Abstract
MicroRNA-21 (miR-21) is overexpressed virtually in all human cancers and displays oncogenic activity in a transgenic murine model. Similarly, the p53 tumor suppressor gene is the most frequently mutated gene in human cancer, and its loss or mutation leads to tumor formation in mice. To ascertain the role of miR-21 in the p53 pathway in vivo and to characterize their interaction in tumorigenesis, we intercrossed the miR-21 −/− and Trp53 −/− mice. We found that Trp53 −/− miR-21 −/−mice develop tumors at a slightly later age, yet show a similar tumor spectrum and survival curve as Trp53 −/− mice. When subjected to genotoxic agents, tissues from Trp53 −/− miR-21 −/− mice have a higher percentage of apoptotic cells. We extracted mouse embryonic fibroblast cells (MEFs) to examine the impact of miR-21 loss on p53-regulated cellular processes in Trp53 −/− cells. Higher cellular apoptosis and senescence were found in Trp53 −/− miR-21 −/− MEFs than in Trp53 −/− MEFs. In addition, loss of miR-21 sensitizes transformed Trp53 −/− cells to DNA damage-induced apoptosis through elevation of Pten expression. These data suggest that inhibition of miR-21 would be beneficial in apoptosis-inducing cancer therapies directed against p53-deficient tumors.
Introduction
One of the most frequently mutated genes in human cancer is the tumor suppressor p53 gene, which is widely regarded as the ‘guardian of the genome’ (1). p53 is a transcription factor that regulates the expression of a battery of genes that control cell cycle progression and apoptosis in response to DNA damage or cellular stress. There are two homologs of p53: p63 (TP73L) and p73 (TP73), which have a similar tumor suppressor activity as p53 (2). Mice lacking p53 (also known as Trp53) are prone to spontaneous tumor formation and develop lymphoma and sarcoma with a short latency (3,4). Restoring p53 function alone is sufficient to cause regression of several different tumors in mice with Trp53 deletion (5–7), supporting the possibility of p53 as a therapeutic target. However, over 30 years after the discovery of p53, no effective anticancer treatment based on p53 has yet been developed.
MicroRNAs (miRs) suppress their target genes through binding the 3′ untranslated regions of messenger RNAs. Several miRs were reported to be involved in the p53 pathway, either regulated by p53 or acting directly to repress the expression of p53 or its downstream effectors, suggesting the importance of miRs in the p53 pathway (8–14). miR-21 is overexpressed in virtually all solid tumors (15–17), and recent studies have also shown that miR-21 is highly expressed in hematological malignancies such as leukemia (18), lymphoma (19) and multiple myeloma (20). Several studies using cell lines reveal that knockdown of miR-21 results in reduced tumor cell growth (21), cell cycle arrest (22) and cell apoptosis (23). Three independent groups corroborated the role of miR-21 in carcinogenesis using genetically engineered mouse models. Overexpression of miR-21 leads to pre-B malignant lymphoid-like phenotype (24) and enhances Kras-driven lung tumorigenesis (25), whereas genetic deletion of miR-21 partially reduced the incidence and growth of Kras-driven lung tumors (25) and genotoxic carcinogen-induced skin carcinogenesis (26). Collectively, these data suggest that miR-21 is an oncomiR (an oncogenic miR).
There is no miR-21 binding site in the p53 3′ untranslated region in either human or mouse, yet there are several reports demonstrating that miR-21 represses the expression of multiple genes in the p53 network. Downregulation of miR-21 in glioblastoma cells leads to elevated expression of p63 and activators of the p53 pathway, including JMY, TOPORS, TP53BP2 and DAXX, causing cell cycle arrest and increasing apoptosis (27). In breast cancer cells, miR-21 inhibition induces the expression of several genes regulated by p53 at the messenger RNA level, including FAM3C, ACTA2, APAF1, BTG2, FAS, CDKN1A(p21) and SESN1 (28). These genes are required for p53 activity, suggesting that miR-21 overexpression could impair the tumor-suppressive function of the p53 pathway. A recent study reports that miR-21 expression is elevated in human lung tumors bearing mutated p53 and displaying distant metastases and that augmented expression of miR-21 confers increased invasive properties in Trp53-deficient mouse tumors (29). However, the functional interaction between p53 and miR-21 in vivo has remained elusive. In this study, we intercrossed miR-21 −/− and Trp53 −/− mice to study the interplay between miR-21 and the p53 pathway in tumorigenesis. In addition, we used cultured mouse embryonic fibroblast cells (MEFs) to characterize the functional overlap between miR-21 and the p53 pathway. This work deepens our understanding of the association between a critical oncomiR and the p53 tumor suppressor network and offers new strategies for cancer therapy.
Materials and methods
Generation of Trp53−/−miR-21−/− mice
Generation of miR-21 −/− mice in the C57BL/6×129S hybrid background has been described (26) and these mice were crossed with C57BL/6J twice. Trp53 −/− mice (B6.129S2-Trp53 tm1Tyj/J) in C57BL/6J background were purchased from Jackson laboratory (Bar Harbor, ME). Trp53 +/+ miR-21 −/− females were crossed with Trp53 −/− miR-21 +/+male mice to generate Trp53 +/– miR-21 +/– mice. And Trp53 +/– miR-21 +/– were then bred with Trp53 +/+ miR-21 −/− to generate Trp5 +/–miR-21 −/− mice, which were intercrossed to generate Trp53 −/− miR-21 −/−. It was estimated that all mice had >90% C57BL/6 genetic background (C57BL/6×129S hybrid background with three backcross). All experimental procedures involving animals in this study were approved by the institutional animal care and use committee at the University of Louisville.
MEF preparation
MEFs were prepared according to an established protocol (30). MEFs were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% non-essential amino acids and incubated at 37°C in a 5% CO2 atmosphere. To measure cell growth, MEFs were seeded at 1×105 cell density, which was designated as day 1 (d1) and cells were counted at days 4, 6, 8 and 10.
Flow cytometry
MEFs were plated in six-well plates with the density of 5×105 cells per well and cultured in medium until 80% confluence is obtained. For cell cycle analysis, MEFs were synchronized by growing in DMEM supplemented with 10% FBS for 3 days and then in DMEM supplemented with 0.1% FBS for 4 days. Cells were split into six-well plates with the density of 5×105 cells per well in DMEM supplemented with 10% FBS. After 6h, MEFs were treated with 0.2 µg/ml doxorubicin. Cells were collected 12h later and washed twice with phosphate-buffered saline (PBS) containing 0.1% Tween-20 and resuspended in ice-cold 70% ethanol. After ethanol fixation, cells were washed with PBS, and then incubated with propidium iodide solution (50 µg/ml propidium iodide and 10 µg/ml ribonuclease A in PBS). After 15min of propidium iodide labeling, MEFs were subjected to flow cytometry and analyzed using the FlowJo software.
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling assay
Wild-type (WT), miR-21 −/−, Trp53 −/− and Trp53 −/− miR-21 −/− male or female mice (8–10 weeks of age, 6 mice per group) were used for in vivo apoptosis study. Doxorubicin was dissolved in dimethyl sulfoxide at 10mg/ml and administered intraperitoneally at 5mg/kg. After 24h, mice were killed. Spleens were collected and fixed with 10% neutral-buffered formalin. Deparaffinized sections were incubated with 20 µg/ml protease K for 15min at room temperature, and then washed with PBS and incubated with terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) reaction mixture (Roche Diagnostics, Indianapolis, IN) for 60min at 37°C in humidified atmosphere. For MEF apoptosis analysis, cells were treated with 1 µmol/l (0.58 µg/ml) doxorubicin; cells were collected and fixed with 4% paraformaldehyde, and then incubated with 3% H2O2, 0.1% Triton X-100 and TUNEL reaction mixture. Tissue sections or fixed cells were finally incubated with converter-POD (with antifluorescein antibody) and subsequently with the diaminobenzidene substrate. Images were acquired using Olympus IX51 microscope and cellSens Dimension software. Nuclei with condensed or fragmented chromatin (brown color) were considered apoptotic death; at least 5000 cells were counted under microscopy with 20× objective lens in each group.
Cell senescence
MEFs were split into six-well plates with a density of 1×105 cells every passage. And at passage 2 (P2), P8 and P12, MEFs were collected and senescence was determined by measuring β-galactosidase with Senescence β-Galactosidase Staining Kit (Cell Signaling, Danvers, MA) according to the manufacturer’s protocols.
Cell transformation and focus-forming assay
P2 MEFs were infected with a retroviral vector pBabe H-Ras (#1768; Addgene, Cambridge, MA) containing an activated H-rasV12 complementary DNA and puromycin resistance gene. After 2 µg/ml puromycin selection, 2000 cells were cultured in six-well plates. On day 21, the cells were fixed with 70% ethanol and stained with crystal violet. The foci were counted under microscopy with 4× objective lens.
Western blotting
Cells were lysed with RIPA buffer (Cell Signaling) supplemented with protease inhibitor (Calbiochem, San Diego, CA). Soluble proteins were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis before transferred to polyvinylidene difluoride membranes. The following antibodies Pdcd4 (D29C6, 1:1000), PTEN (D4.3, 1:1000), Bax (1:1000), Puma (1:1000) and Caspase-3 (1:1000) purchased from Cell Signaling and p63 (ab53039, 1:1000), p73 (ab22045, 1:1000), p21 (ab7960, 1:1000), Spry2 (ab50317, 1:2400), FasL (ab68338, 1:500), Btg2 (ab80322, 1:500), RhoB (ab68827, 1:1000) and Peli1 (ab13812, 1:1000) obtained from Abcam (Cambridge, MA) were used. β-actin primary antibody was purchased from Sigma–Aldrich (St Louis, MO).
Statistical analysis
Average data were expressed as mean and standard deviation. Statistical analysis was carried out using the SPSS16.0 software (SPSS, Chicago, IL). The one-way analysis of variance test was performed and statistical significance was set at *P < 0.05 or **P < 0.01.
Results
Generation of Trp53−/−miR-21−/− mice
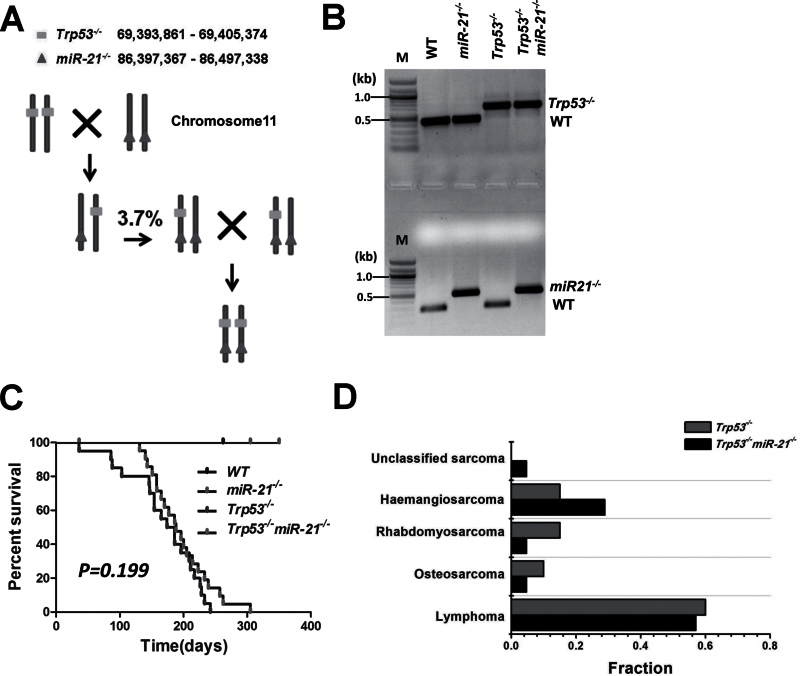
To study the interaction between miR-21 and p53 in tumorigenesis, we intercrossed miR-21 −/− and Trp53 −/− mice to get Trp53 −/− miR-21 −/− mice. The miR-21 gene, located within the 3′ untranslated region of the vacuole membrane protein 1 gene (VMP1), resides on the same chromosome 11 as Trp53. Thus, chromosome crossover during meiosis is needed to generate a recombinant chromosome containing both mutant loci. After screening 54 offspring of the Trp53 +/– miR-21 +/– and miR-21 −/− intercross, we obtained two Trp53 +/– miR-21 −/− mice, which suggests a frequency of ~3.7% for generation of a recombinant chromosome with both mutant alleles in cis (Trp53 and miR-21 are ~17Mb apart). This is comparable with the frequency (~1%) of generating a recombinant chromosome containing the DPH1 (Ovca1-2) and Trp53 alleles (~5.6Mb apart (31)). Trp53 +/– miR-21 −/− mice were then intercrossed to generate Trp53 −/− miR-21 −/− mice (Figure 1A), and PCR amplification was used to verify the genotypes (Figure 1B). Trp53 −/− miR-21 −/− mice were born at the predicted Mendelian frequency, indicating that there is no embryonic lethality due to concomitant loss of both miR-21 and Trp53.
Fig. 1.
Generation of transgenic mice, survival curves and tumor spectra of Trp53 −/− and Trp53 −/− miR-21 −/− mice. (A) The locations of Trp53 and miR-21 on chromosome 11 and the breeding scheme to generate Trp53 −/− miR-21 −/− mice. (B) Genotyping PCR results for each genotype. M: DNA molecular markers. (C) Survival curves of WT (n = 20), miR-21 −/− (n = 20), Trp53 −/− (n = 20) and Trp53 −/− miR-21 −/− (n = 21) mice. (D) Tumor spectrum in Trp53 −/− and Trp53 −/− miR-21 −/− mice.
Analysis of survival and spontaneous tumor development
Mice with the four genotypes were maintained to determine spontaneous tumor development and survival. WT and miR-21 −/− mice were monitored for over 18 months and no tumors were found. The average life span of Trp53 −/− mice was previously reported to be ~160 days (32) and was 169 days in our animal cohort. We found that the average survival for Trp53 −/− miR-21 −/− mice was longer (195 days), but this was not statistically significant (P = 0.199; Figure 1C). We did note that three Trp53 −/− mice died within 3 months, yet no Trp53 −/− miR-21 −/− mice died within the same period. Histological analysis showed that the tumor spectra of Trp53 −/− and Trp53 −/− miR-21 −/− mice were similar, with similar frequencies for primarily lymphoid tumors (60% compared with 57%) and soft tissue sarcoma (40% compared with 43%; Figure 1D and Supplementary Table W1, available at Carcinogenesis Online).
Cell proliferation potential, senescence and cell cycle of MEFs
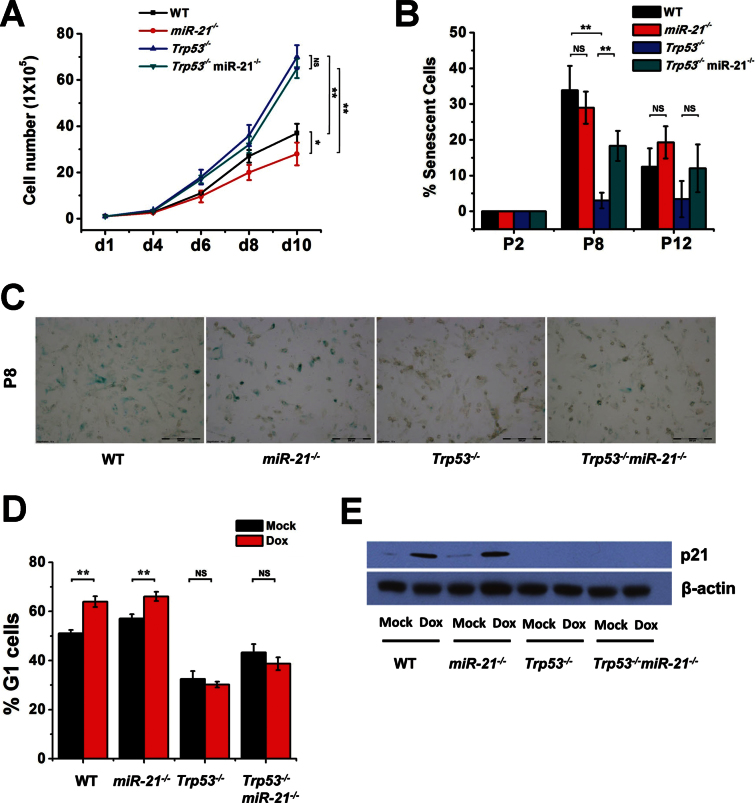
A previous study showed that Trp53 −/− MEFs grow faster than WT MEFs (33), so we compared the growth rates of MEFs with each genotype to determine whether deletion of miR-21 impacts MEF growth with or without p53. P2 MEFs were seeded in six-well plates and cell numbers were counted until day 10. After 4 days, Trp53 −/− and Trp53 −/− miR-21 −/− MEFs started to grow more rapidly than WT and miR-21 −/− MEFs, whereas WT cells grew faster than miR-21 −/− MEFs. Trp53 −/− miR-21 −/− MEFs did not show significant differences in growth rate compared with the Trp53 −/− MEFs (Figure 2A). These data suggest that deletion of miR-21 does not affect cell proliferation of Trp53 −/− MEFs, yet miR-21 loss did reduce cell proliferation of WT MEFs.
Fig. 2.
Proliferation, senescence and cell cycle in MEFs. (A) Cell proliferation curves of MEFs. WT, miR-21 −/−, Trp53 −/− and Trp53 −/− miR-21 −/− MEFs (P2) were plated on day 1 and counted at the indicated times. (B) Bar graphs of cell senescence of MEFs (WT, miR-21 −/−, Trp53 −/− and Trp53 −/− miR-21 −/−), which were stained with senescence β-galactosidase at the indicated passages and positive-staining cells were counted. (C) Representative images of cell senescence from (B). (D) Cell cycle analysis of MEFs treated with doxorubicin. (E) Western blot analysis of p21 expression in MEFs treated with doxorubicin. All experiments were performed using two or more independent MEF preparations. *P < 0.05; **P < 0.01; NS, not significant.
p53 plays a crucial role in cellular senescence, which is a permanent form of cell cycle arrest in primary cultured cells. Unlike WT MEFs, MEFs lacking p53 do not undergo cellular senescence (34). We measured spontaneous senescence of MEFs at different passages using β-galactosidase staining. As expected, there was no senescence of Trp53 −/− MEFs from P8 to P12. At P8, miR-21 −/− MEFs underwent senescence at a level similar to WT cells. Loss of miR-21 led to obvious senescence of Trp53 −/− MEFs at P8, yet by P12, cell senescence was virtually undetectable in both Trp53 −/− and Trp53 −/− miR-21 −/− MEFs (Figure 2B and C).
We next evaluated transient cell cycle arrest of MEFs in response to doxorubicin. Doxorubicin induced G1 arrest in WT MEFs, but failed to do so in Trp53 −/− MEFs (Figure 2D), which is in agreement with a previous study (35). There was no significant difference in G1 arrest between miR-21 −/− and WT MEFs either when untreated or upon doxorubicin treatment. Similarly, Trp53 −/− miR-21 −/− MEFs showed no significant difference in G1 arrest compared with Trp53 −/− MEFs. The p53 transactivational target p21 was not expressed in Trp53 −/− miR-21 −/− or Trp53 −/− MEFs, and its expression and induction were comparable in WT and miR-21 −/− MEFs (Figure 2E). These data suggest that miR-21 did not significantly impact G1 cell arrest induced by DNA damage.
Apoptosis in vivo and in vitro
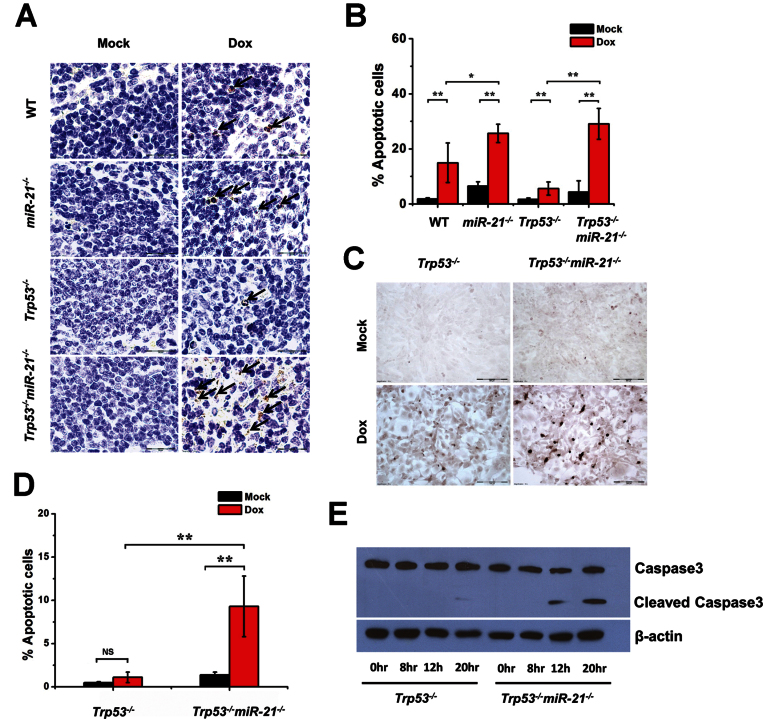
p53 plays a key role in antilymphomagenesis through apoptosis (36), and miR-21 performs its oncomiR function by inhibiting apoptosis (24). We determined the frequency of cellular apoptosis in mouse tissues responding to DNA damage. Spleen is one of the most sensitive organs to DNA damage-induced cell apoptosis (37). Mice were treated with doxorubicin for 24h and apoptosis in the spleen was analyzed using TUNEL. For each group, more apoptotic cells were found in the spleens of mice receiving doxorubicin treatment than in untreated mice (Figure 3A and B). As expected, a low level of apoptosis was observed in the spleens of p53-null mice upon treatment than in other groups. There were more apoptotic cells in miR-21 −/− spleens with or without doxorubicin treatment than in WT spleens. Without DNA damage (no treatment), there were few apoptotic cells observed in the spleens of Trp53 −/− miR-21 −/− mice; however, more apoptotic cells were found upon doxorubicin treatment than in their Trp53 −/− counterparts (Figure 3A and B). These in vivo data support the notion that miR-21 deletion sensitizes cells to apoptosis induced by DNA-damaging agents, either in the presence or in the absence of p53.
Fig. 3.
Apoptosis analysis of miR-21-null tissues and MEFs. (A) Representative images from the TUNEL assay for spleens from mice injected with doxorubicin. (B) Bar graphs of TUNEL assays quantifying the numbers of apoptotic cells in the spleen after treatment with doxorubicin in WT, miR-21 −/−, Trp53 −/− and Trp53 −/− miR-21 −/− mice (n = 6 per group). (C) Representative images from the TUNEL assay on Trp53 −/− and Trp53 −/− miR-21 −/− MEFs treated with doxorubicin. (D) Bar graph quantification of the frequency of apoptotic nuclei in MEFs from (C). (E) Western blot analysis of caspase 3 in Trp53 −/− and Trp53 −/− miR-21 −/− MEFs treated with doxorubicin. *P < 0.05; **P < 0.01; NS, not significant.
We next examined the apoptosis response using MEFs. We found more apoptotic cells for Trp53 −/− miR-21 −/− than for Trp53 −/− MEFs treated with doxorubicin (Figure 3C and D). This increasing apoptosis was accompanied by more caspase-3 cleavage products in Trp53 −/− miR-21 −/− MEFs upon DNA damage than in Trp53 −/− MEFs (Figure 3E). Collectively, these data suggest that miR-21 has an antiapoptosis role in cells lacking p53, which is in line with previous reports that miR-21 deletion increases cellular apoptosis in cells with the WT p53 gene (24–26).
Deletion of miR-21 inhibits Ras-induced transformation in p53-null MEFs
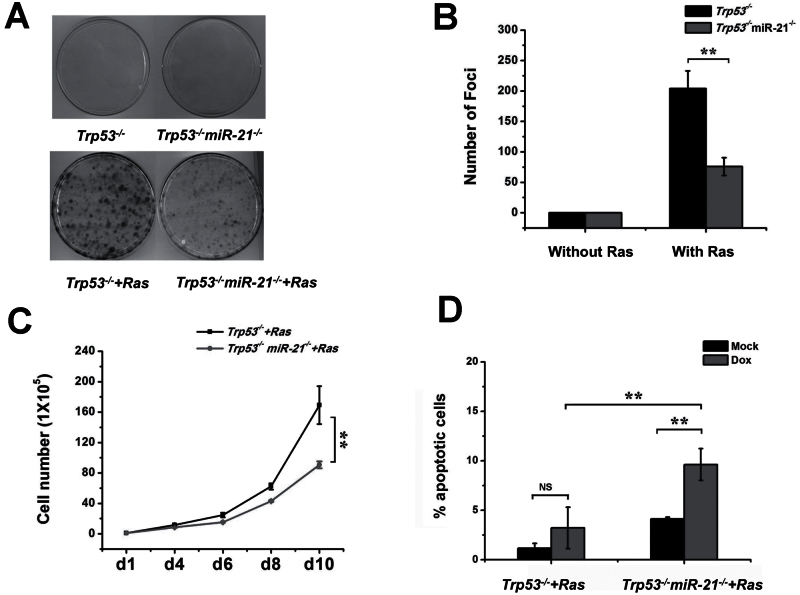
MEFs lacking p53 have been reported to evade Ras-induced senescence (38), whereas miR-21 deletion suppresses SV40LT and Ras-driven transformation (25). We next explored the role of miR-21 in Ras-driven transformation in the absence of p53. We introduced a retroviral vector, pBabe H-Ras, which contains an activated H-rasV12 complementary DNA, into Trp53 −/− and Trp53 −/− miR-21 −/− MEFs. Trp53 −/− cells with Ras formed more foci than Trp53 −/− miR-21 −/− cells with Ras, whereas Trp53 −/− and Trp53 −/− miR-21 −/− MEFs induced with the vector control showed no foci (Figure 4A and B). In addition, Trp53 −/− miR-21 −/− cells transformed with Ras showed slower cell proliferation than the Trp53 −/− cells (Figure 4C). These findings support the idea that miR-21 deficiency inhibits Ras-driven cell transformation in Trp53 −/− MEFs. Next, we analyzed the interplay between p53 and miR-21 in apoptosis of transformed cells. When Trp53 −/− cells and Trp53 −/− miR-21 −/− cells with Ras were exposed to doxorubicin, we observed more apoptosis in Ras-transformed Trp53 −/− miR-21 −/− cells than in Ras-transformed Trp53 −/− cells, confirming that miR-21 loss sensitizes cells with p53 depletion and Ras activation to apoptosis (Figure 5D).
Fig. 4.
Transformation of miR-21-null MEFs with or without p53. (A) A focus formation assay of MEFs transformed with Ras activation. Low passages of MEFs were transduced with a virus carrying H-rasV12. Foci were stained after 21 days of growth. (B) Bar graphs of cell transformation from (A). (C) Cell proliferation of Trp53 −/− and Trp53 −/− miR-21 −/− MEFs transformed by Ras activation. (D) Apoptosis analysis of Trp53 −/− and Trp53 −/− miR-21 −/− MEFs transformed with Ras were treated with doxorubicin. Apoptotic cells were analyzed by TUNEL assay and counted by microscopy. **P < 0.01.
Fig. 5.
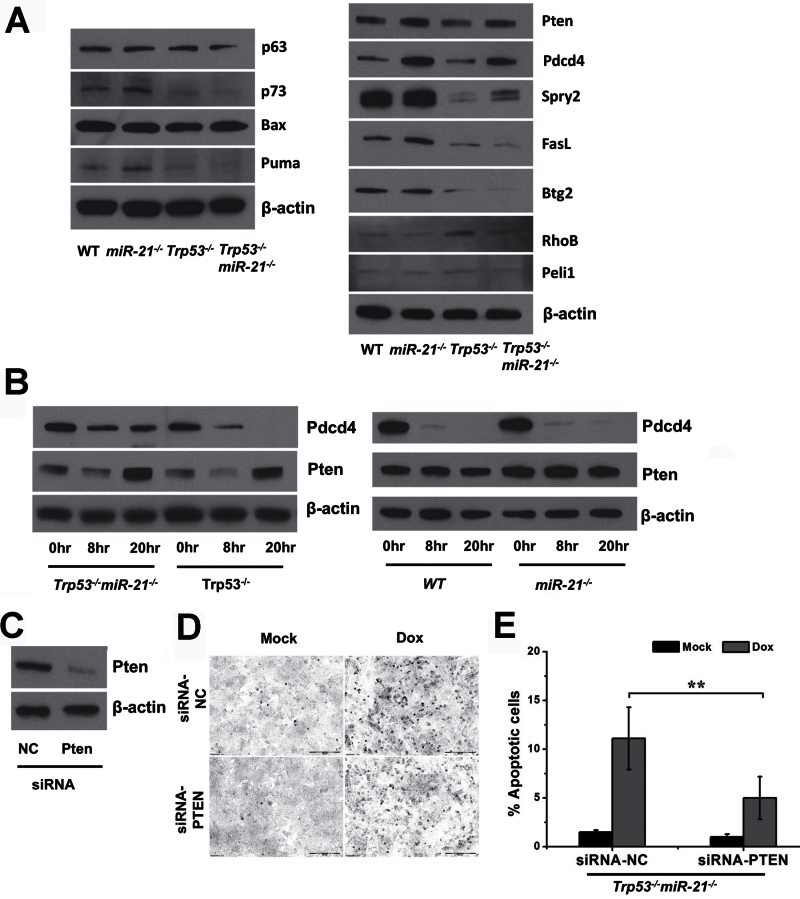
Pten involvement in p53-independent apoptosis. (A) Western blot analysis of genes in the p53 pathway and target genes of miR-21. (B) Western blot analysis of Pdcd4 and Pten in MEFs treated with doxorubicin. (C) Downregulation of Pten with siRNAs in Trp53 −/− miR-21 −/− MEFs transformed with Ras. (D) The TUNEL assay on Trp53 −/− miR-21 −/− cells transfected with siRNAs against PTEN and treated with doxorubicin. (E) Bar graphs of the percentage of apoptotic cells induced by doxorubicin in (D). **P < 0.01.
The convergence of miR-21 with the p53 pathway in apoptosis
To further explore the interplay between p53 and miR-21 in apoptosis, we analyzed the expression of p53 transactivational genes Puma and Bax (which are well known to participate in apoptosis), p63 and p73 (which are homologs of the p53 protein) and several reported miR-21 target genes. Although we did observe that p73 and Puma were downregulated when p53 was deleted, miR-21 ablation did not appear to affect the expression of genes in the p53 pathway (left panel, Figure 5A). Among miR-21 targets, Pten and Pdcd4 were upregulated upon miR-21 deletion, regardless of p53 status (right panel, Figure 5A).
Next, we treated MEFs with doxorubicin and analyzed the protein levels of Pten and Pdcd4. We found that Pdcd4 expression levels were higher in Trp53 −/− miR-21 −/− than in Trp53 −/− cells whether untreated or with treatment (left panel, Figure 5B). However, lower levels of Pdcd4 were found in Trp53 −/− miR-21 −/− cells treated with doxorubicin than in untreated cells. Similar to Pdcd4, Pten protein levels were higher in Trp53 −/− miR-21 −/− cells than in Trp53 −/− cells with or without treatment. We noted that Pten was expressed at a slightly lower level at the 8h time point, and its expression was at the highest level when Trp53 −/− miR-21 −/− cells were treated with doxorubicin for 20h (left panel, Figure 5B). In the presence of p53, Pten levels changed little, whereas Pdcd4 levels were downregulated significantly in WT and miR-21 −/− cells upon doxorubicin treatment (right panel, Figure 5B). These data suggest that Pten is the likely miR-21 target that participates in changes in apoptosis, as its expression levels follow that of cellular apoptosis when p53 is absent. Finally, we inhibited Pten expression in Trp53 −/− miR-21 −/− cells using small interfering RNA (siRNAs) (Figure 5C) to determine whether reduced Pten expression would rescue the increased apoptosis upon miR-21 loss. We found fewer apoptotic cells when Pten was downregulated (Figure 5D and E), which confirms the critical role of Pten in linking miR-21 with p53-independent apoptosis.
Discussion
Recent clinical and laboratory studies have implicated miR-21 in the pathogenesis of various diseases, especially in many cancers (39). miR-21 overexpression is observed in most, if not all, types of carcinomas and hematological malignancies, and it is considered to be one of the most promising oncomiRs for targeted cancer therapy. p53 is a major tumor suppressor in cancer, as it is mutated in about half of all human malignancies. In this study, we have investigated whether miR-21 plays a role in mouse tumorigenesis mediated by loss of p53. We found that there is no significant difference in animal survival and tumor spectrum between Trp53 −/− and Trp53 −/− miR-21 −/− mice (P = 0.199), which are comparable with Kras-induced lung tumorigenesis (25). In that model, miR-21 overexpression causes more tumors and miR-21 deletion leads to fewer tumors in the lung, yet miR-21 overexpression does not significantly impact animal survival (P = 0.357). miR-21 deletion reduces the number of Kras-induced lung tumors, though the survival curve for miR-21-deficient mice with Kras activation compared with the control has not been reported (25). These data suggest that p53 knockout or Kras activation may be a tumorigenesis driver that is too strong for miR-21 deletion to override and thereby reduce tumor burden.
We next analyzed the cellular processes regulated by p53. Lack of p53 leads to defective cell cycle arrest, spontaneous immortalization and attenuated apoptosis (40). Our data obtained from Trp53 −/− miR-21 −/− MEFs show that ablation of miR-21 does not impact G1 arrest, yet it slightly enhances senescence of MEFs at P8. However, the expression of p21, an executor of cell senescence, is not altered by miR-21 deletion, suggesting that other unknown genes regulated by miR-21 may play a role in p53-independent cellular senescence.
Apoptosis is a critical cellular process induced by p53, which functions as a tumor suppressor gene in response to DNA damage (41). All miR-21 transgenic or knockout murine models reveal that miR-21 exercises its oncogenic function through decreased apoptosis (24–26). In several human cancer cell lines, miR-21 inhibition leads to increased cellular apoptosis (15,27,42–45). Analysis of the spleens from Trp53 −/− miR-21 −/− mice indicates a discernible increase in apoptosis compared with Trp53 −/− mice following doxorubicin treatment. In addition, Trp53 −/− miR-21 −/− MEFs and Ras-transformed cells show a stronger response to DNA damage-induced apoptosis than their Trp53 −/− counterparts. Thus, loss of miR-21 appears to have a substantial effect on the propensity of p53-deficient cells to undergo apoptotic death.
We next investigated which miR-21 target is responsible for the increased apoptosis, particularly in cells without the Trp53 gene. Pten has been verified as a miR-21 target in hepatocellular cancer, squamous cell carcinoma and pancreatic cancer (46–48). Interestingly, Pten is transactivated by p53 and is required for p53-mediated apoptosis in immortalized MEFs (49), yet we did not observe significant downregulation of Pten when p53 was deleted (Figure 5A). Our data in this and a previous study show that Pten is upregulated in miR-21 −/− cells compared with WT MEFs (26). In addition, Pten is expressed at a higher level in Trp53 −/− miR-21 −/− cells than in Trp53 −/− cells with or without DNA damage. Furthermore, we confirmed the role of Pten in p53-independent apoptosis when miR-21 is lost, as Pten knockdown by siRNAs significantly reduces cellular apoptosis in Trp53 −/− miR-21 −/− cells.
Doxorubicin (also known as hydroxydaunorubicin) is commonly used to treat many solid tumors and hematological malignancies alone or in combination with other chemotherapeutic agents. One of the main anticancer actions of doxorubicin is DNA damage-induced cell apoptosis, yet tumor resistance to apoptosis remains a key impediment to effective treatment with doxorubicin and other cytotoxic drugs. It is reported that miR-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting Pten (50) and that leukemia cells with elevated miR-21 expression and decreased Pten levels are more resistant to daunorubicin (an analog to doxorubicin), whereas Pten inhibition re-sensitizes cells to this chemotherapeutic agent (51). Our data in this study explicitly demonstrate that miR-21 loss increases doxorubicin-induced apoptosis, in which Pten plays a pivotal role, even when p53, a critical tumor suppressor that regulates apoptosis, is deleted. This result implies that miR-21 targeting is a potential strategy for overcoming chemoresistance in cancer with a loss-of-function p53 mutation, which comprise about half of all human cancers.
Supplementary material
Supplementary Table W1 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute/National Institutes of Health (R01 CA138688); the Diabetes and Obesity Center at University of Louisville funded by National Institutes of Health (P20 RR024489); National Natural Science Foundation of China (81270547 to X.M.).
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations:
- DMEM
Dulbecco’s modified Eagle’s medium
- FBS
fetal bovine serum
- miR-21
microRNA-21
- MEFs
mouse embryonic fibroblast cells;
- P2
passage 2
- PBS
phosphate-buffered saline
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- siRNA
small interfering RNA
- WT
wild-type.
References
- 1. Vogelstein B., et al. (2000). Surfing the p53 network. Nature, 408, 307–310 [DOI] [PubMed] [Google Scholar]
- 2. Stiewe T. (2007). The p53 family in differentiation and tumorigenesis. Nat. Rev. Cancer, 7, 165–168 [DOI] [PubMed] [Google Scholar]
- 3. Donehower L.A., et al. (1992). Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature, 356, 215–221 [DOI] [PubMed] [Google Scholar]
- 4. Jacks T., et al. (1994). Tumor spectrum analysis in p53-mutant mice. Curr. Biol., 4, 1–7 [DOI] [PubMed] [Google Scholar]
- 5. Martins C.P., et al. (2006). Modeling the therapeutic efficacy of p53 restoration in tumors. Cell, 127, 1323–1334 [DOI] [PubMed] [Google Scholar]
- 6. Ventura A., et al. (2007). Restoration of p53 function leads to tumour regression in vivo . Nature, 445, 661–665 [DOI] [PubMed] [Google Scholar]
- 7. Xue W., et al. (2007). Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature, 445, 656–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jin L., et al. (2011). MicroRNA-149*, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc. Natl. Acad. Sci. U.S.A., 108, 15840–15845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Afanasyeva E.A., et al. (2011). MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ., 18, 974–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu W., et al. (2010). Negative regulation of tumor suppressor p53 by microRNA miR-504. Mol. Cell, 38, 689–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamakuchi M., et al. (2010). P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc. Natl. Acad. Sci. U.S.A., 107, 6334–6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suzuki H.I., et al. (2009). Modulation of microRNA processing by p53. Nature, 460, 529–533 [DOI] [PubMed] [Google Scholar]
- 13. Le M.T., et al. (2009). MicroRNA-125b is a novel negative regulator of p53. Genes Dev., 23, 862–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He L., et al. (2007). A microRNA component of the p53 tumour suppressor network. Nature, 447, 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan J.A., et al. (2005). MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res., 65, 6029–6033 [DOI] [PubMed] [Google Scholar]
- 16. Iyevleva A.G., et al. (2012). High level of miR-21, miR-10b, and miR-31 expression in bilateral vs. unilateral breast carcinomas. Breast Cancer Res. Treat., 131, 1049–1059 [DOI] [PubMed] [Google Scholar]
- 17. Kulda V., et al. (2010). Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet. Cytogenet., 200, 154–160 [DOI] [PubMed] [Google Scholar]
- 18. Fulci V., et al. (2007). Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood, 109, 4944–4951 [DOI] [PubMed] [Google Scholar]
- 19. Lawrie C.H., et al. (2007). MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int. J. Cancer, 121, 1156–1161 [DOI] [PubMed] [Google Scholar]
- 20. Pichiorri F., et al. (2008). MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl. Acad. Sci. U.S.A., 105, 12885–12890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Si M.L., et al. (2007). miR-21-mediated tumor growth. Oncogene, 26, 2799–2803 [DOI] [PubMed] [Google Scholar]
- 22. Wang P., et al. (2009). microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res., 69, 8157–8165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li Y., et al. (2010). Anti-miR-21 oligonucleotide enhances chemosensitivity of leukemic HL60 cells to arabinosylcytosine by inducing apoptosis. Hematology, 15, 215–221 [DOI] [PubMed] [Google Scholar]
- 24. Medina P.P., et al. (2010). OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature, 467, 86–90 [DOI] [PubMed] [Google Scholar]
- 25. Hatley M.E., et al. (2010). Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell, 18, 282–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma X., et al. (2011). Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc. Natl. Acad. Sci. U.S.A., 108, 10144–10149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Papagiannakopoulos T., et al. (2008). MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res., 68, 8164–8172 [DOI] [PubMed] [Google Scholar]
- 28. Frankel L.B., et al. (2008). Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem., 283, 1026–1033 [DOI] [PubMed] [Google Scholar]
- 29. Bornachea O., et al. (2012). EMT and induction of miR-21 mediate metastasis development in Trp53-deficient tumours. Sci. Rep., 2, 434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu C., et al. (2001). Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol., 19, 971–974 [DOI] [PubMed] [Google Scholar]
- 31. Chen C.M., et al. (2004). Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev., 18, 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peacock J.W., et al. (2009). PTEN loss promotes mitochondrially dependent type II Fas-induced apoptosis via PEA-15. Mol. Cell. Biol., 29, 1222–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lang G.A., et al. (2004). Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell, 119, 861–872 [DOI] [PubMed] [Google Scholar]
- 34. Harvey D.M., et al. (1991). p53 alteration is a common event in the spontaneous immortalization of primary BALB/c murine embryo fibroblasts. Genes Dev., 5(12B), 2375–2385 [DOI] [PubMed] [Google Scholar]
- 35. Attardi L.D., et al. (2004). Activation of the p53-dependent G1 checkpoint response in mouse embryo fibroblasts depends on the specific DNA damage inducer. Oncogene, 23, 973–980 [DOI] [PubMed] [Google Scholar]
- 36. Schmitt C.A., et al. (2002). Dissecting p53 tumor suppressor functions in vivo . Cancer Cell, 1, 289–298 [DOI] [PubMed] [Google Scholar]
- 37. Gudkov A.V., et al. (2010). Pathologies associated with the p53 response. Cold Spring Harb. Perspect. Biol., 2, a001180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rangarajan A., et al. (2004). Species- and cell type-specific requirements for cellular transformation. Cancer Cell, 6, 171–183 [DOI] [PubMed] [Google Scholar]
- 39. Liu M.F., et al. (2010). Physiological and pathological functions of mammalian microRNAs. In McQueen C.A. (ed.), Comprehensive Toxicology. 2nd edn. Elsevier Science, London, UK, pp. 427–446 [Google Scholar]
- 40. Kenzelmann Broz D., et al. (2010). In vivo analysis of p53 tumor suppressor function using genetically engineered mouse models. Carcinogenesis, 31, 1311–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Haupt S., et al. (2003). Apoptosis - the p53 network. J. Cell. Sci., 116(Pt 20), 4077–4085 [DOI] [PubMed] [Google Scholar]
- 42. Buscaglia L.E., et al. (2011). Apoptosis and the target genes of microRNA-21. Chin. J. Cancer, 30, 371–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li T., et al. (2009). MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem. Biophys. Res. Commun., 383, 280–285 [DOI] [PubMed] [Google Scholar]
- 44. Quintavalle C., et al. (2012). Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in glioma cells. Oncogene. doi:10.1038/onc.2012.410 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 45. Seike M., et al. (2009). MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. U.S.A., 106, 12085–12090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meng F., et al. (2007). MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology, 133, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bao B., et al. (2011). Anti-tumor activity of a novel compound-CDF is mediated by regulating miR-21, miR-200, and PTEN in pancreatic cancer. PLoS ONE, 6, e17850 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48. Darido C., et al. (2011). Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell, 20, 635–648 [DOI] [PubMed] [Google Scholar]
- 49. Stambolic V., et al. (2001). Regulation of PTEN transcription by p53. Mol. Cell, 8, 317–325 [DOI] [PubMed] [Google Scholar]
- 50. Wang Z.X., et al. (2011). MicroRNA-21 modulates chemosensitivity of breast cancer cells to doxorubicin by targeting PTEN. Arch. Med. Res., 42, 281–290 [DOI] [PubMed] [Google Scholar]
- 51. Bai H., et al. (2011). Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett., 585, 402–408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.