin R, Giguère NJ, Saksouk N, Perreault T, Meloche J, et al. Role for miR-204 in human pulmonary arterial hypertension. J Exp Med. 2011;208:535–48. doi: 10.1084/jem.20101812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haghikia A, Stapel B, Hoch M, Hilfiker-Kleiner D. STAT3 and cardiac remodeling. Heart Fail Rev. 2011;16:35–47. doi: 10.1007/s10741-010-9170-x. [DOI] [PubMed] [Google Scholar]