Abstract
STAT5 proteins are activated by tyrosine phosphorylation, but recently further post-translation modifications such as serine/threonine phosphorylation, acetylation at lysine residues or sumoylation in close vicinity of the critical tyrosine residue have been reported. Here, we discuss new findings on impaired STAT5 signaling in lymphocytes isolated from a SUMO-specific protease knockout mouse (SENP1−/−), which results in sumoylated STAT5 and abolishes tyrosine phosphorylation. Van Nguyen and colleagues examined acetylation and sumoylation of STAT5 and found that both modifications act antagonistically to control tyrosine phosphorylation of STAT5.
Keywords: acetylation, crosstalk, sumoylation, STAT5A, STAT5B, tyrosine phosphorylation
STATs are important growth factor- and cytokine-induced transcription factors. STATs1–6 are evolutionarily conserved,1 and fall into six domains (Fig. 1). Extracellular ligands specifically bind to membrane-bound receptors, which subsequently trigger one or more of the four Janus tyrosine kinases (JAKs) to activate STATs by tyrosine phosphorylation at their C-terminal domains.2 JAK2 was also shown to contain serine/threonine kinase activity within its pseudokinase domain, which is the most frequently mutated kinase module in the human kinome.3 Phosphorylated STATs enter the nucleus and activate target genes.4 Beyond tyrosine and serine phosphorylation, lysine ubiquitinylation, neddylation, glycosylation, sumoylation and acetylation were recognized as modulators of STAT signaling.2,5-7
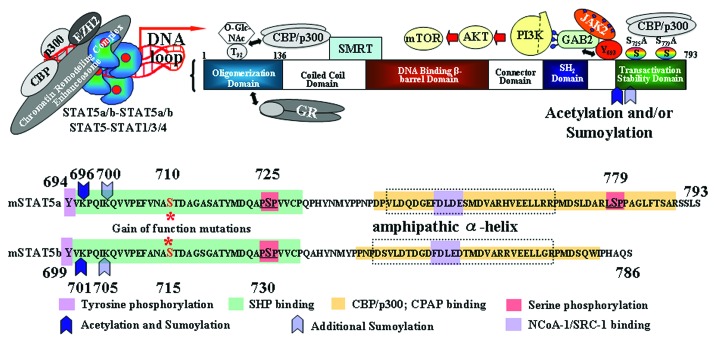
Figure 1. Protein-protein interactions of STAT5 domains with major signaling pathways and chromatin remodeling proteins. Schematic overview of the chromatin remodeling capability of STAT5, exerted through the N- or C-terminal domains. The N-terminus of STAT5 can form oligomers on DNA, resulting in DNA loop formation and interaction with other STAT oligomers forming a transcriptional enhanceosome. Two interaction motifs for the HATs CBP/p300 have been mapped to the extreme N- or C-terminus. The oligomerization domain of STAT5A was also shown to be O-Gluc-NAc modified at threonine position 92. The second CBP/p300 binding interface is at the C-terminus, where particular serine phosphorylation of STAT5A at amino acid position 725 and 779 were shown to boost transcriptional activity and myeloid transformation. Activated and tyrosine phosphorylated STAT5 was also shown to interact with the PI3K-AKT-mTOR pathway through binding to the GAB2 scaffold protein through non-canonical JAK-STAT signaling in the cytoplasm. Shown is also the mapped repressor interaction with SMRT in the coiled coil domain. Moreover, we illustrate a recently mapped interaction of STAT5 oligomers with EZH2 on chromatin. EZH2 is the PcG enhancer of zeste homolog 2, the catalytic component of the polycomb repressive complex 2 that serves to trimethylate histone H3 lysine 27 (H3K27me3). The lower panel shows the C-terminal amino acid compositions of murine STAT5A, from the critical tyrosine residue at position 694 to the carboxyl terminal end of amino acid position 793. Also shown is STAT5B from the critical tyrosine residue at position 699 to the carboxyl terminal end of amino acid position 786 with mapped C-terminal phosphosites, interaction motifs for cofactor (p300/CBP, NCoA-1/SRC-1, CPAP) or phosphatase (SHP) interaction. Very recently, acetylation and sumoylation were mapped to the next lysine at position 696 of STAT5A or 701 of STAT5B following the critical phosphotyrosine site. Moreover, there is an additional sumoylation site found at position 700 of STAT5A and 705 of STAT5B. The C-termini differ in the essential serine phosphorylation site S779 only present in STAT5A. Gain of function mutations at amino acid position 710 of STAT5A or 715 of STAT5B helped to reveal functional insights into STAT5 signaling, which most likely result in a conformational change that allows enhanced and prolonged tyrosine phosphorylation of both STAT5 isoforms when mutated to phenylalanine.
The two genes STAT5A and STAT5B are located next to each other on human chromosome #17 or mouse chromosome #11. Both genes were shown to be essential for hematopoietic development and are aberrantly activated in many hematopoietic cancers.8 They are in close proximity to STAT3 with which they share several functional aspects, like control of G1/S progression and cellular survival. Remarkably, STAT5 is a negative prognostic marker in myeloid leukemias, but a positive one in breast cancer.8-10
STAT5 deletion ablates disease maintenance in chronic myelogenous leukemia and polycythemia vera.11,12 To generalize, STAT5 is essential for generation and/or maintenance of myeloproliferative neoplasms, where it controls proliferation and survival of neoplastic cells.6,8,13-15 These findings define STAT5 as a candidate target for therapy,10 but the exact role of STAT5 for lymphoid neoplasms or carcinomas is incompletely understood and often discussed controversially.8 An important observation from mouse models is that genetic deletion of STAT5 is surprisingly well tolerated in normal hematopoiesis and mainly affects platelet counts.11 Transformation through STAT5 in myeloid cells requires acquisition of complementary oncogenic functions in the nucleus where oligomers of STAT5 activate genes involved in proliferation and survival.13,14 In the cytoplasm of neoplastic myeloid cells the abundant presence of oncogenic STAT5 promotes association with the PI3K-AKT-mTOR pathway to induce cell growth and survival.16,17 Furthermore, STAT5 is a chromatin modifier and it was found to associate with chromatin remodeling proteins including EZH2 or p300/CBP (Fig. 1).8,18 Conditional deletion of STAT5A and STAT5B in adult mouse hematopoietic stem cells revealed a dual role for STAT5 signaling. On the one hand it is a mitogenic factor for most hematopoietic cell lineages including progenitors,8,14 but it is also a key transcriptional regulator that maintains quiescence of hematopoietic stem cells (HSCs) during steady-state hematopoiesis.19
The article “SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation,”20 reveals novel aspects on the control of STAT5 by post-translational modifications. This work examined the phenotype of SENP1 knockout (KO) mice with a detailed biochemical analysis for sumoylation, acetylation and tyrosine phosphorylation of STAT5. Sumoylation of STAT5 occurs on two lysine residues adjacent to the critical tyrosine moiety, where the first lysine residue can also be acetylated (Fig. 1). The authors present a model in which acetylation and sumoylation antagonistically control tyrosine phosphorylation of STAT5.
Acetylation of ε-NH2 residues of lysine moieties is modulated by the opposing enzymatic activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Zinc- or NAD-dependent deacetylation of STATs by HDACs 1, 2, 3 and 4, SIRT1 and acetyl-CoA-dependent acetylation of STATs by CBP, p300, GCN5 and PCAF were found in adherent and hematopoietic cells.5,21-24 Covalent attachment of a small ubiquitin-related modifier (SUMO) to lysine residues can also alter biological functions. The interplay between an activating enzyme (E1), a sole SUMO-conjugase UBC9 (E2) and SUMO-ligases (E3s) catalyzes sumoylation of target proteins. The various E3s promote this post-translational modification.25 SUMO-specific proteases (SENPs1–8, aka sentrin-specific proteases) remove SUMO in order to allow highly dynamic on/off sumoylation-desumoylation-cycles. In contrast to poly-ubiquitinylation, sumoylation does usually not culminate in proteasomal degradation. Sumoylation rather alters the complex formation, localization and activity of proteins.25,26
Van Nguyen and colleagues used a SENP1 KO primary cell system to analyze whether sumoylation has an impact on early T and B cell development.20 This idea was based on the fact that the hematopoietic phenotype of SENP1 KO mice was strikingly similar and overlapping to STAT5 deletion using either complete KO or conditional deletion of STAT5 in the lymphoid lineage via Lck-Cre conditional targeting.11,20,27,28 A putative defect on IL-7 signaling components in fetal livers of SENP1−/− animals was excluded since the lymphoid defects in these mice were more reminiscent of the phenotype of STAT5 null animals.14 Thus, the authors speculated that SENP1 may target sumoylation of STAT5, which was tested by immunoblotting whole cell lysates from lymphoid and myeloid cells. Depending on the number of sumoylation sites, shifts in increments of ~15 to 20 kDa can be observed, i.e., post-translational modification with SUMO causes slower migration of proteins in SDS-PAGE gels. STAT5 has a molecular weight of ~92 to 95 kDa on SDS Page gels. A ~40 kDa slower migrating band of STAT5 was detected when lysates from SENP1−/− B and T cells were analyzed with anti-STAT5A/B antibodies by immunoblotting.
To corroborate sumoylation of STAT5 in SENP1 null cells, STAT5 was precipitated with anti-STAT5A/B antibodies from B cell lysates. These lysates were prepared under stringent conditions to exclude that associated sumoylated proteins cause false-positive results. The obtained precipitates were analyzed with antibodies against SUMO1 or SUMO2/SUMO3. Western blot signals obtained with this method showed modification of STAT5A and STAT5B by SUMO2/SUMO3. Thus, Van Nguyen et al. discovered a SENP1-sensitive sumoylation of STAT5 in primary B cells. Overexpression experiments verified modification of STAT5A/B with SUMO2 and that catalytically active SENP1 removes this modification, which was enhanced by the RING E3 SUMO ligase PIAS3. Such experiments are important as they exclude the possibility that higher migrating bands are splice variants or cross-reactive species. Interestingly, one sumoylated STAT5 species was found in lymphoid cells, but two bands above the expected molecular weight of STAT5 were observed in ectopic expression experiments using STAT5 and SUMO2. Perhaps this finding indicates differential use of STAT5 sumoylation sites in different cell types, which awaits further investigation.
Apparently, T and B cells lacking SENP1 use both sumoylation sites in the STAT5 C-terminus, which explains the shift of ~40 kDa in SDS-PAGE. Sumoylation of STAT5 was not detected in myeloid cells,20 but as discussed above STAT5 activation is a key driver for myeloid neoplasm maintenance. Why myeloid cells do not display STAT5 sumoylation is enigmatic and needs clarification. The identification of STAT5 isoform-specific sumoylation was not clearly demonstrated in lymphocytes, but isoform-specific expression in COS-1 cells (that do not contain significant levels of endogenous STAT5 proteins) suggests that both STAT5A and STAT5B can be tagged with SUMO. The authors used a pan anti-STAT5A/B rabbit polyclonal antibody serum as stated in the supplementary information and this brings down the heterodimers of STAT5A/STAT5B allowing no further distinction for sumoylation for respective STAT5 isoforms. Nonetheless, the fact that expression of the STAT5 target gene Bcl2 was suppressed at mRNA and protein levels in SENP1−/− mice clearly suggests a biological impact of sumoylation on STAT5 signaling.
Sumoylation often occurs at lysine residues within the consensus sequence ΨKxE (Ψ, large hydrophobic amino acid; K, lysine subject to sumoylation; x, any amino acid; E, glutamic acid residue).25 However, mutation of such sites to arginine residues (which cannot be sumoylated due to their mesomerically stabilized guanidine groups) did not prevent sumoylation of overexpressed STAT5. The fact that STAT1 has a sumoylation site (K703) close to the phosphorylated tyrosine 70129 prompted Van Nguyen and colleagues to compare the STAT5 protein sequences from Homo sapiens, Mus musculus and Rattus norvegicus. They found that across species two STAT5A/STAT5B lysine residues adjacent to tyrosine 694 are conserved and sumoylated. Sumoylation of single (K696R or K700R) or double mutants (K696 and 700R) of STAT5A was drastically reduced. Thus, lysine residues 696 and 700 are the major sites for sumoylation of STAT5A. The major sumoylation sites in STAT5B are lysine residues 701 and 705. Their mutation significantly reduced conjugation of ectopically expressed STAT5A with SUMO2. Furthermore, phosphorylation at Y694 is required for the sumoylation of STAT5A and only the nuclear portion of STAT5 was found to be sumoylated in primary B cells. These data suggest that before becoming sumoylated, STAT5 must enter the nucleus as a phosphorylated protein. There is still the possibility that sumoylation of STAT5 happens only when bound to DNA or when present in the nucleus. Here, the STAT5 proteins are very important chromatin remodeling proteins through their N-termini and N-terminal deletion of STAT5 largely abolished lymphocyte functions and development.6,8 Thus, it will be interesting to see if an N-terminally truncated STAT5 molecule can still be sumoylated at the C-terminus or if DNA binding defective mutants also allow for sumoylation. Clearly, STAT5 tyrosine phosphorylation is a prerequisite for sumoylation and it remains to be clarified how fast this takes place after nuclear translocation. Does sumoylation take place immediately when STAT5 enters the nucleus, and does it thereby immediately prevent all transcriptional activity of STAT5? Inducible expression of SENP1, e.g., by using SENP1-ER fusions, might allow such studies.
STAT1 is sumoylated at K703 and mutations impairing sumoylation of STAT1 hardly alter STAT1-dependent transcriptional and antiviral activity or cause no measurable effect on STAT1 activity.29 In contrast, STAT5 was not tyrosine phosphorylated in T and B cells derived from SENP1 null HSCs treated for long-term with IL-7 and FLT3 ligand. However, total STAT5 protein levels were omitted allowing no clear cut answer to the data shown. Sumoylation often targets only a minor pool of a protein species to serve as a transient modification allowing sumoylation-independent persistent changes.25,26 The paper by Van Nguyen et al. hence suggests that a low percentage of sumoylated nuclear STAT5 has a broader impact on the entire STAT5 protein pool in lymphocytes. This situation is reminiscent of the acetylation-dependent inactivation of STAT1 by a limited number of acetylated STAT1 molecules binding the T cell protein phosphatase.5,21,23,24 An alternative scenario could be that the JAK kinase activity in hematopoietic progenitor cells is affected by SENP1 or that tyrosine phosphatase action is increased upon sumoylation. It hence remains puzzling why only part of the total pool of available STAT5 appears to become sumoylation and what the physiological meaning of this observation is. Perhaps, specific phosphatases are recruited upon modification with SUMO.
Acetylation of STAT5B at residues K359, K694 and K701 was first reported in 2010.30 These authors revealed an acetylation-dependent dimerization of endogenous STAT5 in prolactin-treated breast cancer cells. Van Nguyen et al. hypothesized that sumoylation and acetylation may have opposing effects on the tyrosine phosphorylation of STAT5.20 In contrast to STAT1 which is acetylated by the HAT CBP and not by the HATs p300, GCN5 and PCAF5 overexpressed CBP, p300 and to a lesser extent GCN5 and PCAF also enhance STAT5 acetylation.30 Van Nguyen focused on p300 and noted that acetylation of STAT5A K696R by p300 could not be detected with pan-anti-acetyllysine antibodies. Thus, this site appears as the major site for sumoylation and acetylation of STAT5A. Furthermore, endogenous STAT5B was found to be less acetylated at K701 in SENP1 null cells when immunoblots were performed with a STAT5B AcK701-specific antibody.20 These findings and additional overexpression studies involving wild-type and mutant SENP1 argue for sumoylation as a negative regulator of lysine acetylation and tyrosine phosphorylation (Fig. 1).
It will also be interesting to see the outcome of a STAT5A K→R (not acetylatable) or K→Q (mimics acetylation) knock-in approach or add back experiments with STAT5A or STAT5B K→R or K→Q variants in STAT5-deficient primary lymphocytes. More stringent physiologic tests for the role of STAT5 acetylation or sumoylation have to be performed in specific cell types, where STAT5 was shown to be sumoylated and where it is known to control proliferation, differentiation and survival. Further exciting questions are how sumoylation of STAT5 is achieved and which upstream signals dictate these modifications. Most experiments reveal that sumoylation of STAT5 cannot be detected in wild-type animals. It hence seems that SENP1 is expressed in lymphocytes to keep sumoylation of STAT5 low and to allow acetylation and phosphorylation of STAT5 at a physiologic rate. Additional work needs to reveal which further proteins are desumoylated by SENP1 and how this affects hematopoiesis. Furthermore, one wonders whether overexpression of SENP1 would result in higher STAT5 activity, and whether misexpression of SENP1 is observed in lymphoid malignancies.
JAK-STAT signaling is a central pathway that controls normal functions such as hematopoiesis or immunity, but it is also a core pathway in many different cancer types. While a large body of evidence about linear functions of post-translational modifications controlling STAT-dependent pathways has accumulated, it is time to define how these interact with each other. Increasing evidence for the existence of phosphorylation-acetylation switches controlling STAT proteins has accumulated.5,20,23,24 In summary, the work discussed here provides new and important insights for STAT5 as a key regulator of lymphoid development modified by SUMO2 and SENP1. In the absence of SENP1, SUMO2-modified STAT5 accumulates in early lymphoid precursors, resulting in a block in STAT5 acetylation and impaired early lymphoid development.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/21232
References
- 1.Liongue C, O’Sullivan LA, Trengove MC, Ward AC. Evolution of JAK-STAT pathway components: mechanisms and role in immune system development. PLoS One. 2012;7:e32777. doi: 10.1371/journal.pone.0032777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison DA. The Jak/STAT pathway. Cold Spring Harb Perspect Biol. 2012;4:a011205. doi: 10.1101/cshperspect.a011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ungureanu D, Wu J, Pekkala T, Niranjan Y, Young C, Jensen ON, et al. The pseudokinase domain of JAK2 is a dual-specificity protein kinase that negatively regulates cytokine signaling. Nat Struct Mol Biol. 2011;18:971–6. doi: 10.1038/nsmb.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kiu H, Nicholson SE. Biology and significance of the JAK/STAT signalling pathways. Growth Factors. 2012;30:88–106. doi: 10.3109/08977194.2012.660936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krämer OH, Heinzel T. Phosphorylation-acetylation switch in the regulation of STAT1 signaling. Mol Cell Endocrinol. 2010;315:40–8. doi: 10.1016/j.mce.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 6.Friedbichler K, Hoelbl A, Li G, Bunting KD, Sexl V, Gouilleux F, et al. Serine phosphorylation of the Stat5a C-terminus is a driving force for transformation. Front Biosci. 2012;17:3043–56. doi: 10.2741/3897. [DOI] [PubMed] [Google Scholar]
- 7.Stark GR, Darnell JE., Jr The JAK-STAT pathway at twenty. Immunity. 2012;36:503–14. doi: 10.1016/j.immuni.2012.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferbeyre G, Moriggl R. The role of Stat5 transcription factors as tumor suppressors or oncogenes. Biochim Biophys Acta. 2011;1815:104–14. doi: 10.1016/j.bbcan.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Kotecha N, Flores NJ, Irish JM, Simonds EF, Sakai DS, Archambeault S, et al. Single-cell profiling identifies aberrant STAT5 activation in myeloid malignancies with specific clinical and biologic correlates. Cancer Cell. 2008;14:335–43. doi: 10.1016/j.ccr.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Page BD, Khoury H, Laister RC, Fletcher S, Vellozo M, Manzoli A, et al. Small molecule STAT5-SH2 domain inhibitors exhibit potent antileukemia activity. J Med Chem. 2012;55:1047–55. doi: 10.1021/jm200720n. [DOI] [PubMed] [Google Scholar]
- 11.Hoelbl A, Schuster C, Kovacic B, Zhu B, Wickre M, Hoelzl MA, et al. Stat5 is indispensable for the maintenance of bcr/abl-positive leukaemia. EMBO Mol Med. 2010;2:98–110. doi: 10.1002/emmm.201000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan D, Hutchison RE, Mohi G. Critical requirement for Stat5 in a mouse model of polycythemia vera. Blood. 2012;119:3539–49. doi: 10.1182/blood-2011-03-345215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warsch W, Kollmann K, Eckelhart E, Fajmann S, Cerny-Reiterer S, Hölbl A, et al. High STAT5 levels mediate imatinib resistance and indicate disease progression in chronic myeloid leukemia. Blood. 2011;117:3409–20. doi: 10.1182/blood-2009-10-248211. [DOI] [PubMed] [Google Scholar]
- 14.Friedbichler K, Kerenyi MA, Kovacic B, Li G, Hoelbl A, Yahiaoui S, et al. Stat5a serine 725 and 779 phosphorylation is a prerequisite for hematopoietic transformation. Blood. 2010;116:1548–58. doi: 10.1182/blood-2009-12-258913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G, Miskimen KL, Wang Z, Xie XY, Brenzovich J, Ryan JJ, et al. STAT5 requires the N-domain for suppression of miR15/16, induction of bcl-2, and survival signaling in myeloproliferative disease. Blood. 2010;115:1416–24. doi: 10.1182/blood-2009-07-234963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baumgartner C, Cerny-Reiterer S, Sonneck K, Mayerhofer M, Gleixner KV, Fritz R, et al. Expression of activated STAT5 in neoplastic mast cells in systemic mastocytosis: subcellular distribution and role of the transforming oncoprotein KIT D816V. Am J Pathol. 2009;175:2416–29. doi: 10.2353/ajpath.2009.080953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harir N, Boudot C, Friedbichler K, Sonneck K, Kondo R, Martin-Lannerée S, et al. Oncogenic Kit controls neoplastic mast cell growth through a Stat5/PI3-kinase signaling cascade. Blood. 2008;112:2463–73. doi: 10.1182/blood-2007-09-115477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandal M, Powers SE, Maienschein-Cline M, Bartom ET, Hamel KM, Kee BL, et al. Epigenetic repression of the Igk locus by STAT5-mediated recruitment of the histone methyltransferase Ezh2. Nat Immunol. 2011;12:1212–20. doi: 10.1038/ni.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Li G, Tse W, Bunting KD. Conditional deletion of STAT5 in adult mouse hematopoietic stem cells causes loss of quiescence and permits efficient nonablative stem cell replacement. Blood. 2009;113:4856–65. doi: 10.1182/blood-2008-09-181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Nguyen T, Angkasekwinai P, Dou H, Lin FM, Lu LS, Cheng J, et al. SUMO-specific protease 1 is critical for early lymphoid development through regulation of STAT5 activation. Mol Cell. 2012;45:210–21. doi: 10.1016/j.molcel.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krämer OH, Knauer SK, Greiner G, Jandt E, Reichardt S, Gührs KH, et al. A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 2009;23:223–35. doi: 10.1101/gad.479209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spange S, Wagner T, Heinzel T, Krämer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–98. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 23.Ginter T, Bier C, Knauer SK, Sughra K, Hildebrand D, Münz T, et al. Histone deacetylase inhibitors block IFNγ-induced STAT1 phosphorylation. Cell Signal. 2012;24:1453–60. doi: 10.1016/j.cellsig.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Cudejko C, Wouters K, Fuentes L, Hannou SA, Paquet C, Bantubungi K, et al. p16INK4a deficiency promotes IL-4-induced polarization and inhibits proinflammatory signaling in macrophages. Blood. 2011;118:2556–66. doi: 10.1182/blood-2010-10-313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–56. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 26.Brandl A, Wagner T, Uhlig KM, Knauer SK, Stauber RH, Melchior F, et al. Dynamically regulated sumoylation of HDAC2 controls p53 deacetylation and restricts apoptosis following genotoxic stress. J Mol Cell Biol. 2012 doi: 10.1093/jmcb/mjs013. [DOI] [PubMed] [Google Scholar]
- 27.Hoelbl A, Kovacic B, Kerenyi MA, Simma O, Warsch W, Cui Y, et al. Clarifying the role of Stat5 in lymphoid development and Abelson-induced transformation. Blood. 2006;107:4898–906. doi: 10.1182/blood-2005-09-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–75. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song L, Bhattacharya S, Yunus AA, Lima CD, Schindler C. Stat1 and SUMO modification. Blood. 2006;108:3237–44. doi: 10.1182/blood-2006-04-020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma L, Gao JS, Guan Y, Shi X, Zhang H, Ayrapetov MK, et al. Acetylation modulates prolactin receptor dimerization. Proc Natl Acad Sci U S A. 2010;107:19314–9. doi: 10.1073/pnas.1010253107. [DOI] [PMC free article] [PubMed] [Google Scholar]