Abstract
Sulfiredoxin (Srx) is the enzyme that reduces the hyperoxidized inactive form of peroxiredoxins. To study the function of Srx in carcinogenesis in vivo, we tested whether loss of Srx protects mice from cancer development. Srx null mice were generated and colon carcinogenesis was induced by an azoxymethane (AOM) and dextran sulfate sodium (DSS) protocol. Compared with either wild-type (Wt) or heterozygotes, Srx−/− mice had significantly reduced rates in both tumor multiplicity and volume. Mechanistic studies reveal that loss of Srx did not alter tumor cell proliferation; however, increased apoptosis and decreased inflammatory cell infiltration were obvious in tumors from Srx null mice compared with those from Wt control. In addition to the AOM/DSS model, examination of Srx expression in human reveals a tissue-specific expression pattern. Srx expression was also demonstrated in tumors from colorectal cancer patients and the levels of expression were associated with patients’ clinic stages. These data provide the first in vivo evidence that loss of Srx renders mice resistant to AOM/DSS-induced colon carcinogenesis, suggesting that Srx has a critical oncogenic role in cancer development, and Srx may be used as a marker for human colon cancer pathogenicity.
Introduction
Sulfiredoxin (Srx), or neoplastic progression 3, was initially identified as a preferentially expressed gene of unknown function in transformation sensitive mouse epithelial JB6P+ cells (1). It was then demonstrated as the enzyme that catalyzes the adenosine triphosphate-dependent reduction of the hyperoxidized sulfinic acid inactive form of peroxiredoxins (Prxs) (2,3). Srx is evolutionarily conserved from yeast to other species including cyanobacteria, fungi, arabidopsis and vertebrates. The reaction catalyzed by Srx involves the hydrolysis of adenosine triphosphate and the formation of biochemical intermediates including a phosphoryl sulfinyl anhydride and a covalent thiosulfinate (4–7). This reversible process efficiently reduces the sulfinic acid of hyperoxidized Prxs to the sulfenic form and restores their peroxidase activity. Alternative enzymatic function of Srx has also been reported; for example, Srx is capable of catalyzing the deglutathionylation of protein phosphatase and Prx II (8,9). In contrast to the well-studied biochemical function, the biological significance of Srx in development and diseases, including tumorigenesis and tumor progression, has not been fully explored.
The preferential expression of Srx in transformation sensitive versus resistant JB6P+ cells and subsequent discovery of its function of reducing hyperoxidized Prxs support the hypothesis that Srx may have an oncogenic function that is critical for tumorigenesis and cancer development in vertebrates. We have demonstrated that Srx is a novel downstream target of oncogenic AP-1 activation in a mouse model of skin tumorigenesis; when tumorigenesis is suppressed by the transgenic expression of TAM67, a dominant negative c-Jun, the expression of Srx is also inhibited (10). These findings are substantiated by the study of the regulatory mechanism of Srx expression in cancer cell lines and mouse tissues, and an AP-1/Nrf2 dependent pathway in the transcriptional regulation of Srx has been further elaborated (11,12). Given the importance of AP-1 activation and Nrf2 signaling in a variety of human cancers (13,14), it is our interest to study the functional significance of Srx in tumorigenesis. By screening Srx expression in various tissues, our previous work has demonstrated that Srx is highly expressed in several types of human cancers including those of skin and lung but not in adjacent normal tissues (10,15). In human lung cancer cell lines, ectopic expression of Srx increases cell proliferation (16), whereas depletion of endogenous Srx suppresses cell migration and invasion in culture and inhibits tumor growth and metastasis formation by mouse xenografts (15). In lung cancer patients, expression of Srx is tightly associated with tobacco usage and is significantly associated with a poor prognosis in those treated with chemotherapy or radiation therapy (17). These studies indicate that Srx has a distinct role in promoting tumorigenesis and cancer progression. In vertebrates, carcinogenesis is a complex process that includes stages of tumor initiation, promotion and progression. Whether Srx is required for tumorigenesis in vivo has not been investigated.
Srx is encoded by a single gene and no other isoforms have been identified among species varying from bacteria and plants to vertebrates. Genomic depletion of Srx gene in mouse does not result in adverse effects in embryonic or early development, fertility or adult life under laboratory conditions (18). Based on others and our previous data, we predict that Srx null mice are resistant to tumorigenesis in vivo. Colorectal cancer is the third most common cancer with over a million new patients diagnosed each year worldwide. Whether Srx is required for colon tumorigenesis and cancer progression has not been investigated. Therefore, we explored the functional significance of Srx in colorectal cancer development using Srx null mice and a well-characterized azoxymethane (AOM) and dextran sulfate sodium (DSS)-induced colon carcinogenesis model. Additionally, findings from tumors of colorectal cancer patients reveal that Srx may be used as a valuable marker for colon cancer pathogenicity.
Materials and methods
Cell culture and western blot
Human lung cancer A549 cells were obtained from ATCC and cultured in RPMI medium containing 10% fetal bovine serum under standard conditions. Stable cells that express a control non-target ShRNA, a ShRNA targeting Srx or a construct overexpressing Srx, were established as reported previously (15). For western blot, cells were lysed in RIPA buffer (Santa Cruz Biology, Santa Cruz, CA) and sodium dodecyl sulfate–polyacrylamide gel electrophoresis was performed using the NuPage precast system (Lifetech, Frederick, MD). For staining, cells were cultured on glass chamber slides following manufacturer’s protocol (Lab-Tek provided by Fisher Scientific).
Immunohistochemistry
Several commercial anti-Srx antibodies were obtained and tested for their specificity and sensitivity at series of working dilutions. Human lung cancer A549 cells stably expressing Srx or a lentiviral ShRNA that efficiently depleted endogenous Srx, as established in our previous study, were cultured on glass slides and used as positive and negative control, respectively. Anti-Srx (Proteintech, Chicago, IL) was identified as a highly specific antibody for both western blot and immunohistochemistry (IHC) staining of mouse and human Srx. IHC with hematoxylin counterstaining was performed using the HRP-DAB kit (R&D Systems, Minneapolis, MN). Other IHC antibodies used were anti-Ki67 1:50 (Dako, Carpinteria, CA) and anti-F4/80 1:100 (eBioscience, San Diego, CA). Images of IHC staining were taken using a digital camera attached to a Nikon Eclipse 80i microscope and NIS-Element software. Srx staining intensity was semiquantitated using the image-J software plus color deconvolution plugin.
Mouse genotyping and development of Srx null FVB mice
Srx null B6/129 mice were bred with FVB Wt mice to generate F1 heterozygotes (Srx+/−). After seven generations of backcross to FVB mice, the offspring of Srx+/− sibling breeding were used for AOM/DSS-induced colon carcinogenesis. Mouse genomic DNA was extracted from tail clips using commercial genomic DNA extraction kit (Qiagen, Valencia, CA). PCR-based genotyping was performed as reported previously (18).
AOM/DSS protocol
A randomized, double-blind experimental design was applied to eliminate potential subjective bias on protocol execution and data collection. Mouse procedures were conducted following the Policy on Humane Care and Use of Laboratory Animals (National Institutes of Health) and Guidelines of the Animal Care and Laboratory Animal Welfare (NCI-Frederick ACUC). Briefly, male mice at 6 week age, including Wt (n = 46), heterozygous (n = 51) and nulls (n = 30), were injected intraperitoneally with 10mg/kg of AOM (Sigma–Aldrich, St Louis, MO). After 1 week of recovery, mice were administrated ad libitum with 2% DSS (MP Biomedical, Solon, OH) in drinking water for 1 week and then maintained in normal diet for 20 weeks. During this period, mice with severe prolapse were euthanized. Twenty weeks after DSS, all mice were euthanized. Mouse colons were extracted and examined for tumors. Mouse tissues were fixed in 4% paraformaldehyde and stored in 70% ethanol before proceeding with standard paraffin embedding, sectioning and hematoxylin and eosin staining. Mouse genotyping was performed using mouse tail genomic DNA and a PCR-based method.
Tumor measurement and histopathology
Mice were humanely killed and isolated colons were cleaned by multiple rinses with phosphate-buffered saline. Tumors were macroscopically counted and tumor volume was determined as reported before by measuring the length of two perpendicular axes with a caliper (19). For histology and pathology assessment, a selected region of each colon was step-sectioned for 10 slides and the first slide was stained by hematoxylin and eosin staining. Tumor pathology was then microscopically assessed using similar criteria as reported previously in detail (20). Briefly, lesions with increased number of glands, aberrant hyperchromatic nuclei and mild dysplasia were defined as adenomas; lesions with severe dysplasia, loss of gland architecture, poorly differentiated glandular cells and disruption of submucosa were defined as adenocarcinoma.
In situ apoptosis assay
Mouse colon apoptosis assay was performed using terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay. The TACS2TdT-DAB in situ apoptosis detection kit was commercially obtained and assay was performed as manufacturer suggested (R&D Systems). Samples were counter stained with methyl green, dehydrated and mounted before microscopic visualization using instruments as described above.
Human tissue microarray
Human tissue microarray slides FDA999 (normal) and COC1021 (colon cancer) with defined clinical diagnosis and pathology information were commercially obtained (US Biomax, Rockville, MD). IHC was performed as described above.
Statistical analysis
Quantitative data were presented as means ± standard deviation ( ). Data were analyzed with indicated statistical methods by using GraphPad Prism (Version 5.04) or Microsoft Excel (Version 2010). For calculation of the P value, parameters of two-tailed, 95% confidence interval were used for all analysis. P < 0.05 is considered statistically significant.
). Data were analyzed with indicated statistical methods by using GraphPad Prism (Version 5.04) or Microsoft Excel (Version 2010). For calculation of the P value, parameters of two-tailed, 95% confidence interval were used for all analysis. P < 0.05 is considered statistically significant.
Results
Development of IHC method to detect Srx expression and creation of Srx null FVB mice
An IHC method was developed to measure the expression of Srx in fixed cells/tissues as described in Materials and methods. We identified a highly specific anti-Srx antibody. By western blot, this antibody was capable of recognizing both endogenous and overexpressed Srx (Supplementary Figure 1A, available at Carcinogenesis Online). When applied in IHC, the cytoplasmic staining of Srx in control parental cells (Supplementary Figure 1B, available at Carcinogenesis Online) and overexpression cells (Supplementary Figure 1C, available at Carcinogenesis Online) were distinct from Srx knockdown cells (Supplementary Figure 1D, available at Carcinogenesis Online). The absence of staining in Srx knockdown cells indicates that our method is specific with low or negligible non-specific background staining. Therefore, we established a relative sensitive and reliable IHC method for the detection of Srx expression.
Srx null B6/129 mice, with a neomycin cassette replacing exon II of the Srx gene, were established as described previously (18). After crossbreeding with FVB mice for multiple generations, Srx null FVB mice were generated and maintained. Similar to their ancestors, these mice also have normal phenotype under laboratory conditions. The loss of Srx was validated at the genomic level by PCR-based genotyping (Figure 1A) and at the protein level by western blot of mouse liver lysates (Figure 1B). Additionally, loss of Srx expression is also evident in null mice as demonstrated by IHC staining (Figure 1C).
Fig. 1.
Generation of Srx null FVB mice. (A) Genotyping using a PCR-based amplification of Srx and neomycin sequences in genomic DNA of mouse tail. (B) Loss of Srx protein expression in the liver extracts of the null mice. (C) IHC staining of Srx expression in the liver of Wt but not in null mice. Framed inserts indicate higher magnification. Bar = 100 µm.
Srx null mice are resistant to AOM/DSS-induced colon carcinogenesis
To study the role of Srx in tumorigenesis in vivo, a widely used AOM/DSS-induced mouse colon carcinogenesis protocol was applied (Figure 2A). The majority of mice survived through 20 weeks post-AOM/DSS treatment. Mice with severe colorectal prolapse during this period were euthanized. A previous study indicates that Srx nulls have a reduced time of survival compared with Wt mice after treatment with lethal dose of lipopolysaccharide (18). Therefore, the first question we asked is whether the genetic status of Srx affects mouse survival after AOM/DSS treatment. As shown in Figure 2B, when Srx−/− mice were compared with sibling-matched Wt or heterozygous mice, there is no significant difference in the rate of survival by Kaplan–Meier survival analysis (P > 0.05). These data suggest that genomic loss of Srx does not change the lifespan expectancy of mice under AOM/DSS treatment.
Fig. 2.
Srx null mice are resistant to AOM/DSS-induced colon carcinogenesis. (A) Schematic presentation of the AOM/DSS protocol. (B) Survival curve of mice post-AOM/DSS treatment. The average of Srx null, heterozygous (Het) and Wt mice in colon tumor incidence (C), multiplicity (D), size distribution (E) and volume (F). Tumors were macroscopically examined and tumor volume was measured with a caliper. Statistical methods used were Kaplan–Meier survival analysis (B), Fisher’s exact test (C) and Student’s t-test (D–F). Compared with Wt group, # P > 0.05; *P < 0.05.
Next we examined tumor formation in mouse colons. In Srx−/− group, the tumor incidence indicates a lower trend although it is not statistically significant compared with either Wt (P = 0.14) or heterozygotes (P = 0.15; Figure 2C). However, the average tumor multiplicity in Srx−/− group is significantly reduced by more than 2-fold compared with either −/+ or +/+ mice (both P < 0.05; Figure 2D). In particular, a significant reduction of tumors with diameter smaller than 2mm or larger than 4mm in Srx−/− mice was observed (Figure 2E). The average tumor volume per mouse in Srx−/− group is also significantly less than that of heterozygote or Wt (both P < 0.05; Figure 2F). Comparison of the Srx−/+ group with the Srx+/+ group shows no statistical significance in tumor incidence, size distribution or volume (P > 0.05 in all comparisons). This is consistent with our finding that Srx protein expression levels in heterozygotes are comparable with those found in matched Wt mice (Figure 1B). Therefore, for the rest of the study, we focused only on the comparison between Srx null and Wt counterparts.
Macroscopically, colorectal polyps are mainly distributed in the distal region of the colon, with obvious larger size and clustered tumors in colons of Srx+/+ mice (Figure 3A). For histopathology analysis, we randomly selected six colons with tumors from either Srx−/− or Srx+/+ group and performed sequential sections. Pathology examination reveals that the majority of polyps in Srx−/− mice are tubular adenomas, whereas a significantly higher percentage of polyps in Srx+/+ mice are moderately or poorly differentiated adenocarcinomas (P < 0.05; Figure 3B and C). We also examined the expression of Srx in mouse colon tissues. Srx is not expressed in the normal mouse colon. However, positive staining of Srx was found in tumors from Srx+/+ mice (Figure 3D).
Fig. 3.
Expression of Srx in AOM/DSS-induced mouse colon tumors but not in uninvolved tissue. (A) Representative gross images of colons extracted from Wt and Srx null mice. Histopathological diagnosis of tumors from Srx−/− or Wt mice. Representative hematoxylin and eosin staining images were shown in (B) and data from six of each Wt or Srx−/− group were shown in (C). # P > 0.05; *P < 0.05 (t-test). (D) Representative images of Srx staining in AOM/DSS-induced colon adenocarcinomas in Wt mice but not in normal colon tissue. Arrowheads indicate example areas of positive staining. Framed inserts indicate higher magnification. Bar = 100 µm in all images.
Our above data demonstrate that Srx null mice have a significant reduction in tumor multiplicity, volume and numbers of carcinomas compared with Wt counterparts, which indicates that loss of Srx renders mice resistant to AOM/DSS-induced colon carcinogenesis.
Loss of Srx does not change cell proliferation but leads to increased intratumoral apoptosis
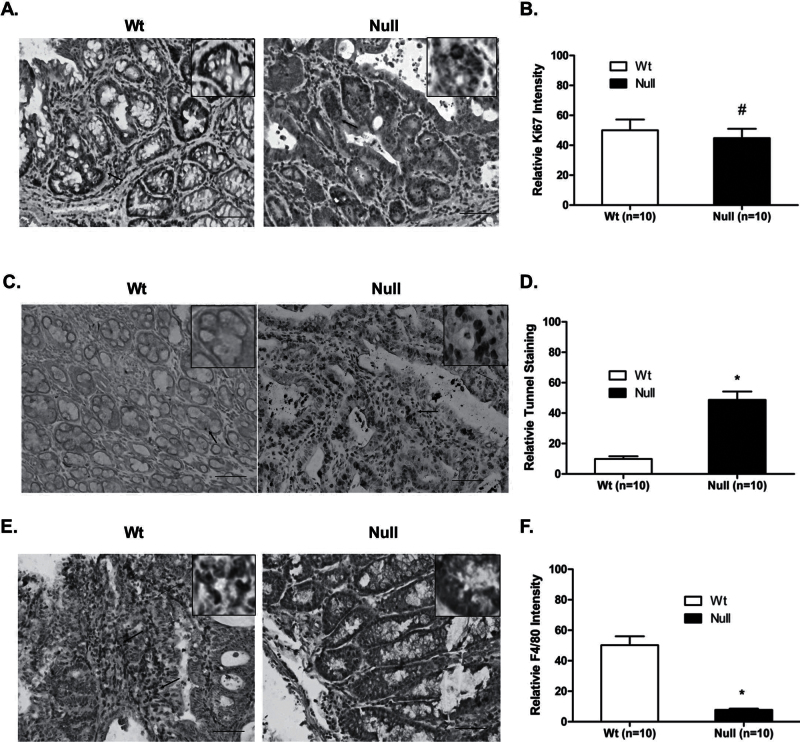
To further understand why depletion of Srx in mice leads to the reduction of colon carcinogenesis, we investigated whether the reduction of tumor multiplicity and volume in Srx−/− mice resulted from alterations in cell proliferation and/or apoptosis. To compare the rate of cell proliferation, colon tissues from Srx+/+ or Srx−/− mice were stained for Ki67, an intracellular marker for cell proliferation. Compared with normal colon epithelium, there is an apparent increase of Ki67 staining in colon adenomas or adenocarcinomas. However, proliferation rates in tumors from Srx−/− and Srx+/+ mice are similar as indicated by the comparable staining intensity of Ki67 in those tissues (P > 0.05; Figure 4A and B). Therefore, the difference in tumor volume between Srx null and Wt mice must not be resulted from potential changes in cell proliferation. Next we examined intratumoral apoptosis using TUNEL staining. In normal colon epithelium, there are few TUNEL+ cells and there is no significant difference between Srx+/+ and Srx−/− mice. In contrast, a significantly increased staining of TUNEL+ cells is demonstrated in tumors from Srx−/− mice compared with those from Wt mice (P < 0.05; Figure 4C and D). Therefore, loss of Srx is associated with increased apoptosis in Srx−/− tumors, suggesting that reduction of tumor multiplicity and volume in Srx−/− mice may be resulted from increased intratumoral apoptosis.
Fig. 4.
Srx−/− tumors have comparable levels of cell proliferation, increased rate of apoptosis and decreased number of macrophage infiltration compared with Wt tumors. (A and B) Similar levels of cell proliferation were found in Srx−/− and Wt tumors by staining of nuclei Ki67. (C and D) TUNEL assay indicates increased rate of apoptosis in Srx−/− tumors. (E and F) Decreased macrophage infiltration in Srx−/− tumors as indicated by staining of F4/80. Arrowheads indicate example nuclei/cells with positive staining. Compared with Wt, # P > 0.05; *P < 0.05 (t-test). Framed inserts indicate higher magnification. Bar = 100 µm in all images.
Tumors from Srx−/− mice are characterized with less inflammatory cell infiltration compared with those from Wt counterparts
Inflammation caused by DSS plays an essential role in the formation and progression of tumors in the mouse AOM/DSS model (21). We next asked whether there is a difference in inflammatory cell infiltration between Srx+/+ and Srx−/− tumors. F4/80 is a transmembrane protein expressed on the cell surface of mouse mature macrophages and is widely used as a reliable marker for macrophage infiltration under inflammatory conditions (22). In mouse colon normal epithelium, we found very few F4/80+ cells and there is no difference between tissues from Srx+/+ or Srx−/− mice. An overall increase of F4/80+ cells was seen in tumor tissues. Moreover, the population of F4/80+ cells in tumors from Srx+/+ mice was significantly higher than that of Srx−/− mice (P < 0.05; Figure 4E and F). These findings suggest that loss of Srx is associated with reduced intratumoral macrophage infiltration, which may contribute to the reduction of tumor multiplicity and volume found in Srx−/− mice.
Expression of Srx in human is tissue specific with non-detectable expression in normal colon epithelium
In our above experiments, we noticed that Srx does not expressed in normal mouse colon except tumors. We then asked whether this observation is human relevant. The expression pattern of Srx in human has not been investigated before. Therefore, we examined the expression of Srx in human normal tissues using the established IHC method. Sample images of Srx staining negative and positive in various human normal tissues were shown (Figure 5). In consistence with lack of Srx expression in mouse normal colon, Srx is also not detected in all three samples of human normal colon, but is weakly expressed in all samples of stomach (3, number of samples examined) and kidney (3), and highly expressed in all samples of pancreas (3) and liver (4). Among other tissues, Srx is not detected in all samples of human adrenal gland (3), breast (3), cardiac and skeletal muscle (6), cerebellum (9), colon (3), endometrium (3), esophagus (3), larynx (3), lung (3), mesothelium (3), ovary (3), peripheral nerves (3), prostate (3), retina (3), small intestine (3), spleen (3), thymus (3), thyroid (3) and tonsil (3). These data indicate that Srx may not be expressed in normal colon but does express in other organs in a tissue-specific manner.
Fig. 5.
Expression of Srx in human normal tissues. Example images show that Srx is not expressed in human breast, cerebellum and stomach, but positively present in kidney, pancreas and liver. Framed inserts indicate higher magnification. Bar = 100 µm in all images.
Srx is highly expressed in tumor specimens from colorectal cancer patients
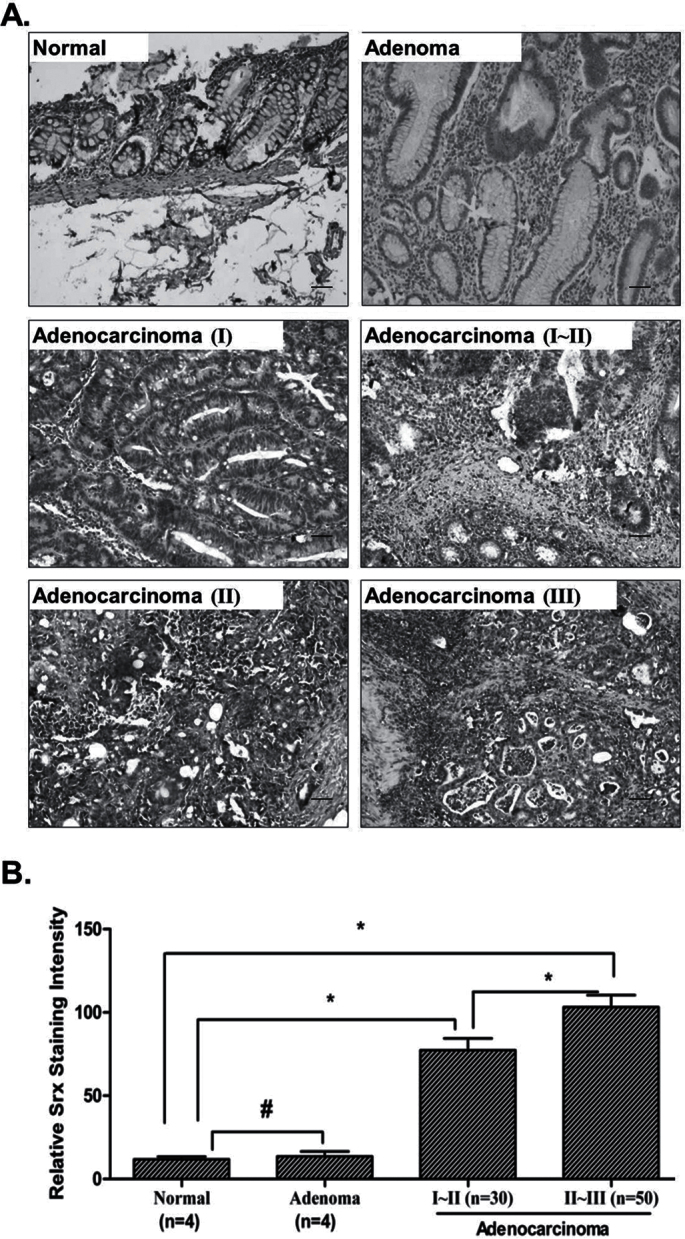
Next we asked whether Srx is expressed in tumor specimens from human colorectal cancer patients. A tissue microarray method based on the established IHC protocol was used to detect Srx expression in tumors from human colorectal cancer patients. Similar to our above findings, Srx is not expressed in human colon normal epithelium and cancer adjacent normal tissues. Srx is also not expressed in all four samples of human colon adenoma. In contrast, Srx is highly expressed in human colon carcinomas (n = 80; Figure 6A and B). In tissues from patients with advanced adenocarcinomas at clinical stages II–III (n = 50), the expression of Srx is also higher (Figure 6B). Therefore, in addition to the increased expression demonstrated in AOM/DSS-induced mouse colon tumors, significant differential expression of Srx is also found in tumors from human colorectal cancer patients, which suggest that Srx may be used as a valuable marker for colon cancer pathogenicity.
Fig. 6.
Increased expression of Srx in tumors from human colon cancer patients by tissue microarray. (A) Representative images of Srx staining in human colon normal, adenoma or adenocarcinoma at different clinical stages. (B) Summary of Srx staining intensity in (A) by semiquantitation. # P > 0.05; *P < 0.05 (t-test). Bar = 100 µm in all images.
Discussion
Although Srx has been indicated previously in the pathogenesis of several types of human cancers including those from skin and lung, the role of Srx in colon cancer has not been investigated. Whether Srx is important for tumorigenesis in vivo is also not clear. In this study, we demonstrated that depletion of Srx renders mice resistant to colon carcinogenesis using a well-characterized AOM/DSS-induced colon carcinogenesis model. The tumor-resistant phenotype of Srx null mice may be associated with increased intratumoral apoptosis and decreased inflammatory cell infiltration. Our observation is patient relevant, as increased expression of Srx is not only found in tumors induced by AOM/DSS but also in tumors from colorectal cancer patients with other causes. Therefore, our study reveals an important role of Srx in colorectal cancer development.
The AOM/DSS model is currently the most commonly used non-hereditary rodent model to mimic colorectal cancer development in human patients. The combination of a single hit of mutagenic AOM with 1 week exposure to tumor promoter DSS in mouse has been proved to be a highly reproducible and potent protocol to recapitulate the aberrant crypt foci–adenoma–carcinoma sequence that occurs in human colorectal cancer (21,23). Within 20 weeks after AOM/DSS treatment, the development of numerous colon tumors in the distal colon and rectum in mouse has been demonstrated in multiple mouse strains. Significant differences in the susceptibility to AOM/DSS-induced colon carcinogenesis have been observed among different mouse strains due to variations in genetic background (24). To minimize strain-dependent variations in tumor development, we established Srx nulls in an FVB inbred background using the original Srx null mouse line (18). It is commonly accepted that there are no gender differences in the susceptibility of mice to AOM/DSS (25), except that females of C57BL/6J mice appear to be more susceptible than male controls (26). In the previous study, males of Srx null mice appear to be more susceptible to lipopolysaccharide-induced endotoxemia (18). To exclude potential gender difference that complicates the interpretation of results, only males of Srx null mice were used in our study. Within this context, we demonstrate that Srx null mice have a lower incidence and significantly lower tumor multiplicity and volume, which suggests that depletion of Srx renders mice resistant to AOM/DSS-induced colon carcinogenesis. Whether there are gender differences in the susceptibility of Srx null mice to AOM/DSS needs to be further investigated in the future.
In our efforts to understand the molecular basis of Srx in colon tumorigenesis, we demonstrate that loss of Srx does not cause apparent defects in animal growth and breeding under laboratory conditions. However, under stress conditions such as severe endotoxic shock, Srx null mice appears to have a delayed/prolonged inflammatory response that is characterized by upregulated expression of genes involved in adaptive and innate immunity (18). It is still unknown how the defect in the adaptive and/or innate immunity may affect the process of colon tumorigenesis in Srx null mice. In our study, early temporary bloody diarrhea and colon prolapse associated with AOM/DSS treatment have been found in both Wt and Srx null mice. There is no significant difference in survival between Srx null and Wt mice post-AOM/DSS treatment. Therefore, the differences we observed in colon tumor multiplicity, size distribution and volume may reflect an intrinsic, long-term accumulative effect of AOM/DSS-associated distinctions between Srx null and Wt mice.
Compared with those from Wt mice, tumors from Srx nulls are characterized with unchanged cell proliferation and increased rate of apoptosis. Although Srx is not expressed in normal colon epithelium, it is expressed in tumors induced by AOM/DSS in the Wt mice. Based on the primary function of Srx to reduce the hyperoxidized Prxs, expression of Srx in tumor cells may increases cells’ ability to scavenge hydrogen peroxide and enhance their capability of surviving through oxidative stress. This has been demonstrated previously in various cell lines including embryonic fibroblasts (18,27), neurons (28,29) and mouse skin epithelial cells (10). Therefore, tumors derived from Srx null mice are likely to be more sensitive to oxidative injury and apoptosis as evidenced by TUNEL assay in our study, which may at least partially explain the reduced tumor multiplicity and volume found in Srx null mice.
Our study also reveals that tumors from Srx nulls have less inflammatory cell infiltration compared with those from Wt counterparts. Although somewhat controversial, the sinister role of inflammatory cells, especially macrophages in tumorigenesis, has been demonstrated in many studies and intensively reviewed in literature (30,31). The contribution of macrophages may involve in multiple aspects of cancer development. For example, macrophages produce reactive oxygen and nitrogen species that react with DNA to cause mutagenic events, as evidenced in infection-associated tumorigenesis (30). Tumor-associated macrophages (TAMs) may also secret growth factors such as epithelial growth factor, basic fibroblast growth factor and colony-stimulating factor-1 to stimulate tumor growth. Moreover, TAMs produce vascular endothelial growth factor, IL-1, IL-8 and other cytokines and chemokines to stimulate angiogenesis (32). Furthermore, various proteases produced by TAMs, including matrix metalloproteinase such as MMP2, MMP7 and MMP9, can promote tumor cell invasion and metastasis (33). Therefore, functional intact TAMs are essential regulators of tumorigenesis and cancer progression. Based on these studies, we speculate that reduced macrophage infiltration found in Srx−/− tumors may contribute to the tumor-resistant phenotype of Srx null mice. Indeed, Srx is expressed in macrophages and its expression is stimulated by inflammatory cytokines such as interferon through a nitric oxide-mediated signaling pathway (34). Expression of Srx enhances the Prx-dependent antioxidant capacity of macrophages, which may also involve in host response to inflammation (34). In contrast, loss of Srx affects the expression of cytokines, chemokines, interleukins and various inflammatory genes in Srx null mice (18). Depletion of Srx may thus disrupt the nitric oxide–Srx–Prx axis, compromise macrophages’ ability to infiltrate and modify tumor microenvironment. Taken together, these factors eventually lead to the reduction of colon carcinogenesis in Srx null mice.
Besides the AOM/DSS-induced colon carcinogenesis model, we also examined the expression of Srx in normal human tissues and tumor specimens from colorectal cancer patients. We found a tissue-specific expression pattern of Srx in human organs and a significant increased expression of Srx in tumors from colorectal cancer patients. The association of Srx with patient clinic stages may indicate that Srx can be used as a potential therapeutic target or diagnostic molecular indicator for human colon cancer pathogenesis. In summary, we demonstrate that loss of Srx renders mice resistant to AOM/DSS-induced colon tumorigenesis and is relevant to human colorectal cancer pathogenesis. In the future, it will be helpful to further clarify molecular mechanism of Srx in colon tumorigenesis, to test whether Srx is involved in the progression of colon tumors from benign adenomas to malignant carcinomas and to investigate whether targeting Srx can be used as an effective therapeutic strategy for human colon cancer treatment.
Supplementary material
Supplementary Figure 1 can be found at http://carcin.oxfordjournals.org/
Funding
National Cancer Institute Intramural Research Program (Z01B C010025 to N.H.C.).
Supplementary Material
Acknowledgements
We thank Ms Jen Wise and Mr Dan Logsdon for mouse maintenance and experimental help, and Dr Yinling Hu (NCI-Frederick) for instrument support. We also thank members of the CGS advisory committee and members of the Laboratory of Cancer Prevention (NCI-Frederick) for valuable discussion. Q.W. is a Cancer Research Training Award (CRTA) fellow, Cancer Genetics & Signaling (CGS) fellow and the recipient of a National Cancer Institute Pathway to Independence Award.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- AOM
azoxymethane
- DSS
dextran sulfate sodium
- Prx
peroxiredoxin
- Srx
sulfiredoxin
- TAM
tumor-associated macrophage
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- Wt
wild-type.
References
- 1. Sun Y., et al. (1994). Molecular cloning of five messenger RNAs differentially expressed in preneoplastic or neoplastic JB6 mouse epidermal cells: one is homologous to human tissue inhibitor of metalloproteinases-3. Cancer Res., 54, 1139–1144 [PubMed] [Google Scholar]
- 2. Biteau B., et al. (2003). ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature., 425, 980–984 [DOI] [PubMed] [Google Scholar]
- 3. Chang T.S., et al. (2004). Characterization of mammalian sulfiredoxin and its reactivation of hyperoxidized peroxiredoxin through reduction of cysteine sulfinic acid in the active site to cysteine. J. Biol. Chem., 279, 50994–51001 [DOI] [PubMed] [Google Scholar]
- 4. Roussel X., et al. (2011). The rate-limiting step of sulfiredoxin is associated with the transfer of the γ-phosphate of ATP to the sulfinic acid of overoxidized typical 2-Cys peroxiredoxins. FEBS Lett., 585, 574–578 [DOI] [PubMed] [Google Scholar]
- 5. Jonsson T.J., et al. (2008). Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature., 451, 98–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roussel X., et al. (2008). Evidence for the formation of a covalent thiosulfinate intermediate with peroxiredoxin in the catalytic mechanism of sulfiredoxin. J. Biol. Chem., 283, 22371–22382 [DOI] [PubMed] [Google Scholar]
- 7. Jonsson T.J., et al. (2009). Protein engineering of the quaternary sulfiredoxin.peroxiredoxin enzyme.substrate complex reveals the molecular basis for cysteine sulfinic acid phosphorylation. J. Biol. Chem., 284, 33305–33310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Findlay V.J., et al. (2006). A novel role for human sulfiredoxin in the reversal of glutathionylation. Cancer Res., 66, 6800–6806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park J.W., et al. (2009). Deglutathionylation of 2-Cys peroxiredoxin is specifically catalyzed by sulfiredoxin. J. Biol. Chem., 284, 23364–23374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei Q., et al. (2008). Sulfiredoxin is an AP-1 target gene that is required for transformation and shows elevated expression in human skin malignancies. Proc. Natl Acad. Sci. U.S.A., 105, 19738–19743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soriano F.X., et al. (2009). Transcriptional regulation of the AP-1 and Nrf2 target gene sulfiredoxin. Mol. Cells., 27, 279–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim H., et al. (2010). Redox regulation of lipopolysaccharide-mediated sulfiredoxin induction, which depends on both AP-1 and Nrf2. J. Biol. Chem., 285, 34419–34428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lau A., et al. (2008). Dual roles of Nrf2 in cancer. Pharmacol. Res., 58, 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colburn N.H., et al. (2008). Targeting transcription factors for cancer prevention—the case of Nrf2. Cancer Prev. Res. (Phila.), 1, 153–155 [DOI] [PubMed] [Google Scholar]
- 15. Wei Q., et al. (2011). Sulfiredoxin-peroxiredoxin IV axis promotes human lung cancer progression through modulation of specific phosphokinase signaling. Proc. Natl Acad. Sci. U.S.A., 108, 7004–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lei K., et al. (2008). Protein cysteine sulfinic acid reductase (sulfiredoxin) as a regulator of cell proliferation and drug response. Oncogene., 27, 4877–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Merikallio H., et al. (2011). Nuclear factor erythroid-derived 2-like 2 (Nrf2) and DJ1 are prognostic factors in lung cancer. Hum. Pathol., 43, 577–584 [DOI] [PubMed] [Google Scholar]
- 18. Planson A.G., et al. (2011). Sulfiredoxin protects mice from lipopolysaccharide-induced endotoxic shock. Antioxid. Redox Signal., 14, 2071–2080 [DOI] [PubMed] [Google Scholar]
- 19. Euhus D.M., et al. (1986). Tumor measurement in the nude mouse. J. Surg. Oncol., 31, 229–234 [DOI] [PubMed] [Google Scholar]
- 20. Cui X., et al. (2010). Mechanistic insight into the ability of American ginseng to suppress colon cancer associated with colitis. Carcinogenesis., 31, 1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Robertis M., et al. (2011). The AOM/DSS murine model for the study of colon carcinogenesis: from pathways to diagnosis and therapy studies. J. Carcinog., 10, 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirsch S., et al. (1981). Expression of the macrophage-specific antigen F4/80 during differentiation of mouse bone marrow cells in culture. J. Exp. Med., 154, 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neufert C., et al. (2007). An inducible mouse model of colon carcinogenesis for the analysis of sporadic and inflammation-driven tumor progression. Nat. Protoc., 2, 1998–2004 [DOI] [PubMed] [Google Scholar]
- 24. Suzuki R., et al. (2006). Strain differences in the susceptibility to azoxymethane and dextran sodium sulfate-induced colon carcinogenesis in mice. Carcinogenesis., 27, 162–169 [DOI] [PubMed] [Google Scholar]
- 25. Rosenberg D.W., et al. (2009). Mouse models for the study of colon carcinogenesis. Carcinogenesis., 30, 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boddicker R.L., et al. (2011). Early lesion formation in colorectal carcinogenesis is associated with adiponectin status whereas neoplastic lesions are associated with diet and sex in C57BL/6J mice. Nutr. Cancer., 63, 1297–1306 [DOI] [PubMed] [Google Scholar]
- 27. Baek J.Y., et al. (2012). Sulfiredoxin protein is critical for redox balance and survival of cells exposed to low steady-state levels of H2O2. J. Biol. Chem., 287, 81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu C.L., et al. (2012). c-Jun-dependent sulfiredoxin induction mediates BDNF protection against mitochondrial inhibition in rat cortical neurons. Neurobiol. Dis., 46, 450–462 [DOI] [PubMed] [Google Scholar]
- 29. Nagashima S., et al. (2011). CRMP5-associated GTPase (CRAG) protein protects neuronal cells against cytotoxicity of expanded polyglutamine protein partially via c-Fos-dependent activator protein-1 activation. J. Biol. Chem., 286, 33879–33889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Qian B.Z., et al. (2010). Macrophage diversity enhances tumor progression and metastasis. Cell., 141, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fulton A.M., et al. (1984). Mutagenic activity of tumor-associated macrophages in Salmonella typhimurium strains TA98 and TA 100. Cancer Res., 44, 4308–4311 [PubMed] [Google Scholar]
- 32. Carmeliet P., et al. (2000). Angiogenesis in cancer and other diseases. Nature., 407, 249–257 [DOI] [PubMed] [Google Scholar]
- 33. Condeelis J., et al. (2006). Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell., 124, 263–266 [DOI] [PubMed] [Google Scholar]
- 34. Diet A., et al. (2007). Regulation of peroxiredoxins by nitric oxide in immunostimulated macrophages. J. Biol. Chem., 282, 36199–36205 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.